Abstract
Background:
Population-based studies of breast cancer have estimated that at least some PALB2 mutations are associated with high breast cancer risk. For women carrying PALB2 mutations, knowing their carrier status could be useful in directing them towards effective cancer risk management and therapeutic strategies. We sought to determine whether morphological features of breast tumours can predict PALB2 germline mutation status.
Methods:
Systematic pathology review was conducted on breast tumours from 28 female carriers of PALB2 mutations (non-carriers of other known high-risk mutations, recruited through various resources with varying ascertainment) and on breast tumours from a population-based sample of 828 Australian women diagnosed before the age of 60 years (which included 40 BRCA1 and 18 BRCA2 mutation carriers). Tumour morphological features of the 28 PALB2 mutation carriers were compared with those of 770 women without high-risk mutations.
Results:
Tumours arising in PALB2 mutation carriers were associated with minimal sclerosis (odds ratio (OR)=19.7; 95% confidence interval (CI)=6.0–64.6; P=5 × 10−7). Minimal sclerosis was also a feature that distinguished PALB2 mutation carriers from BRCA1 (P=0.05) and BRCA2 (P=0.04) mutation carriers.
Conclusion:
This study identified minimal sclerosis to be a predictor of germline PALB2 mutation status. Morphological review can therefore facilitate the identification of women most likely to carry mutations in PALB2.
Keywords: PALB2, hereditary, breast cancer, tumour morphology
PALB2, a partner and localiser of BRCA2, is crucial for proficient homologous recombination repair of DNA double-strand breaks through its regulation of BRCA2 and its interaction with BRCA1 (Xia et al, 2006; Sy et al, 2009; Zhang et al, 2009). Bi-allelic inactivating mutations in PALB2 underlie Fanconi anaemia subtype N and have been shown to be associated with high risk of childhood cancers (Reid et al, 2007; Xia et al, 2007). Heterozygous germline loss-of-function mutations in PALB2 have been associated with increased risk of breast cancer (Rahman et al, 2007).
The first study that reported an association between PALB2 mutations and breast cancer risk involved familial breast cancer cases and unaffected controls from the United Kingdom. Using only some information obtained from just 10 families, and under strong modelling assumptions, the average relative risk associated with 5 protein-truncating PALB2 mutations was estimated indirectly to be 2.3-fold (95% confidence interval (CI)=1.4–3.9) (Rahman et al, 2007). Subsequent population-based studies estimated the risk associated with PALB2 mutations to be higher (Erkko et al, 2008; Southey et al, 2010). For example, PALB2 c.1592delT was identified in 18 out of 1918 (0.9%) Finnish breast cancer cases unselected for family history compared with 6 out of 2501 (0.2%) unaffected controls (odds ratio (OR)=3.94; 95% CI=1.5–12.1). Using the family histories of the case carriers, PALB2 c.1592delT was estimated to be associated with a 40% (95% CI=17–77%) risk of breast cancer to the age of 70 years (Erkko et al, 2008). Similarly, PALB2 c.3113G>A was identified in 5 out of 1403 (0.4%) unselected Australian breast cancer cases and 0 out of 764 (0%) unaffected controls (Southey et al, 2010). Using the family histories of the five carrier cases, the estimated cumulative risk for PALB2 c.3113G>A was 91% (95% CI=44–100%) to the age of 70 years. Therefore, population-based studies of breast cancer that have directly used the family history data have estimated that at least some PALB2 mutations are associated with a breast cancer risk (penetrance) comparable to that of the average pathogenic mutation in BRCA2: 45% (95% CI=31–56%) (Antoniou et al, 2003).
Mutations in PALB2 are rare (varying from 0.1% to 1.5% depending upon the population) (Foulkes et al, 2007; Rahman et al, 2007; Tischkowitz et al, 2007; Dansonka-Mieszkowska et al, 2010; Papi et al, 2010; Southey et al, 2010; Bogdanova et al, 2011; Casadei et al, 2011; Ding et al, 2011; Hellebrand et al, 2011; Teo et al, 2013a, 2013b) but for women carrying them, and their relatives who might also be mutation carriers, knowing their mutation status has the potential to be clinically important as carriers are at high risk of breast cancer. Identified mutation carriers could be informed of optimal, risk appropriate clinical screening and treatment. Potential therapies could include those that target homologous DNA repair dysfunction (Buisson et al, 2010). As PALB2 mutations have also been associated with increased risk of developing a second breast cancer (Tischkowitz et al, 2012), risk reducing surgery and treatment might also be considered by PALB2 mutation carriers. The integration of PALB2 mutation testing into clinical practice is still in progress and strategies that effectively identify potential PALB2 mutation carriers could help facilitate this important process.
