The mechanism underlying the selective packaging of genomic RNA into HIV-1 virions is not known. This paper provides important new biophysical insights into the nature of protein–RNA interactions responsible for HIV-1 genome packaging by quantifying the electrostatic and hydrophobic contributions to specific and nonspecific RNA.
Keywords: Gag, HIV, Psi, UTR, packaging signal
Abstract
Despite the vast excess of cellular RNAs, precisely two copies of viral genomic RNA (gRNA) are selectively packaged into new human immunodeficiency type 1 (HIV-1) particles via specific interactions between the HIV-1 Gag and the gRNA psi (ψ) packaging signal. Gag consists of the matrix (MA), capsid, nucleocapsid (NC), and p6 domains. Binding of the Gag NC domain to ψ is necessary for gRNA packaging, but the mechanism by which Gag selectively interacts with ψ is unclear. Here, we investigate the binding of NC and Gag variants to an RNA derived from ψ (Psi RNA), as well as to a non-ψ region (TARPolyA). Binding was measured as a function of salt to obtain the effective charge (Zeff) and nonelectrostatic (i.e., specific) component of binding, Kd(1M). Gag binds to Psi RNA with a dramatically reduced Kd(1M) and lower Zeff relative to TARPolyA. NC, GagΔMA, and a dimerization mutant of Gag bind TARPolyA with reduced Zeff relative to WT Gag. Mutations involving the NC zinc finger motifs of Gag or changes to the G-rich NC-binding regions of Psi RNA significantly reduce the nonelectrostatic component of binding, leading to an increase in Zeff. These results show that Gag interacts with gRNA using different binding modes; both the NC and MA domains are bound to RNA in the case of TARPolyA, whereas binding to Psi RNA involves only the NC domain. Taken together, these results suggest a novel mechanism for selective gRNA encapsidation.
INTRODUCTION
Amidst the vast excess of cellular RNAs, assembling human immunodeficiency virus type 1 (HIV-1) virions selectively package two copies of the viral RNA genome (gRNA). During assembly, viral Gag proteins coalesce at the plasma membrane and bud off to form new virions ∼100 nm in size (for review, see Sundquist and Krausslich 2012). The Gag precursor protein consists of the matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), and p6 domains, which are cleaved into freestanding proteins during virus maturation (Coffin et al. 1997; Fields et al. 2007). During assembly of immature virions, Gag binds to gRNA via NC and to the plasma membrane via the MA domain. In addition, the CA domain forms CA–CA contacts critical for assembly (for review, see Ganser-Pornillos et al. 2012; Sundquist and Krausslich 2012).
The 5′ untranslated region (UTR) of the HIV-1 viral RNA contains motifs required for gRNA packaging commonly referred to as the Psi (ψ) packaging signal (Lever et al. 1989; Wilkinson et al. 2008), which consists of a series of stem–loops (SL1–SL4). Herein ψ refers to the packaging signal in the virus, and Psi RNA refers to the RNA construct containing SL1–SL3 (Fig. 1), which we have examined in this report. For some retroviruses, such as murine leukemia virus (MLV) and Rous sarcoma virus, the minimal ψ packaging element is well defined (Lu et al. 2011b, and references therein), but in HIV-1, in addition to SL1–SL4, other elements in the 5′ UTR contribute either directly or indirectly to packaging (for review, see Lu et al. 2011b) (Skripkin et al. 1994; Russell et al. 2003; Parkash et al. 2012). Some specific cellular RNAs that lack ψ are also packaged into virions (Houzet et al. 2007; Kleiman et al. 2010; Keene and Telesnitsky 2012), and in the absence of ψ-containing RNAs, selectively packaged cellular RNA levels increase while additional cellular RNAs are also abundantly and nonselectively incorporated into virions (Muriaux et al. 2001; Rulli et al. 2007).
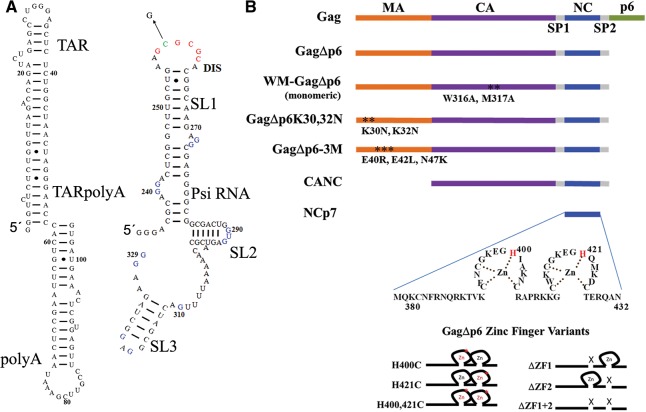
FIGURE 1.
Protein and RNA constructs used in this work. (A) The TARPolyA RNA and Psi RNA are derived from the NL4-3 virus isolate. A single G residue was appended to the 5′ end of TARPolyA, and three G residues were appended to the 5′ end of Psi RNA to improve in vitro transcription. Psi RNA contains the dimerization initiation site (DIS) shown in red with the residue C258 (green) mutated to G to generate monomeric Psi. The 12 residues changed to A in the Psi-12M variant are shown in blue. (B) GagΔp6 protein constructs examined in this work with full-length Gag (top) shown for comparison. The amino acid sequence of NC is shown with zinc ions chelated by the invariant CCHC motif of each zinc finger. Zinc finger variants studied herein are depicted in cartoon form, with single H-to-C point mutations indicated by a red asterisk and with deletions indicated by an X.
Aside from the ψ stem–loops, the 5′ UTR of the HIV-1 genome also contains the transactivation response (TAR) stem–loop critical for viral RNA transcription, the polyadenylation site-containing PolyA stem–loop (PolyA), and the primer binding site (PBS), which harbors the sequence that anneals to primer tRNALys3 for reverse transcription initiation (Fig. 1A). Within ψ, SL1 contains the dimerization initiation site (DIS), a palindromic sequence (GCGCGC in the NL4-3 isolate) responsible for dimerization of the two copies of packaged gRNA (Skripkin et al. 1994; for review, see Moore and Hu 2009; Lu et al. 2011b). SL2 contains the splice donor site, critical for splicing of the viral RNA for translation of viral accessory proteins, and SL3 is a sequence shown to contribute to gRNA packaging (Clever and Parslow 1997; McBride and Panganiban 1997; Russell et al. 2003) and high-affinity NC binding (De Guzman et al. 1998; Athavale et al. 2010). In HIV-1, genome packaging and dimerization are hypothesized to be coordinated processes, as virions contain only dimeric gRNA (Moore and Hu 2009; Nikolaitchik et al. 2013). A model for the switch between translation and packaging has been proposed based on NMR spectroscopy data showing that the DIS loop of SL1 folds back and interacts with an upstream region in the viral RNA, thereby preventing dimerization of SL1 and promoting translation (Lu et al. 2011a). In MLV, dimerization of the gRNA exposes high-affinity binding sites for the NC domain of Gag (D’Souza and Summers 2004; Gherghe et al. 2010; Miyazaki et al. 2010), suggesting that packaging and Gag binding to ψ are coordinated with dimerization. In HIV-1, a 159-nt “core encapsidation signal” was recently described that binds NC with similar affinity as the full-length 5′ leader sequence (Heng et al. 2012). However, there may be important differences in the mechanisms by which the discrete NC domain and the full-length Gag protein bind to gRNA with high affinity.