Characterisation of the morphology of breast cancers arising in PALB2 mutation carriers and non-carriers offers the possibility of identifying tumour morphological features predictive of an underlying germline PALB2 mutation, as they have been shown for underlying BRCA1 mutations (Lakhani et al, 1998; Southey et al, 2011; Hopper et al, 2012). This could be conducted at the time of diagnosis and therefore, be used to facilitate personalised treatment strategies, as well as enabling identification of those relatives who have also inherited a similar high breast cancer risk.
Breast cancer tumour morphology can be suggestive of underlying familial, if not heritable, risk. We recently reported that, in a population-based sample of 375 women with early-onset breast cancer cases with no known high-risk mutation in a breast cancer susceptibility gene, minimal sclerosis, presence of circumscribed growth, extensive intraductal carcinoma and lobular growth patterns were independent predictors of increased breast cancer risk for their first-degree female relatives (2.0-fold to 3.3-fold increased risk for relatives, P<0.02 for all listed features). Relatives of the 128 (34%) index cases with none of these 4 features were at population risk (standardised incidence ratio=1.03, 95% CI=0.57–1.85), while relatives of the 37 (10%) index cases with two or more features were at high risk (standardised incidence ratio=5.18, 95% CI=3.22–8.33) (Dite et al, 2012).
Breast cancer morphological features can also be used to identify women most likely to carry germline mutations in breast cancer susceptibility genes. It has been known for some time that some morphological features are more common in cancers arising in BRCA1 mutation carriers (Lakhani et al, 1998). These features have been identified by studying carriers across a wide range of ages at diagnosis and ascertained either because of their strong family cancer history or through population-based sampling. Lack of oestrogen receptor (ER) and progesterone receptor (PR) expression has also been reported to improve prediction of BRCA1 mutation status based on family history (Lakhani et al, 2002; James et al, 2006; Mavaddat et al, 2010). Using a population-based sample of 452 young women with breast cancer, we found that just two breast tumour morphological features (trabecular growth pattern and high mitotic index) were sufficient to identify 28 of the 29 BRCA1 mutation carriers in the study (Southey et al, 2011). Moreover, prediction of mutation status using these two features was more sensitive and specific than using family history alone, and when combined, the area under the receiver operator curve was in excess of 0.9.
A detailed analysis of the morphological features of PALB2 mutation-associated breast cancers has not been previously conducted. Some information about the general morphology of breast tumours arising in PALB2 mutation carriers is available from work studying breast tumours carrying the Finnish founder mutation PALB2 c.1592delT. Mutation carriers with a family history of breast cancer were more likely to have ‘triple negative' tumours (negative for ER, PR, and human epidermal growth factor receptor 2 (HER2) expression) when compared with familial non-PALB2 mutation-associated breast cancers (54.5% and 12.2%, respectively; P<0.0001). The PALB2 c.1592delT-associated tumours were reported to be more often of higher grade and to have greater expression of Ki67, which is a cellular marker for proliferation than tumours arising in non-carriers of the mutation. Carrying this PALB2 mutation was also reported to be associated with reduced survival; comparing affected PALB2 mutation carriers, negative for HER2 expression, with a family history of breast cancer with affected non-carriers of BRCA1, BRCA2, or PALB2 mutations, the hazard ratio was estimated to be 4.57 (95% CI=1.96–10.64; P=0.0004) (Heikkinen et al, 2009).
In this study, we conducted a standardised pathology review of 28 invasive breast cancers arising in women who carry a germline loss-of-function PALB2 mutation. The morphological characteristics of these 28 tumours were compared with those of a population-based sample of 770 unselected breast tumours that had undergone the same standard pathology review.
Materials and methods
Subjects
The women in this study were participants in three breast cancer research resources: the Breast Cancer Family Registry (BCFR) (John et al, 2004), in particular the Australian BCFR; the Victorian Familial Breast Cancer Cohort (VFBCC) (Sawyer et al, 2012); and the Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) (Mann et al, 2006). All participants provided written informed consent to participate in these research programs that were approved by the relevant ethics committees, including the Cancer Council Victoria and the New South Wales Cancer Council, and all participating sites/centres of the BCFR, kConFab, and the VFBCC. This study was approved by the Human Research Ethics Committee of The University of Melbourne.
PALB2 mutation carriers
A total of 28 women with invasive breast cancers who had been found to carry a PALB2 germline mutation were included in this study. This included 24 women who carried PALB2 c.3113G>A (5 from the Australian BCFR, 2 from the Ontario BCFR, 1 from the Utah BCFR, 5 from the VFBCC, and 11 from kConFab). The remaining four women were from kConFab; one was a carrier of PALB2 c.196C>T, another carried PALB2 c.1947_1948insA, and two were carriers of PALB2 c.2982_2983insT.