Recent reports have shown that HIV-1 Gag interacts with nucleic acids via both the MA and NC domains. MA interactions with RNA are secondary to its binding to phosphatidylinositol-(4, 5)-bisphosphate (PIP2)-containing lipids once it reaches the plasma membrane, whereas NC prefers to bind to RNA (Alfadhli et al. 2009; Chukkapalli et al. 2010; Jones et al. 2011). MA is myristoylated and also contains a basic patch, which enhances its binding to PIP2 (Chukkapalli et al. 2008, 2010). NC is a highly basic protein containing two CCHC zinc fingers (Fig. 1) that are critical for RNA-binding specificity, nucleic acid chaperone activity (Levin et al. 2010; Darlix et al. 2011), and gRNA packaging (Gorelick et al. 1999b; Kafaie et al. 2008). Investigations into the RNA-binding specificity of Gag have previously studied the binding of the NC protein to RNA and DNA (Dannull et al. 1994; Fisher et al. 1998, 2006; Vuilleumier et al. 1999; Avilov et al. 2009; Athavale et al. 2010), and more recently the binding of Gag to short oligonucleotides has also been investigated (Stephen et al. 2007; Wu et al. 2010; Jones et al. 2011). In this study, we investigate the binding of Gag and NC to longer ∼100-nt RNAs derived from the gRNA 5′ UTR. To better understand how Gag-ψ binding contributes to specific gRNA packaging, two model RNAs were chosen: Psi RNA, which contains SL1–SL3, and a non-Psi construct containing the TAR and PolyA stem–loops (TARPolyA) (Fig. 1). We used a salt titration approach, which allowed determination of both the electrostatic and nonelectrostatic contributions to binding. Our findings suggest that zinc finger-dependent Gag-ψ interaction results in a specific binding mode that facilitates selective packaging of gRNA.
RESULTS
Direct binding assays show salt-resistant binding of Gag to Psi RNA
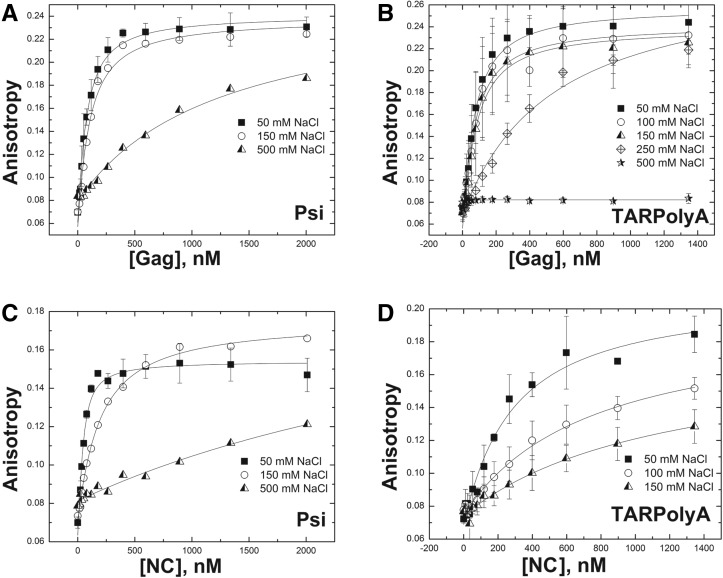
To examine the interactions between Gag and RNAs derived from the HIV-1 genome, we first performed direct binding measurements in which either fluorescently labeled TARPolyA or Psi RNA (Fig. 1) was incubated with various amounts of Gag or NC protein. The recombinant GagΔp6 protein (referred to as WT Gag or Gag) used herein is not myristoylated and lacks the C-terminal p6 domain (Fig. 1). Binding was detected via fluorescence anisotropy (FA) using fluorescently labeled RNAs, and assays were performed at various [NaCl] concentrations (50–500 mM). Although binding affinities of Gag to Psi RNA and TARPolyA were similar at 50 mM NaCl (Kd ∼ 50 nM), at 0.5 M NaCl, Gag still binds Psi RNA (Kd = 300 nM), whereas no binding was observed to TARPolyA (Fig. 2A,B). NC showed robust binding to Psi RNA at 50 mM and 150 mM NaCl (Kd = 50 nM and Kd = 74 nM, respectively) and weaker binding at 0.5 M NaCl (Kd > 2000 nM) (Fig. 2C). NC binding to TARPolyA was even more salt sensitive with very weak binding observed at 150 mM NaCl (Fig. 2D). These assays showed a dramatic difference in the salt dependence of Gag and NC binding to ψ vs. non-ψ RNAs. The apparent salt independence of Kd’s under low-salt conditions is an artifact of the RNA concentration used (∼30–40 nM), which was similar to the Kd values determined. To overcome this technical issue and to obtain additional insights into the mechanism of binding, a salt titration assay was used.
FIGURE 2.
Direct binding assays using fluorescence anisotropy equilibrium measurements. Binding of GagΔp6 to (A) Psi RNA and (B) TARPolyA at various NaCl concentrations are shown. Binding of NC to (C) Psi RNA and (D) TARPolyA at various NaCl concentrations are shown.
Gag binds Psi RNA with high affinity and specificity
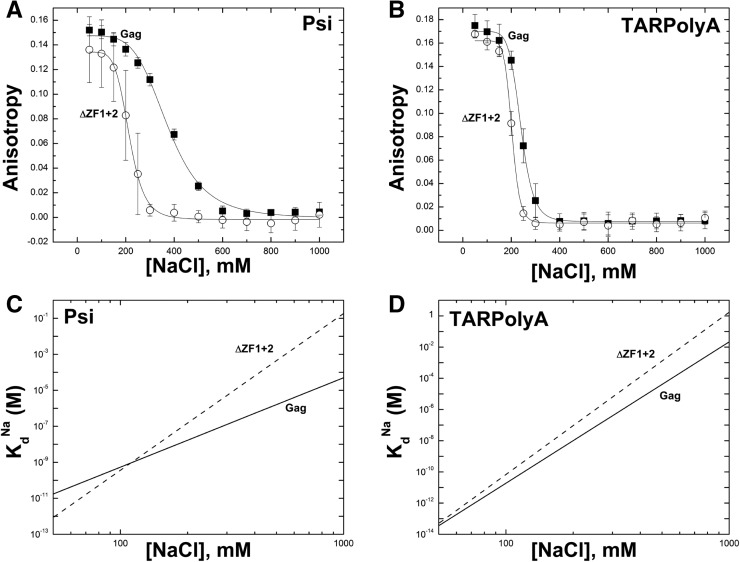
To gain additional insights into ψ vs. non-ψ RNA binding of Gag and NC, a salt titration assay was performed in which a fixed concentration of Gag or NC protein was bound to RNA (∼30 nM) and FA was measured as a function of [NaCl]. In this experiment, as [NaCl] increases over a range of from 50 mM to 1 M, Gag was gradually displaced from the labeled RNA (either Psi or TARPolyA), leading to a decrease in the FA signal. A protein concentration of 400 nM was chosen based on the direct binding experiments (Fig. 2); however, similar results were obtained when experiments were performed at 750 nM protein (Supplemental Information; Supplemental Fig. S1). Figure 3 shows the results of salt titration assays for Gag (Fig. 3A) and NC (Fig. 3B) binding to TARPolyA and Psi.
FIGURE 3.
Salt titration of Gag and NC binding to Psi RNA and TARPolyA. (A) Prebound Gag–Psi RNA or Gag–TARPolyA complexes were titrated with increasing amounts of NaCl, leading to a decrease in the anisotropy. (B) A similar set of titrations was performed with NC–Psi RNA and NC–TARPolyA complexes. (C) Data from A and B are regraphed in a log–log plot showing the dependence of the apparent binding affinity (Kd) on NaCl concentration according to 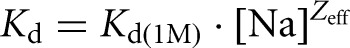 . The slope of each line is the Zeff value and the value of the Kd at 1 M NaCl is the Kd(1M). The Zeff and Kd(1M) parameters were obtained by fitting the experimental salt titration data. A complete description of this method and subsequent analysis are available in the Materials and Methods and the Supplemental Methods.
. The slope of each line is the Zeff value and the value of the Kd at 1 M NaCl is the Kd(1M). The Zeff and Kd(1M) parameters were obtained by fitting the experimental salt titration data. A complete description of this method and subsequent analysis are available in the Materials and Methods and the Supplemental Methods.
Dramatic differences in the salt dependence of binding to each RNA were observed. Data from salt titrations were fit to determine the nonelectrostatic component of binding (i.e., protein–RNA dissociation constant at 1 M salt, Kd(1M)) and the effective charge (Zeff) of the protein interface involved in direct RNA binding as described in the Supplemental Information. The fits were graphically represented by plotting Kd vs. [NaCl] (Fig. 3C), and the Kd(1M) and Zeff values, calculated according to equation 1 (see Materials and Methods), are reported in Table 1. Because the salt effect on cationic protein binding is largely insensitive to the identity of the cation, given the same net positive charge (i.e., Na+ vs. K+ or Mg2+ vs. Ca2+) (Rouzina and Bloomfield 1997; Vo et al. 2006; Athavale et al. 2010), using this analysis Kd may be predicted for a particular protein–RNA pair under different buffer conditions. Kd(1M) reflects the binding under conditions in which all electrostatic interactions are screened out by salt, and the only contribution to binding comes from specific contacts such as aromatic residue stacking with unpaired nucleic acid bases or specific hydrogen bonds. Moreover, the measured binding is to the highest affinity RNA site, as discussed in the Supplemental Information. Zeff is a measure of the number of Na+ ions displaced from RNA upon protein binding and reflects the number of the positively charged groups at the protein surface directly interacting with the RNA phosphates (Record et al. 1976; Rouzina and Bloomfield 1997). In solution, ∼70% and ∼90% of the phosphate backbone is occupied by Na+ ions for ssDNA and dsDNA, respectively (Record et al. 1976). Because the Na+ occupancy of the structured RNAs studied here (Fig. 1) likely falls between these two numbers, our reported Zeff values have not been normalized to account for partial occupancy. We ensured that the Zeff and Kd(1M) values calculated were insensitive to the number of protein-binding sites on each RNA (Supplemental Table S1). In addition to the fitted parameters Zeff and Kd(1M), we also determined ΔA, the total change in anisotropy upon protein binding. This value depends on the change in size between free and bound RNA/protein complex and is distinct for every protein–RNA pair. Finally, for each protein/RNA interaction, we also report Na1/2 values, which are the titration midpoints, or the amount of NaCl required to displace 50% of bound protein. The Na1/2 and Anisotropy vs. [Na] slope at Na1/2 are related to binding parameters Zeff, Kd(1M), and ΔA, as discussed in the Supplemental Information, and described by equations S12 and S13, respectively.
TABLE 1.
Binding parameters for GagΔp6 variants
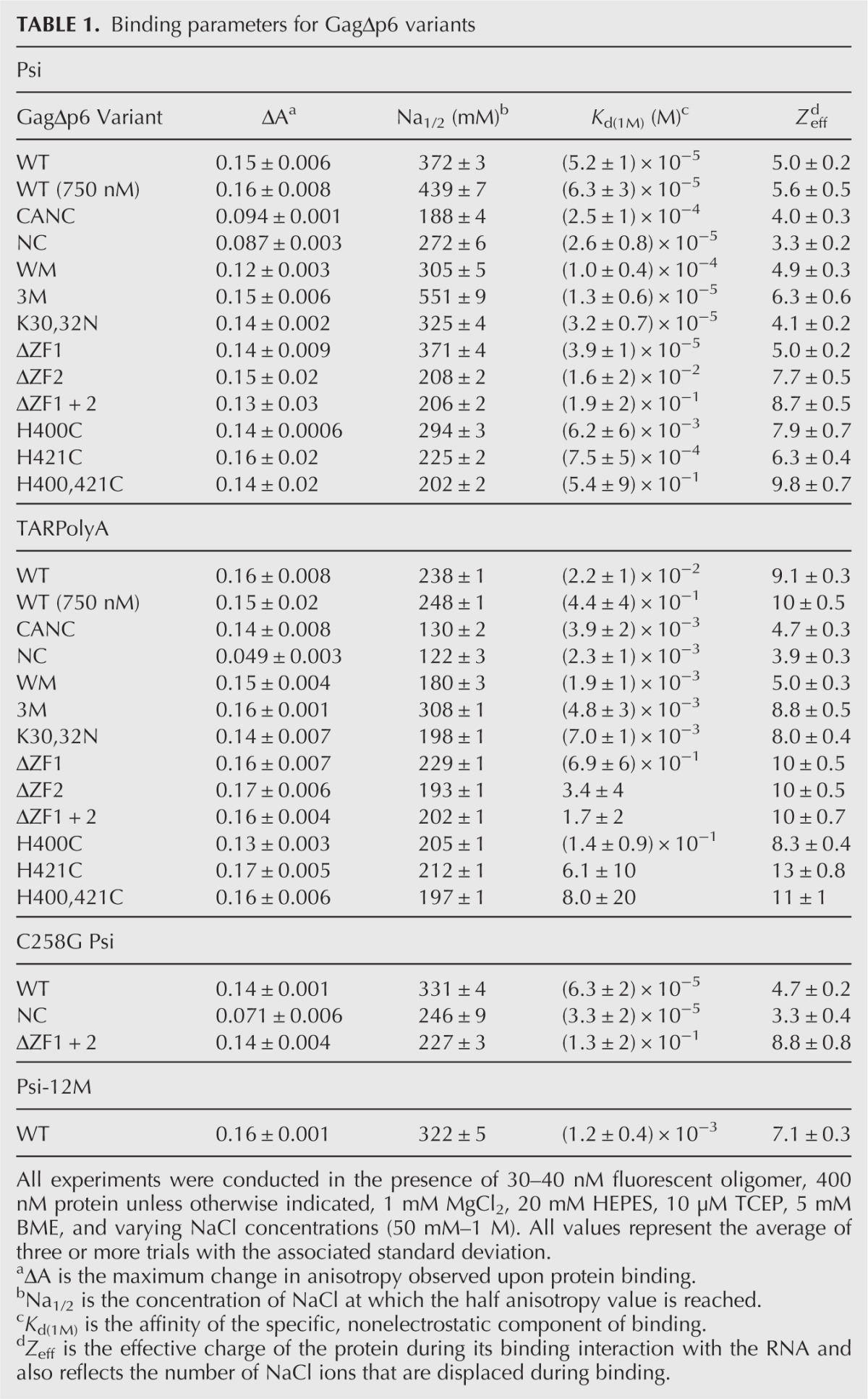
NC binding to TARPolyA (Fig. 3B) was characterized by a Kd(1M) of 2.3 × 10−3 M, which is ∼100-fold weaker than NC binding to Psi RNA (Kd(1M) ∼ 2.6 × 10−5 M). This finding is in agreement with previous studies showing ∼100-fold affinity range for the Kd(1M) values for NC binding to 6-nt ssDNA oligos (Vuilleumier et al. 1999). NC/TARPolyA and NC/Psi RNA interactions were both characterized by a Zeff of ∼3–4, in agreement with previous studies (Vuilleumier et al. 1999; Vo et al. 2009a; Athavale et al. 2010). Furthermore, the nonelectrostatic binding component of Gag, Kd(1M) was even more selective for Psi RNA relative to TARPolyA (∼1000-fold) than NC (Table 1). Moreover, Gag bound to TARPolyA with an approximately twofold greater Zeff of 9 relative to Psi RNA (Zeff = 5) (Table 1)—suggesting that Gag interacts with TARPolyA with four additional positive charges relative to Psi RNA. As shown in Figure 2, A and B, to displace Gag from TARPolyA or Gag from Psi, a greater amount of NaCl was required than for the corresponding NC interactions. This finding is reflected by higher Na1/2 values for Gag/RNA interactions (Table 1) and suggests that Gag domains outside of NC contribute to binding. Importantly, at physiological NaCl (∼150 mM), the estimated Kd’s for Gag binding to TARPolyA RNA and Psi RNA differ by approximately sixfold. Surprisingly, Gag binds to TARPolyA with a higher affinity than to Psi RNA, suggesting that selective packaging of Psi-containing RNAs is unlikely to be due to stronger equilibrium binding affinity. In summary, Gag binds Psi RNA with reduced charge interactions and much stronger nonelectrostatic interactions relative to TARPolyA binding.