The PALB2 mutation carriers in the Australian BCFR, kConFab, and the VFBCC have been reported previously (Southey et al, 2010; Teo et al, 2013a, 2013b). The PALB2 c.3113G>A carriers in the Ontario BCFR and Utah BCFR were identified via Taqman assay as described in Southey et al (2010) and Teo et al (2013a) by screening 1831 and 68 probands from these BCFRs, respectively. Probands from the California (n=2052), New York (n=849), and Philadelphia (n=403) BCFRs had also been genotyped for PALB2 c.3113G>A using Taqman assay but no carriers were identified. The 28 PALB2 mutation carriers were from 21 participating families as described in Table 1.
Table 1. Basic demographics of 28 PALB2 mutation carriers with tumour material available for pathology review.
| Mutation | Probands | Relatives | Ages of diagnosis | Resource | Reference |
|---|---|---|---|---|---|
|
PALB2 c.3113G>A |
3 |
2 |
28, 35, 37, 42, 47 |
Australian BCFR |
Southey et al (2010) |
| |
2 |
0 |
45, 57 |
Ontario BCFR |
— |
| |
1 |
0 |
48 |
Utah BCFR |
— |
| |
7 |
4 |
32, 40, 41, 47, 47, 48, 49, 49, 54, 61, 63 |
kConFab |
Southey et al (2010) |
| |
5a |
0 |
33, 38, 42, 44, 45 |
VFBCC |
Teo et al (2013b) |
|
PALB2 c.196C>T |
1 |
0 |
43 |
kConFab |
Teo et al (2013a) |
|
PALB2 c.1947_1948insA |
1 |
0 |
42 |
kConFab |
Teo et al (2013a) |
| PALB2 c.2982_2983insT | 1 | 1 | 47, 54 | kConFab | Teo et al (2013a) |
Abbreviations: BCFR=Breast Cancer Family Registry; kConFab=Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer; VFBCC=Victorian Familial Breast Cancer Cohort.
One proband in the VFBCC was a relative of a proband participating in the Australian BCFR, and both were carriers of PALB2 c.3113G>A.
The diagnostic haematoxylin and eosin pathology slides, blocks, or digital images of the haematoxylin and eosin sections for each of the 28 PALB2 mutation carriers were retrieved from the diagnostic centres. A pathology review was conducted by an expert breast pathologist (EP) using a standardised pathology review tool (described below). Data on ER, PR, and HER2 status of the PALB2 mutation-associated tumours were collected, if available, from diagnostic laboratories and pathology reports. The HER2 status was considered to be positive if immunohistochemical test results were ranked 3+ (higher than normal amount of HER2 protein was present) or if tested as positive via fluorescence in situ hybridisation. An immunohistochemical test result of 1+ (normal amount of HER2 protein was present) was classified as negative for HER2 expression while an immunohistochemical test result of 2+ (moderate amount of HER2 protein was present) without a confirmatory fluorescence in situ hybridisation test was classified as equivocal.
Non-PALB2 mutation carriers: population-based sample
The Australian BCFR used population-based sampling to recruit 1485 population-based probands between 1993 and 1999. The DNA derived from the Australian BCFR probands diagnosed before the age of 40 years (n=692) was screened for genetic mutations in the coding and flanking intronic regions of PALB2 using high-resolution melt analysis (Southey et al, 2010). The Australian BCFR probands diagnosed at ages 40 or older (n=793) were genotyped for PALB2 c.3113G>A using Taqman assay (Southey et al, 2010). First, primary invasive breast tumours from 836 (56%) of these probands were retrieved from diagnostic centres and systematically reviewed by pathologists as described below and elsewhere (John et al, 2004; Southey et al, 2011; Dite et al, 2012). Among the breast tumours that were reviewed, 40 (5%) were from BRCA1 mutation carriers, 18 (2%) were from BRCA2 mutation carriers, 1 (0.1%) was from a carrier of ATM c.7271T>G and 4 (0.5%) were from TP53 mutation carriers (Southey et al, 1999; Andrulis et al, 2002; Chenevix-Trench et al, 2002; Apicella et al, 2003; Dite et al, 2003; Bernstein et al, 2006; Smith et al, 2007; Neuhausen et al, 2009; Mouchawar et al, 2010; Dite et al, 2012). Three breast tumours were from PALB2 mutation carriers (Southey et al, 2010) and were included in the PALB2 mutation carrier group (Table 1). The remaining 770 (93%) tumours were from women not found to carry a mutation in BRCA1, BRCA2, ATM, PALB2, or TP53 after extensive screening (Southey et al, 1999; Dite et al, 2003; Mouchawar et al, 2010).