Both MA and NC domains contribute to Gag/TARPolyA binding
To dissect the difference between Psi and TARPolyA binding at a molecular level, salt titration assays were performed with Gag variants CANC and W316A,M317A-GagΔp6 (WM-Gag). The latter variant contains changes in CA residues that are critical for Gag–Gag dimerization (Datta et al. 2007). Binding of either of these proteins to TARPolyA or Psi RNA is characterized by Zeff values of ∼4–5, similar to the value obtained for WT Gag binding to Psi RNA (Table 1; Supplemental Fig. S2). Thus, deletion of the MA domain removes additional positive charges involved in TARPolyA binding, but not Psi RNA binding. Interestingly, the Kd(1M) for TARPolyA binding by CANC was the same as for NC, and this value is ∼10-fold smaller compared with WT Gag, suggesting that removal of MA allows optimization of nonelectrostatic NC interactions with RNA. The similarity between CANC and NC in binding to TARPolyA suggests that CA–CA interactions only play a minor role in Gag–RNA binding. Moreover, this observation excludes the possibility that the Zeff of ∼9 measured for Gag binding to TARPolyA is due to dimerization of Gag. Consistent with the notion of weak Gag–RNA binding cooperativity was also the observation that Psi RNA binds to Gag, CANC, and NC with similar Zeff values and with Kd(1M) values, differing only by approximately twofold for Gag and NC. Zeff reflects all ions displaced upon nucleic acid binding—ions displaced by direct contacts between the RNA and protein and those displaced by protein–protein contacts occurring upon RNA binding. Because the difference between Gag and NC binding comes from additional CA–CA interactions in Gag, we conclude that the CA–CA dimer interactions are weak and primarily nonelectrostatic, consistent with their primarily hydrophobic nature (for review, see Ganser-Pornillos et al. 2012).
It was surprising to find that eliminating dimerization leads to a major change in Gag binding to TARPolyA, as mentioned above; WM-Gag has a significantly lower Zeff (∼5) compared with WT Gag (∼9). Thus, eliminating Gag dimerization has a similar effect on non-Psi RNA binding as deleting MA. Indeed, WM-Gag and CANC appeared very similar in their RNA-binding properties (Supplemental Fig. S2). Both proteins bound Psi and TARPolyA with Zeff ∼5, and their Kd(1M) values were ∼10-fold smaller to Psi RNA than to TARPolyA, reflecting the RNA sequence specificity of the NC domain. The reduction in positive charges interacting with TARPolyA in the case of WM-Gag suggests that the loss of Gag dimerization favors an NC-only binding mode.
To further test the contribution of Gag’s MA domain to TARPolyA and Psi binding, we examined two previously described Gag variants (Jones et al. 2011) in which the MA domain is mutated to be either more positively charged (E40R,E42L,N47K GagΔp6, or Gag-3M) or more neutral (K30,32N GagΔp6). Binding to both RNAs by Gag-3M was characterized by higher Na1/2 values compared with any of the other proteins tested (551 mM for Psi RNA and 308 mM for TARPolyA) (Table 1; Supplemental Fig. S3). The Kd(1M) was reduced for Psi RNA binding (approximately fourfold) and TARPolyA binding (approximately fivefold) (Table 1), reflecting additional nonelectrostatic RNA contacts with Gag-3M, possibly due to the E42L mutation. Relative to WT Gag, the Zeff is increased for Gag-3M binding to Psi RNA (5 vs. 6.3), consistent with a contribution to electrostatic binding from the more positive MA domain in this variant. In contrast, Gag-3M bound to TARPolyA with an effective charge similar to WT Gag, suggesting that the additional charged residues in the MA domain of Gag-3M do not contribute to binding to TARPolyA.
Binding by Gag-K30,32N to both RNAs was characterized by a slightly lower Na1/2 value relative to WT Gag, but greater relative to CANC (Table 1; Supplemental Fig. S3). The effects of the neutralizing mutations also reduce the effective charge of binding to TARPolyA (Zeff ∼8), which falls between the value for WT Gag and CANC (Zeff = 9.1 and 4.7, respectively). Nonelectrostatic binding by Gag-K30,32N to TARPolyA was largely unchanged relative to WT Gag, which would be expected, as the deletion of the MA domain only reduced the Kd(1M) by approximately sixfold.
Taken together, the results of our studies of MA mutants suggest that both the MA and NC domains are involved in TARPolyA binding, but MA does not contribute to Psi RNA binding, as shown by lower Zeff values in MA variants. However, changes to Kd(1M) in these Gag variants are modest when compared with the zinc finger variants described below.
The zinc fingers in Gag are required for high-affinity Psi RNA binding
Previous studies have shown that NC binds RNAs in a largely nonspecific manner via electrostatic interactions between the nucleic acid backbone and the basic residues in the protein (Levin et al. 2005; Fisher et al. 2006; Vo et al. 2009a; Darlix et al. 2011; Wu et al. 2012). However, binding to certain RNA motifs, especially single-stranded UG sequences (Fisher et al. 1998), or exposed G residues in loop regions such as SL3 (De Guzman et al. 1998), involves high-affinity binding in which the zinc finger (ZF) aromatic residues stack with the G base in a hydrophobic pocket formed between the two ZFs (De Guzman et al. 1998; Darlix et al. 2011). To examine the contribution of the ZFs to Psi RNA and TARPolyA binding, we examined single and double-mutant Gag variants, in which either one or both critical ZF His residues were mutated to Cys (Fig. 1). These H-to-C mutations do not disrupt the tetrahedrally chelated zinc ion, but alter the zinc finger fold (Julian et al. 1993), and both viral infectivity and RNA packaging are severely reduced in the double H-to-C mutant (Gorelick et al. 1999a; Kafaie et al. 2008). We also examined ZF deletion variants in which either one or both ZFs were deleted (Fig. 1). These more severe changes also remove basic and aromatic residues within the ZFs and elicit defects in gRNA packaging (Houzet et al. 2008; Kafaie et al. 2008).