Pathology review
The haematoxylin and eosin-stained breast tumour tissue was reviewed and scored for morphology features by one or more trained pathologists using a standardised tool as previously applied (Armes et al, 1998; Southey et al, 2011; Dite et al, 2012) and validated (Longacre et al, 2006). Briefly, tumour grade was scored using the modified system of Bloom-Richardson by assessing mitotic rate, nuclear pleomorphism, and tubular differentiation (Elston et al, 1999). Tumours were typed into primary growth pattern (representing 75% or more of the tumour or ∼60% of the tumour if a secondary pattern was present) and secondary pattern (representing ∼40% of the tumour) using the World Health Organization breast carcinoma classification with minor modifications (Page et al, 1987). The carcinomas were categorised into 17 histological types: infiltrating ductal not otherwise specified, tubular, cribriform, micropapillary, mucinous (colloid), secretory, medullary (classical), medullary (atypical), adenoid cystic, metaplastic, lobular (classical), lobular (trabecular), lobular (alveolar), lobular (solid), tubulo (lobular), pleomorphic lobular, or other. Tumours were classified as having a primary histological type with no secondary type if >70% of the tumour presented with features characteristic of the histological type. Tumours were also classified as having both a primary (60%) and a secondary histological type (40%) if the tumour presented with features characteristic of two histological types. Sclerosis of the tumour was defined as fibrosis composed of fibroblasts and/or collagen that is devoid of tumour cells (Van den Eynden et al, 2008; Dite et al, 2012). The presence of extensive sclerosis is similar to a fibrotic focus as defined by Van den Enden et al (2008), which has been shown to be easily assessable and reproducible morphological feature in breast cancer (Van den Eynden et al, 2007). A tumour was defined to have minimal sclerosis if ⩽20% of the tumour volume contained sclerosis and defined to contain extensive sclerosis if >20% of the tumour volume consisted of sclerosis. Information of the remaining tumour features from the pathology reviews was extracted as ‘present' or ‘absent' for statistical analysis as presented in Table 2.
Table 2. Classification criteria of standardised pathology review tool to assess tumour features in invasive breast cancer.
| |
Criteria for classification |
|
|---|---|---|
| Present | Absent | |
| Nuclear grade |
Malignant |
Bland/intermediate |
| Minimal tubule formation |
Tubule formation observed in <10% of tumour |
Tubule formation observed in ⩾10% of tumour |
| Number of mitotic cells ⩾20 |
⩾20 Mitotic cells identified per 10 high powered fields |
<20 Mitotic cells identified per 10 high powered fields |
| Syncytial growth pattern |
⩾75% of the tumour was observed to consist of broad sheets of tumour cells with indistinct cell borders |
Absent |
| Pushing margins |
>50% of tumour border observed to be well defined by a continuous pushing front of tumour cells |
Absent |
| Circumscribed growth pattern |
>50% of tumour border observed to be well defined |
Absent |
| Lymphocytic infiltration site |
Diffuse within tumour |
Absent or observed to be at the border of the tumour |
| Lymphocytic infiltration level |
Intense |
Absent/minimal/moderate |
| Minimal sclerosis |
Minimal: ⩽20% of tumour is observed to contain sclerosis |
Extensive: >20% of tumour consists of sclerosis which is defined as fibrosis composed of fibroblasts and/or collagen that is devoid of tumour cells |
| Necrosis |
Present |
Absent/uncertain |
| Apoptosis |
Intense |
Absent/minimal/moderate |
| Lymphovascular invasion |
Cancerous cells observed in blood and/or lymphatic vessels |
Uncertain or absence of cancerous cells in blood and lymphatic vessels |
| Acinar growth pattern |
Present |
Absent |
| Lobular growth pattern |
Present |
Absent |
| Trabecular growth pattern |
Present |
Absent |
| Tubular growth pattern |
Present |
Absent |
| Atypical lobular hyperplasia |
Present |
Absent |
| Atypical ductal hyperplasia |
Present |
Absent |
| Lobular carcinoma in situ |
Present |
Absent |
| Ductal carcinoma in situ | Present | Absent |
The ER and PR status were obtained from immunohistochemical testing of tumour tissues or from histopathology reports held by cancer registries (Armes et al, 1999) or diagnostic laboratories (McCredie et al, 2003). The ER and PR status were available for ∼90% of the retrieved tumours of non-carriers of PALB2 mutations (746 and 745 tumours, respectively).
Statistical analyses
Missing data for tumour morphology features (average 4 (0.5%) missing per feature) were estimated using multiple imputation, made possible by the correlations between different morphological features (see Southey et al, 2011 and Dite et al, 2012). Multiple linear logistic regression was used to estimate the OR and 95% CI for associations between each of the morphological features and carrier status (PALB2 mutation carriers vs non-carriers of high-risk mutations, PALB2 mutation carriers vs BRCA1 mutation carriers and PALB2 mutation carriers vs BRCA2 mutation carriers), after adjusting for the number of affected first-degree relatives, number of affected second-degree relatives and age at diagnosis. These adjustments were necessary given that sampling of some carriers was from cases selected specifically because they had a family history and/or an early age at diagnosis. For the multivariate models, the best-fitting model was identified by stepwise selection, starting with the most significant variable and testing the addition of each of the remaining variables. All analyses were performed with Stata Version 11 (StataCorp, 2009). Following convention, all statistical tests were two-sided and P-values<0.05 were considered as nominally statistically significant. The positive and negative predictive values of a morphological feature for unselected cases were calculated based on the prevalence of PALB2 c.3113G>A affected carriers in a population-based study (0.36%) (Southey et al, 2010) and the prevalence of the morphological feature in the Australian BCFR breast cancer cases recruited by population-based sampling.
Results
Tumour morphological features associated with PALB2 mutation status
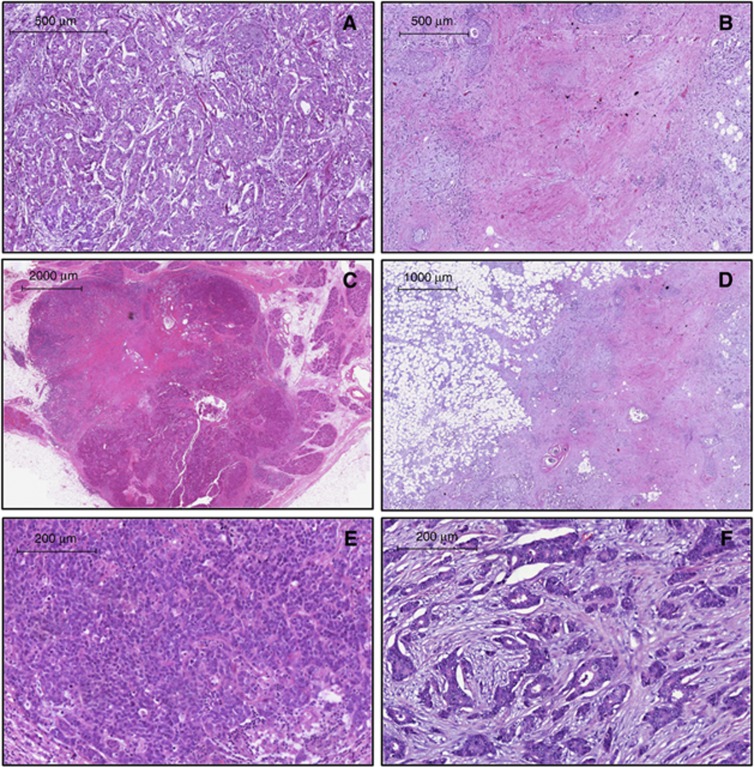
Table 3 shows that having minimal sclerosis was associated with PALB2 mutation status (OR=19.7; 95% CI=6.0–64.6; P=5 × 10−7). This association of minimal sclerosis remains strongly significant even after correcting for multiple comparisons (Bonferroni correction). There was marginal evidence for an association between PALB2 mutation status and having minimal tubule formation (OR=5.6; 95% CI=1.3–24.2; P=0.02), having lobular carcinoma in situ (OR=5.7; 95% CI=1.1–29.4; P=0.04), having circumscribed growth (OR=2.9; 95% CI=1.0–8.5; P=0.05), and being ER positive (OR=3.9; 95% CI=0.95–16.3; P=0.06). There was no evidence that any of the other tumour morphological features was associated with PALB2 mutation status. Figure 1 shows examples of tumours with and without sclerosis, circumscribed growth, and tubule formation. After adjusting for having minimal sclerosis, no other feature was significantly associated with PALB2 mutation status.
Table 3. Morphological features of PALB2 mutation-associated tumours compared with those of non-carriers of high-risk genetic mutations.