For proteins in which both zinc fingers were disrupted, nonelectrostatic binding to Psi was severely reduced (Fig. 4; Table 1), with a Kd(1M) ∼10−1 M for ΔZF1 + 2 and H400,421C, values that were dramatically higher (∼10,000-fold) than WT Gag. Thus, intact native ZFs are required for high-affinity nonelectrostatic binding. Importantly, in the double ZF mutants, Zeff values of ∼9–10 were obtained for Psi RNA binding, suggesting that the loss of binding specificity causes Gag to bind Psi RNA with a larger positive interface likely involving MA. Alternatively, loss of the zinc finger structure could allow for more favorable interactions between RNA and the basic residues in NC that would not be available for binding with intact zinc fingers, as suggested by others (Hargittai et al. 2004). Single ZF mutation variants were characterized by less severe binding defects with Zeff values between 5 and 9, and the Kd(1M) values ∼10- to 100-fold higher than for the WT Gag (Table 1). An exception was ΔZF1, which had a Kd(1M) value similar to WT Gag. Interestingly, for ZF variants higher Zeff values typically correlate with higher Kd(1M) values, indicating that the loss of binding specificity is commensurate with more positive charges of Gag directly contacting the RNAs. In viruses containing zinc finger deletion variants, gRNA packaging is reduced ∼10-fold for ΔZF1 and ΔZF2 and ∼100-fold for ΔZF1 + 2 (Houzet et al. 2008).
FIGURE 4.
Salt titrations in the presence of zinc finger variant ΔZF1 + 2. WT GagΔp6 is also shown for comparison. Binding to (A) Psi RNA and (B) TARPolyA are shown, with corresponding log–log plots shown in C and D, respectively. Plots of the other zinc finger variants are not shown for clarity, but the data from fits are presented in Table 1.
Gag binding to TARPolyA was also affected by ZF mutation, albeit to a lesser extent (Fig. 4; Table 1). ZF variants bound TARPolyA with ∼10−1 to 1 M Kd(1M), which was reduced ∼10- to 100-fold compared with WT Gag. Thus, TARPolyA binding by Gag also has a nonelectrostatic component associated with the ZFs of NC. Zeff was also increased slightly for ZF variants to ∼10–13 for ZF variants, compared with 9 for WT Gag. Therefore, as for Psi binding, the loss of nonelectrostatic binding for these proteins is accompanied by optimization of electrostatic contacts.
Role of Psi RNA dimerization and single-stranded G bases in high-affinity Gag binding
We next tested the contribution of Psi RNA dimerization to binding by introducing the C258G mutation to the DIS loop in SL1 of Psi RNA (Fig. 1A). This mutation reduces dimerization in virions by ∼50% (Shen et al. 2000). Native gel analysis showed minimal dimer formation by the C258G variant over the range of salt (Supplemental Fig. S4A) and RNA concentrations (Supplemental Fig. S4B) used in this work. In contrast, WT Psi RNA readily dimerized under the same conditions (Supplemental Fig. S4). The parameters obtained for Psi C258G RNA binding to Gag, NC, and ΔZF1 + 2 were the same, within error, as binding to WT Psi RNA (Table 1; Supplemental Fig. S5), suggesting that Psi RNA dimerization is not essential for high specificity of Gag binding to the Psi RNA construct examined here. According to one model (Lu et al. 2011a), a structural switch in the RNA coordinates dimerization and packaging, and Gag binding may possibly induce this switch. Noting that the C258G mutant bound identically to WT Psi RNA, there appears to be no preference for Gag to bind to dimerized RNA using this in vitro assay. However, Gag binding to full-length monomeric and dimeric vRNA could be altered by additional sequence elements not contained within our construct (Chamanian et al. 2013).
We next investigated whether there were additional sequence features of Psi that enhance Gag binding. We introduced 12 point mutations to Psi (Psi-12M) to remove single-stranded G bases in loops and bulges that have been proposed to be high-affinity NC binding sites based on SHAPE footprinting (Wilkinson et al. 2008). GagΔp6 binding to Psi-12M was characterized by an ∼25-fold weaker nonelectrostatic (i.e., specific) binding (Kd(1M) ∼ 1.2 × 10−3) (Table 1) relative to WT Psi. This interaction is ∼18-fold stronger than binding to TARPolyA, suggesting that some Gag-Psi RNA binding specificity resides in interactions beyond those that occur with the unpaired G bases. Additionally, the Zeff of binding to Psi-12M was ∼7, which falls between the Zeff obtained for WT GagΔp6 binding to Psi (∼5) and TARPolyA (∼9). Comparison of Gag binding to Psi and Psi-12M in Supplemental Figure S6 suggests that mutation of sites within Psi leads to the loss of most, but not all, of the Gag-Psi RNA binding specificity, implying that additional elements of primary and/or secondary structure contribute to Psi RNA recognition.
DISCUSSION
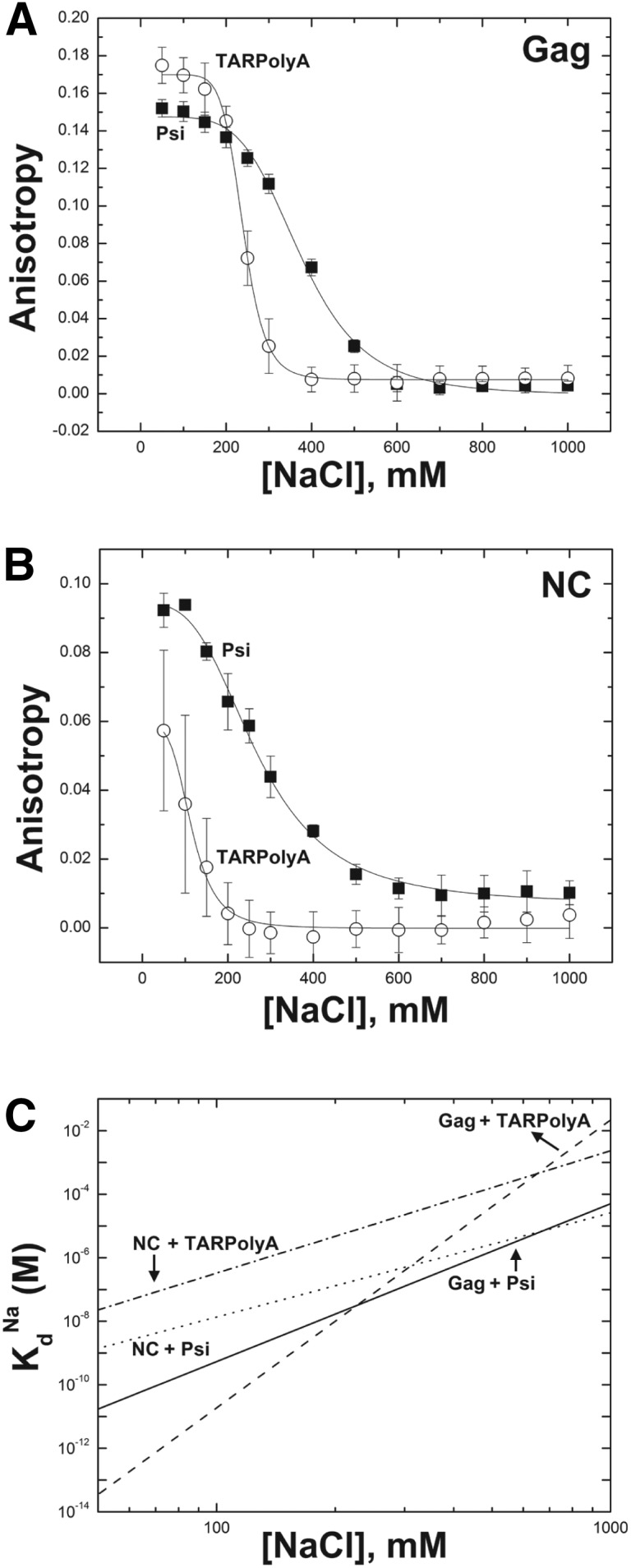
In this study we examined properties of recombinant HIV-1 Gag and NC binding to ∼100 nt RNAs derived from the 5′ UTR using an FA-based salt titration method to extract electrostatic and nonelectrostatic components of the strongest protein–RNA binding interaction. Surprisingly, we found that Gag bound Psi RNA and non-Psi RNA (i.e., TARPolyA) with markedly different biophysical characteristics. Psi RNA binding is optimized for specific, salt-independent interactions, and TARPolyA binding maximizes the total number of positive Gag charges binding to RNA at the expense of specific contacts. This finding is consistent with the model that HIV-1 Gag is a highly flexible polyprotein with two cationic RNA-binding sites in NC and MA domains (for review, see Rein et al. 2011). Specifically, Gag interacts with an RNA derived from the ψ portion of the genome with a Zeff and a Kd(1M) that are twofold and 100-fold lower than the values measured for TARPolyA binding. The higher Zeff observed in Gag TARPolyA binding suggests that an additional cationic binding interface in the MA domain is involved. The data also suggest that MA plays a role in binding TARPolyA, but not Psi RNA, which is consistent with the finding that CANC, a MA deletion variant, binds TARPolyA with a Zeff similar to that of NC (∼4) (Table 1). Surprisingly, the monomeric WM-Gag variant also bound to TARPolyA with reduced Zeff, suggesting that CA dimerization facilitates simultaneous NC and MA binding to non-Psi RNA.