|
PALB2 |
Non-carrier |
PALB2
vs
non-carrier |
|||||
|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | 95% CI | P-value | |
|
Malignant nuclear grade | |||||||
| Present | 20 | 71 | 603 | 78 | 0.58 | 0.21–1.61 | 0.3 |
| Absent | 8 | 29 | 165 | 21 | |||
| Missing |
0 |
0 |
2 |
0.3 |
|
|
|
|
Minimal tubule formation | |||||||
| Present | 25 | 89 | 525 | 68 | 5.56 | 1.28–24.18 | 0.02 |
| Absent | 3 | 11 | 243 | 32 | |||
| Missing |
0 |
0 |
2 |
0.3 |
|
|
|
|
Number of mitotic cells ⩾20 | |||||||
| Present | 10 | 36 | 237 | 31 | 2.34 | 0.85–6.39 | 0.1 |
| Absent | 16 | 57 | 530 | 69 | |||
| Missing |
2 |
7.1 |
4 |
0.5 |
|
|
|
|
Syncytial growth pattern | |||||||
| Present | 1 | 4 | 42 | 6 | 0.62 | 0.06–5.99 | 0.7 |
| Absent | 27 | 96 | 723 | 94 | |||
| Missing |
0 |
0 |
5 |
0.7 |
|
|
|
|
Pushing margins | |||||||
| Present | 2 | 7 | 17 | 2 | 2.81 | 0.41–19.27 | 0.3 |
| Absent | 26 | 93 | 744 | 97 | |||
| Missing |
0 |
0 |
9 |
1.2 |
|
|
|
|
Circumscribed growth pattern | |||||||
| Present | 8 | 29 | 100 | 13 | 2.92 | 1.00–8.51 | 0.05 |
| Absent | 20 | 71 | 661 | 86 | |||
| Missing |
0 |
0 |
9 |
1.2 |
|
|
|
|
Lymphocytic infiltration site | |||||||
| Present | 8 | 29 | 258 | 34 | 0.75 | 0.27–2.11 | 0.6 |
| Absent | 20 | 71 | 502 | 65 | |||
| Missing |
0 |
0 |
10 |
1.3 |
|
|
|
|
Lymphocytic infiltration level | |||||||
| Present | 5 | 18 | 117 | 15 | 0.6 | 0.14–2.56 | 0.5 |
| Absent | 23 | 82 | 639 | 83 | |||
| Missing |
0 |
0 |
14 |
1.8 |
|
|
|
|
Minimal sclerosis | |||||||
| Present | 14 | 50 | 30 | 4 | 19.68 | 6.00–64.59 | 5 × 10−7 |
| Absent | 14 | 50 | 734 | 95 | |||
| Missing |
0 |
0 |
6 |
0.8 |
|
|
|
|
Necrosis | |||||||
| Present | 7 | 25 | 224 | 29 | 1.12 | 0.38–3.34 | 0.8 |
| Absent | 21 | 75 | 541 | 70 | |||
| Missing |
0 |
0 |
5 |
0.7 |
|
|
|
|
Apoptosis | |||||||
| Present | 17 | 61 | 563 | 73 | 0.89 | 0.33–2.45 | 0.8 |
| Absent | 11 | 39 | 206 | 27 | |||
| Missing |
0 |
0 |
1 |
0.1 |
|
|
|
|
Lymphovascular invasion | |||||||
| Present | 6 | 21 | 237 | 31 | 1.18 | 0.37–3.79 | 0.8 |
| Absent | 21 | 75 | 531 | 69 | |||
| Missing |
1 |
3.6 |
2 |
0.3 |
|
|
|
|
Atypical lobular hyperplasia | |||||||
| Present | 0 | 0 | 9 | 1 | N/A | ||
| Absent | 28 | 100 | 758 | 98 | |||
| Missing |
0 |
0 |
3 |
0.4 |
|
|
|
|
Atypical ductal hyperplasia | |||||||
| Present | 2 | 7 | 25 | 3 | 1.43 | 0.24–8.63 | 0.7 |
| Absent | 26 | 93 | 741 | 96 | |||
| Missing |
0 |
0 |
4 |
0.5 |
|
|
|
|
Lobular carcinoma
in situ | |||||||
| Present | 2 | 7 | 30 | 4 | 5.65 | 1.09–29.38 | 0.04 |
| Absent | 26 | 93 | 734 | 95 | |||
| Missing |
0 |
0 |
6 |
0.8 |
|
|
|
|
Ductal carcinoma
in situ | |||||||
| Present | 17 | 61 | 206 | 27 | 0.5 | 0.19–1.36 | 0.2 |
| Absent | 11 | 39 | 562 | 73 | |||
| Missing |
0 |
0 |
2 |
0.3 |
|
|
|
|
Acinar growth pattern | |||||||
| Present | 22 | 79 | 671 | 87 | 0.53 | 0.17–1.70 | 0.3 |
| Absent | 6 | 21 | 99 | 13 | |||
| Missing |
0 |
0 |
0 |
0 |
|
|
|
|
Lobular growth pattern | |||||||
| Present | 9 | 32 | 283 | 37 | 0.86 | 0.31–2.39 | 0.8 |
| Absent | 19 | 68 | 487 | 63 | |||
| Missing |
0 |
0 |
0 |
0 |
|
|
|
|
Trabecular growth pattern | |||||||
| Present | 8 | 29 | 121 | 16 | 1.8 | 0.60–5.35 | 0.3 |
| Absent | 20 | 71 | 649 | 84 | |||
| Missing |
0 |
0 |
0 |
0 |
|
|
|
|
Tubular growth pattern | |||||||
| Present | 4 | 14 | 125 | 16 | 0.65 | 0.17–2.44 | 0.5 |
| Absent | 24 | 86 | 645 | 84 | |||
| Missing |
0 |
0 |
0 |
0 |
|
|
|
|
Lobular/pleomorphic lobular | |||||||
| Present | 5 | 17.9 | 132 | 17 | 1.05 | 0.39–2.81 | 0.9 |
| Absent |
23 |
82.1 |
638 |
83 |
|
|
|
|
Oestrogen receptor | |||||||
| Present | 17 | 61 | 444 | 58 | 3.93 | 0.95–16.25 | 0.06 |
| Absent | 2 | 7 | 246 | 32 | |||
| Missing |
9 |
32.1 |
80 |
10.4 |
|
|
|
|
Progesterone receptor | |||||||
| Present | 11 | 39 | 465 | 60 | 0.91 | 0.28–2.90 | 0.9 |
| Absent | 8 | 29 | 224 | 29 | |||
| Missing | 9 | 32.1 | 81 | 10.52 | |||
Abbreviations: CI=confidence interval; OR=odds ratio.
N/A: unable to be analysed due to zero observations of atypical lobular hyperplasia.
Figure 1.
Morphological characteristics of PALB2 tumours. (A) Minimal sclerosis ( × 5 magnification), (B) extensive sclerosis ( × 5 magnification), (C) circumscribed growth ( × 1 magnification), (D) absence of circumscribed growth ( × 2 magnification), (E) minimal tubule formation ( × 10 magnification), and (F) intermediate tubule formation ( × 10 magnification).
With respect to the immunohistochemistry of tumours arising in PALB2 mutation carriers, information on ER and PR expression was available for 19 PALB2 mutation carriers; 11 (58%) were ER+/PR+, 6 (32%) were ER+/PR−, and only 2 (11%) were ER−/PR−. This distribution was different to that for non-carriers from the Australian BCFR (P=0.002). Of the non-carriers from the Australian BCFR with information available on ER and PR expression, 387 (56%) were ER+/PR+, 56 (8%) were ER+/PR−, 78 (11%) were ER−/PR+, and 167 (24%) were ER−/PR−. Expression status of HER2 was available for five PALB2 mutation-associated tumours (data not shown), and only one of these tumours had the triple negative (ER−/PR−/HER2−) phenotype. The Australian BCFR does not currently have data on HER2 expression.
For unselected cases, the positive and negative predictive values of minimal sclerosis as a predictive feature of the carrier status of PALB2 c.3113G>A were 2.5% and 99.9%, respectively.
Comparison with breast tumours arising in carriers of high-risk mutations in other breast cancer susceptibility genes
Table 4 presents the individual associations of minimal sclerosis with PALB2, BRCA1, and BRCA2 mutation-associated tumours when compared with tumours of non-carriers of high-risk mutations.
Table 4. Minimal sclerosis in BRCA1, BRCA2, and PALB2 mutation-associated tumours and in non-carriers of high-risk mutations.
| |
Minimal sclerosis |
|
||
|---|---|---|---|---|
| Present N (%) | Absent N (%) | Odds ratio (95% CI) | P-value | |
| Non-carriers |
30 (3.9) |
734 (95.3) |
|
|
|
PALB2 mutation carriers |
14 (50) |
14 (50) |
PALB2 vs non-carriers |
|
| |
|
|
19.7 (6–64.6) |
5 × 10−7 |
|
BRCA1 mutation carriers |
9 (22.5) |
31 (77.5) |
BRCA1 vs non-carriers |
|
| |
|
|
3.15 (1.3–7.7) |
0.01 |
|
BRCA2 mutation carriers |
2 (11.1) |
16 (88.9) |
BRCA2 vs non-carriers |
|
| 1.29 (0.27–6.17) | 0.8 | |||
Abbreviation: CI=confidence interval.
When compared with tumours arising in PALB2 mutation carriers, those arising in BRCA1 mutation carriers were more likely to have a high mitotic count (>50; P=0.004), extensive sclerosis (OR=0.21; 95% CI=0.05–0.99, P=0.05), and necrosis (P=0.01), be ER negative (P=0.001) and PR negative (P=0.03), and less likely to have a lobular growth pattern (P=0.02). When compared with tumours arising in PALB2 mutation carriers, those arising in BRCA2 mutation carriers were more likely to have extensive sclerosis (OR=0.06, 95% CI=0.004–0.88, P=0.04).
Discussion
This report brings together several lines of evidence that support the relevance of genetic information about PALB2 to breast cancer clinical genetics services. Is it now time for this information to be made available to women who are seeking advice and explanation for their person and family history of breast cancer?
The appropriate translation of new genetic information requires clear evidence and cost-benefit analysis. In the specific example of PALB2, there are several characterised genetic epidemiological features of the mutation spectrum that need to be considered and managed in the process of translation.