The zinc fingers of NC are the source of Gag’s binding specificity to ψ, as shown by biochemical assays (Dannull et al. 1994), cell-based assays (Lever et al. 1989; Gorelick et al. 1990), high-resolution structures (De Guzman et al. 1998; Amarasinghe et al. 2000), and experiments described herein (Fig. 4). Binding of double ZF variants to Psi RNA is characterized by a Kd(1M) that is 10,000-fold greater than the value measured for WT Gag. Interestingly, the parameters determined for the ZF variants binding to Psi RNA are similar to those of WT Gag binding to TARPolyA (Table 1), strongly supporting the conclusion that the ZF structures are critical for binding specificity. In the NMR structures of NC bound to SL3, G320 of the SL3 tetraloop (Fig. 1) interacts with the first ZF of NC via residues Val13, Phe16, Ile24, and Ala25, and G318 of the SL3 tetraloop contacts the second zinc finger via residues Trp37, Gln45, and Met46 (De Guzman et al. 1998). These primarily nonelectrostatic contacts of Gag’s zinc fingers with Psi RNA are likely responsible for the ∼1000-fold smaller value of Kd(1M) measured for Gag binding to Psi RNA relative to TARPolyA. Likewise, the much higher Kd(1M) observed for binding of Psi-12M RNA to WT Gag suggests that removal of single-stranded G residues from Psi RNA largely eliminates this RNA’s enhanced binding specificity (Supplemental Fig. S6). Gag also bound Psi-12M with a higher Zeff, suggesting involvement of MA in addition to NC, a binding mode that resembles binding to TARPolyA.
The TAR stem–loop has a well-documented role in binding to the HIV-1 Tat protein, which stimulates transcription of the full-length viral RNA (Ott et al. 2011). In addition, NC binding to TAR RNA and its role in destabilizing the hairpin during the minus-strand transfer step of reverse transcription is well established (Kanevsky et al. 2005, 2011; Vo et al. 2009b; Levin et al. 2010; Heng et al. 2012). In an HIV-1 variant that does not depend on Tat–TAR interaction for transcription activation, TAR is not required for gRNA packaging (Das et al. 2007), but TAR destabilization leads to aberrant RNA dimerization and packaging (Das et al. 2012). Thus, although Gag–TAR interactions are not important for gRNA packaging, they likely occur during virus assembly (for review, see Lu et al. 2011b). One recent study investigating which portions of the 5′ UTR are essential for gRNA packaging found TAR to be dispensable for genome packaging, whereas deletion of the PolyA stem caused an approximately threefold decrease in packaged gRNA (Didierlaurent et al. 2011). This finding may be due, in part, to a long-range interaction proposed to occur between the PolyA stem–loop and a downstream region in the viral RNA (Paillart et al. 2002), which, if disrupted, leads to misfolding of the RNA. Compared with Psi RNA, TARPolyA binding to Gag and NC is characterized by a much higher Kd(1M) for both proteins and higher Zeff in the case of Gag (Table 1). However, Gag’s affinity for TARPolyA is approximately sixfold stronger than for Psi RNA at physiological ionic strength (∼150 mM NaCl) (Fig. 3C). Thus, binding affinity alone is unlikely to explain selective viral RNA packaging.
Live cell microscopy studies of HIV-1 assembly suggest that the number of Gag molecules selecting the gRNA in the cytoplasm is relatively small (i.e., <10) (for review, see Jouvenet et al. 2011). These studies also show that assembly of HIV-1 virus-like particles (VLPs) is nucleated at the plasma membrane, where a small number of Gag molecules are responsible for anchoring viral RNA in membranes. Interestingly, viral genomes are not retained at the plasma membrane when their packaging signals were mutated (Jouvenet et al. 2009). Moreover, in cells where HIV-1 genomic RNA was available for packaging, ∼85% of the observed VLP assembly events resulted in the encapsidation of gRNA. Although similar assembly times were measured in the presence or absence of gRNA, technical limitations did not allow visualization of less than ∼10 Gag molecules, thereby precluding observation of the initial Gag–gRNA association kinetics. Thus, the high probability of gRNA packaging implies that the packaging selectivity likely arises from the binding of the first few Gag molecules to gRNA and is consistent with the hypothesis that specific Gag–gRNA complexes display faster early assembly kinetics.
Based on our results, we hypothesize that the conformation of the first few Gag molecules strongly bound to ψ RNA via zinc finger-specific nonelectrostatic interactions differentiates this complex from Gag bound to ribonucleoprotein complexes containing cellular RNAs (Fig. 5). Whereas ψ–RNA binding involves only the NC domain, non-ψ binding is characterized by electrostatic interactions with both NC and MA, while the nonelectrostatic contacts are reduced. Thus, although the MA domain binds to the plasma membrane irrespective of the identity of the RNA bound to Gag, we propose that Gag bound to RNAs that lack ψ are kinetically hindered in binding to the plasma membrane in comparison to ψ-containing complexes (Fig. 5). In non-ψ complexes, MA is partially occupied by nucleic acid binding via the same interface used for PIP2 binding and, therefore, these complexes have a kinetic barrier to plasma membrane binding that involves MA–RNA dissociation (Shkriabai et al. 2006; Chukkapalli et al. 2010, 2013; Datta et al. 2011; Jones et al. 2011). We propose that the availability of the MA domain for plasma membrane binding in the Gag molecules bound to ψ-containing gRNA could confer a kinetic advantage for gRNA–Gag complexes to initiate assembly (Fig. 5).
FIGURE 5.
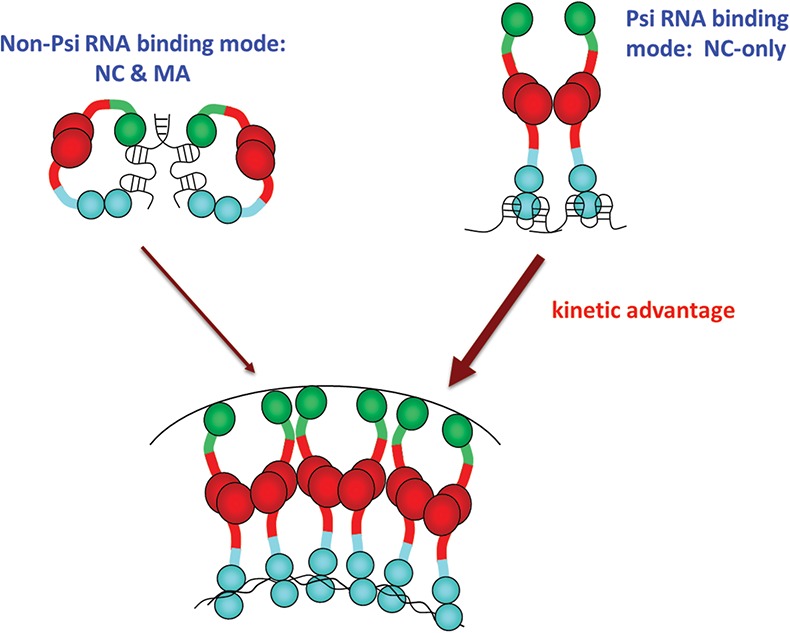
Model for selection and packaging of gRNA by the first few binding Gag molecules. Non-ψ binding is characterized by Gag binding in a NC- and MA-bound conformation, but Gag binds ψ in an NC-only binding mode. The NC-only mode leaves MA free to interact with the membrane and has a kinetic advantage over complexes in which MA is bound to nucleic acids.
In summary, in this work we have explored the effect of multiple mutations and deletions in Gag on the specificity of binding to Psi RNA vs. TARPolyA RNA. We show that Gag can bind to different RNA molecules with distinct binding modes and these mechanistic differences may have important implications for gRNA packaging. Importantly, this mechanism accounts for selective ψ RNA packaging even when the binding affinities of the competing protein–RNA complexes at physiological salt are similar. Our data also suggest that the zinc finger structures in the Gag NC domain and interactions with single-stranded G-rich motifs in ψ confer specific Gag binding. Future salt titration studies of Gag binding to additional Psi RNA mutants, as well as other vRNA-derived and unrelated RNA molecules, will further explore the details of the interactions responsible for Gag’s binding specificity.
MATERIALS AND METHODS
Preparation of proteins and nucleic acids
All Gag proteins used in this study lack the p6 domain. WT GagΔp6 and variants were prepared as previously described (Datta and Rein 2009; Jones et al. 2011). Briefly, the preparation involves ammonium sulfate precipitation and ion-exchange (phosphocellulose resin) purification, followed by size exclusion chromatography. Fractions containing protein were then concentrated, aliquoted, and stored at −80°C. Some of the NC was a gift of Dr. Robert Gorelick (NCI-Frederick) and was purified as previously described (Urbaneja et al. 1999). NC prepared by solid-phase synthesis and reconstituted with zinc (Liu et al. 2005) was also used. TARPolyA, Psi, and PsiC258G RNAs were in vitro transcribed from linearized plasmids, originally cloned from plasmid pMSMΔEnv containing HIV-1 NL4-3 cDNA, a gift of Dr. Kathleen Boris-Lawrie (The Ohio State University). The PsiC258G mutant plasmid was prepared from the WT Psi RNA plasmid by Quik-change site-directed mutagenesis (Stratagene). Psi-12M RNA is derived from WT Psi RNA and contains the following 12 point mutations: G240,241A, G272,273A, G290A, U291A, G292A, G310A, G318,320A, and G328,329A. Psi-12M RNA was transcribed from a PCR-assembled template using the following DNA oligonucleotides purchased from Integrated DNA Technologies:
5′-TAATACGACTCACTATAGGGACGCAAAACTCGGCTTGCTGA-3′,
5′-TTTTCTAGCTTTCGCTAGTAAAAATTTTTGGCGTACTTT-3′,
5′-CGCAAAACTCGGCTTGCTGAAGCGCGCACGGCAAGAAACGAGGGGCGGCGACTGAAAAGTACGCCAAAAATTTTT-3′, and 5′-AAAAATTTTTGGCGTACTTTTCAGTCGCCGCCGCCTCGTTTCTTGCCGTGCGCGCTTCAGCAAGCCGAGTTTTGCG-3′.
RNAs were fluorescently labeled with fluorescein on their 3′ ends as previously described (Pagano et al. 2007; Jones et al. 2013). Concentrations and labeling efficiencies were determined using the following extinction coefficients—fluorescein, 8.5 × 104 M−1 cm−1; WT Psi, 8.7 × 105 M−1 cm−1; PsiC258G, 8.7 × 105 M−1 cm−1; Psi-12M, 8.7 × 105 M−1 cm−1; TARPolyA, 9.26 × 105 M−1 cm−1. After labeling, RNAs were aliquoted and stored at −20°C. Analysis of RNAs by native gel electrophoresis is described in the Supplemental Information.
FA equilibrium binding assays
Direct FA-binding experiments were performed as previously described (Stewart-Maynard et al. 2008). Varying amounts of GagΔp6 were incubated with 30–40 nM 3′-fluorescently labeled RNA in a buffer containing 20 mM HEPES (pH 7.5), 1 mM MgCl2, 10 μM tris-2-carboxy-ethyl phosphine (TCEP), 5 mM β-mercaptoethanol (BME), and varying amounts of NaCl. NC-containing reactions were identical except that they contained 20 μM TCEP and two equivalents of zinc acetate per NC. Reactions were incubated in the dark at room temperature for 30 min prior to FA and fluorescence intensity measurements using a SpectraMax M5 plate reader (Molecular Devices). Fitting of the direct titration curves is described in the Supplemental Information.
FA salt titration assay
Reaction conditions were the same as in direct binding measurements, except that protein concentrations were held constant (400 or 750 nM) and NaCl concentrations were varied from 50 mM to 1 M. This protein concentration was chosen because the RNA binding had reached a plateau according to equilibrium binding measurements performed at 50 mM NaCl (Supplemental Fig. S1). However, only the strongest protein-binding sites on RNA are saturated under these conditions (see Supplemental Information). Therefore, our salt titrations allow characterization and comparison of the highest affinity NC or Gag sites on each RNA molecule studied. To correct for the difference in solution viscosity and/or conformational changes in RNA molecules created by the increasing NaCl concentration, FA values in the absence of protein were subtracted from the FA for protein-containing reactions in each experiment. The corrected data were fit to sigmoidal curves using OriginPro8 Software (OriginLab Corporation), which yielded Na1/2 values or the concentration of NaCl at which 50% protein is dissociated from the labeled RNA. The dissociation constant Kd(Na) varies with [Na] as follows:
| (1) |
Equation 1 is derived in the Supplemental Information and follows from the features of nonlinear screening of nucleic acid charges by cationic proteins or ligands (Manning 1976; Record et al. 1976; Rouzina and Bloomfield 1996a,b,c). Substituting equation 1 into the binding isotherm (Supplemental equation S4) allows fitting of the two basic parameters describing the salt-sensitive protein/RNA binding: Kd(1M) and Zeff, as described in the Supplemental Information.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Rob Gorelick (SAIC-Frederick, Inc., NCI-Frederick) and Dr. Kathleen Boris-Lawrie (The Ohio State University) for HIV-1 NC protein and pMSMΔEnv plasmid, respectively. We thank Dr. George Barany and Dr. Dan Mullen (University of Minnesota) for providing us with synthetic HIV-1 NC protein. This work was supported by NIH GM65056 (K.M.-F.), NIH R01 CA76534 (L.J.P.), ARRA supplement to CA76534-12S (L.J.P. and K.M.-F.), and by a fellowship to C.P.J. from The Ohio State University Center for RNA Biology.
REFERENCES
- Alfadhli A, Still A, Barklis E 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol 83: 12196–12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. Implications for genome recognition. J Mol Biol 301: 491–511 [DOI] [PubMed] [Google Scholar]
- Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN 2010. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry 49: 3525–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilov SV, Godet J, Piemont E, Mely Y 2009. Site-specific characterization of HIV-1 nucleocapsid protein binding to oligonucleotides with two binding sites. Biochemistry 48: 2422–2430 [DOI] [PubMed] [Google Scholar]
- Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, Le Grice SF, Arts EJ 2013. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 13: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol 82: 2405–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Oh SJ, Ono A 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci 107: 1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Inlora J, Todd GC, Ono A 2013. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J Virol 87: 7155–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever JL, Parslow TG 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol 71: 3407–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus H 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Dannull J, Surovoy A, Jung G, Moelling K 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J 13: 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol 410: 565–581 [DOI] [PubMed] [Google Scholar]
- Das AT, Harwig A, Vrolijk MM, Berkhout B 2007. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol 81: 7742–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Vrolijk MM, Harwig A, Berkhout B 2012. Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SA, Rein A 2009. Preparation of recombinant HIV-1 gag protein and assembly of virus-like particles in vitro. Methods Mol Biol 485: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A 2007. Conformation of the HIV-1 Gag protein in solution. J Mol Biol 365: 812–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SA, Heinrich F, Raghunandan S, Krueger S, Curtis JE, Rein A, Nanda H 2011. HIV-1 Gag extension: Conformational changes require simultaneous interaction with membrane and nucleic acid. J Mol Biol 406: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279: 384–388 [DOI] [PubMed] [Google Scholar]
- Didierlaurent L, Racine PJ, Houzet L, Chamontin C, Berkhout B, Mougel M 2011. Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles. Nucleic Acids Res 39: 8915–8927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza V, Summers MF 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431: 586–590 [DOI] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM 2007. Fields virology. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol 72: 1902–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, et al. 2006. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res 34: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Pornillos O 2012. Assembly and architecture of HIV. Adv Exp Med Biol 726: 441–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW Jr, Gorelick RJ, Rein A, Weeks KM 2010. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci 107: 19248–19253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Nigida SM Jr, Bess JW Jr, Arthur LO, Henderson LE, Rein A 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol 64: 3207–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Benveniste RE, Gagliardi TD, Wiltrout TA, Busch LK, Bosche WJ, Coren LV, Lifson JD, Bradley PJ, Henderson LE, et al. 1999a. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain mne produce virions that are replication defective in vitro and in vivo. Virology 253: 259–270 [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO 1999b. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: Characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256: 92–104 [DOI] [PubMed] [Google Scholar]
- Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K 2004. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol 337: 951–968 [DOI] [PubMed] [Google Scholar]
- Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF 2012. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol 417: 224–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M 2007. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res 35: 2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M 2008. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res 36: 2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K 2011. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J Virol 85: 1594–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K 2013. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 19: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci 106: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD 2011. Visualizing HIV-1 assembly. J Mol Biol 410: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian N, Demene H, Morellet N, Maigret B, Roques BP 1993. Replacement of His23 by Cys in a zinc finger of HIV-1 NCp7 led to a change in 1H NMR-derived 3D structure and to a loss of biological activity. FEBS Lett 331: 43–48 [DOI] [PubMed] [Google Scholar]
- Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375: 592–610 [DOI] [PubMed] [Google Scholar]
- Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fosse P 2005. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J Mol Biol 348: 1059–1077 [DOI] [PubMed] [Google Scholar]
- Kanevsky I, Chaminade F, Chen Y, Godet J, Rene B, Darlix JL, Mely Y, Mauffret O, Fosse P 2011. Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res 39: 8148–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene SE, Telesnitsky A 2012. cis-Acting determinants of 7SL RNA packaging by HIV-1. J Virol 86: 7934–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman L, Jones CP, Musier-Forsyth K 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett 584: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A, Gottlinger H, Haseltine W, Sodroski J 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol 63: 4085–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286 [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7: 754–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF 2005. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys J 89: 3470–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. 2011a. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Summers MF 2011b. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410: 609–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning GS 1976. On the application of polyelectrolyte limiting laws to the helix-coil transition of DNA. V. Ionic effects on renaturation kinetics. Biopolymers 15: 1333–1343 [DOI] [PubMed] [Google Scholar]
- McBride MS, Panganiban AT 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol 71: 2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y, Garcia EL, King SR, Iyalla K, Loeliger K, Starck P, Syed S, Telesnitsky A, Summers MF 2010. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J Mol Biol 396: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Hu WS 2009. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev 11: 91–102 [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A 2001. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci 98: 5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SH, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS 2013. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog 9: e1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q 2011. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 10: 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP 2007. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem 282: 8883–8894 [DOI] [PubMed] [Google Scholar]
- Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R 2002. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J Biol Chem 277: 5995–6004 [DOI] [PubMed] [Google Scholar]
- Parkash B, Ranjan A, Tiwari V, Gupta SK, Kaur N, Tandon V 2012. Inhibition of 5′-UTR RNA conformational switching in HIV-1 using antisense PNAs. PLoS One 7: e49310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record MT Jr, Lohman ML, De Haseth P 1976. Ion effects on ligand-nucleic acid interactions. J Mol Biol 107: 145–158 [DOI] [PubMed] [Google Scholar]
- Rein A, Datta SA, Jones CP, Musier-Forsyth K 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem Sci 36: 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzina I, Bloomfield VA 1996a. Competitive electrostatic binding of charged ligands to polyelectrolytes: Planar and cylindrical geometries. J Phys Chem 100: 4294–4304 [DOI] [PubMed] [Google Scholar]
- Rouzina I, Bloomfield VA 1996b. Influence of ligand spatial organization on competitive electrostatic binding to DNA. J Phys Chem 100: 4305–4313 [Google Scholar]
- Rouzina I, Bloomfield VA 1996c. Macroion attraction due to electrostatic correlation between screening counterions. 1. Mobile surface-adsorbed ions and diffuse ion cloud. J Phys Chem 100: 9977–9989 [Google Scholar]
- Rouzina I, Bloomfield VA 1997. Competitive electrostatic binding of charged ligands to polyelectrolytes: Practical approach using the non-linear Poisson-Boltzmann equation. Biophys Chem 64: 139–155 [DOI] [PubMed] [Google Scholar]
- Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81: 6623–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RS, Hu J, Beriault V, Mouland AJ, Laughrea M, Kleiman L, Wainberg MA, Liang C 2003. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type 1 RNA. J Virol 77: 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Jette L, Liang C, Wainberg MA, Laughrea M 2000. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol 74: 5729–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45: 4077–4083 [DOI] [PubMed] [Google Scholar]
- Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci 91: 4945–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Datta SA, Worthy KM, Bindu L, Fivash MJ, Turner KB, Fabris D, Rein A, Fisher RJ 2007. Measuring the binding stoichiometry of HIV-1 Gag to very-low-density oligonucleotide surfaces using surface plasmon resonance spectroscopy. J Biomol Tech 18: 259–266 [PMC free article] [PubMed] [Google Scholar]
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol 82: 10129–10142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG 2012. HIV-1 Assembly, budding, and maturation. Cold Spring Harb Perspect Med 2: a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: Elucidation of the structural determinants for function. J Mol Biol 287: 59–75 [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K 2006. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol 363: 244–261 [DOI] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K 2009a. Effect of Mg2+ and Na+ on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Implications for reverse transcription. J Mol Biol 386: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K 2009b. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop–loop “kissing” to “zipper”. J Mol Biol 386: 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mely Y 1999. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: A fluorescence study. Biochemistry 38: 16816–16825 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Datta SA, Mitra M, Gorelick RJ, Rein A, Levin JG 2010. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: Biological implications. Virology 405: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ 2012. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication. Virus Res 171: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]