First, is information about PALB2 mutation status clinically relevant? Several reports now provide evidence that the risk of breast cancer associated with at least some PALB2 mutations is of the same magnitude as that associated with ‘high-risk' mutations in other cancer susceptibility genes such as BRCA2 and MSH2 (Antoniou et al, 2003; Erkko et al, 2008; Southey et al, 2010; Win et al, 2012). Risks of this magnitude support the relevance of this information to clinical genetic services, but what use is this information to women who might be carriers of mutations in PALB2 and at high risk of cancer? For affected women, and especially those identified as carriers of PALB2 mutations at the time of diagnosis, there is the potential for treatment that target homologous DNA repair dysfunction (Buisson et al, 2010). There is also the importance of advising on and managing the high risk of breast cancer that could involve risk reducing surgery (for both affected and unaffected carriers) and the potential for gene-specific medical risk reduction.
Second, mutations in PALB2 are very rare and thus, without additional information, application of traditional genetic counselling and testing regimes would be costly and identify very few carriers. We estimated that the positive predictive value of minimal sclerosis for unselected cases would be about 2.5%, but this estimate has a wide CI due to the lack of precise knowledge about the prevalence of PALB2 mutations in such cases. It should be noted, however, that given the high penetrance of PALB2 mutations, they will be more common in cases with a family history of breast cancer, as are BRCA1 and BRCA2 mutations. Therefore, it would be expected that the positive predictive value of minimal sclerosis will be substantially greater for cases with a family history. In the Australian and other settings, there is also the potential to consider testing for specific mutations in PALB2 that are found more commonly in these populations (Rahman et al, 2007; Erkko et al, 2008; Southey et al, 2010; Teo et al, 2013a, 2013b). At present, this might represent some cost saving at the level of genetic testing at the laboratory bench. The increasing introduction of massively parallel sequencing into the diagnostic testing laboratory continues to reduce the cost of testing and expand the genetic distance that can be covered in single instrument runs. This advancement in technology could result in making the detection of PALB2 mutations a natural part of clinical genetic testing, even in contexts other than breast cancer.
Third, this study provides important information that could help translation of genetic information about PALB2 into clinical use. Similar to the way that pathology has been used to facilitate the identification of women who carry germline mutations in BRCA1 and the identification of carriers of mismatch repair genes (Southey et al, 2005, 2011; Hopper et al, 2012), the new information presented here could be used to facilitate the identification of carriers of PALB2 mutations at the time of diagnosis, even irrespective of family history. It is also of note that the key feature associated with carrying a PALB2 mutation (minimal sclerosis in the breast tumour) is also a feature that distinguishes PALB2 mutation carriers from BRCA1 (P=0.05) and BRCA2 (P=0.04) mutation carriers. Moreover, we have previously shown that, even without knowledge of germline PALB2 mutation status, minimal sclerosis is associated with about a five-fold increased risk for relatives of women with early-onset breast cancer (Dite et al, 2012). The presence of central sclerosis is more frequently identified in basal-like breast cancers, and has been associated with a worse prognosis (Fulford et al, 2006; Marginean et al, 2010). Therefore, inclusion of this feature in standard pathology review, particularly for early-onset cases, could help identify families carrying high-risk genetic mutations through means other than conventional approaches based on family cancer history.
Despite the key interactions of PALB2 with both BRCA1 and BRCA2 in the same complex during homologous recombination repair, our results, overall, do not provide evidence of similarities in tumour morphological features between tumours arising in PALB2, BRCA2, or BRCA1 mutation carriers. However, it is interesting that we observed five lobular or pleomorphic lobular carcinomas (observed as primary or as secondary histological type) in women with PALB2 mutations that were diagnosed before the age of 50 years (ranging from 37 years to 47 years) and to note that in a population-based study of early-onset breast cancer (diagnosis under the age of 40 years), tumours arising in BRCA2 mutation carriers were more frequently pleomorphic lobular carcinomas compared with those arising in non-carriers of BRCA1 or BRCA2 mutations (Armes et al, 1998). There has also been consistent evidence that the proportion of ER-negative breast tumours increases with age at diagnosis for BRCA2 mutation carriers (P=1.2 × 10−5 and P=0.02 reported by Mavaddat et al, 2010 and Eerola et al, 2005, respectively).
It is important to note that the majority of tumours (24 out of 28) that have undergone pathology review in this study have been derived from carriers of the PALB2 c.3113G>A mutation. Therefore, it is unclear whether the predictive value of having minimal sclerosis is specific to PALB2 c.3113G>A or whether it could be extended to all PALB2 mutations.
Due to the rarity of PALB2 loss-of-function mutations, an international effort to combine data for a large number of carriers of PALB2 loss-of-function mutations is required to validate tumour morphological features associated with PALB2 mutation status observed in this study. A larger study would also allow for the data to be stratified by age at diagnosis to examine the potential for age-dependent associations with tumour morphology (as is evident for BRCA1 mutation carriers; Hopper et al, 2012) and for some PALB2 mutations to be associated with triple negative breast cancer (Heikkinen et al, 2009; Tischkowitz & Xia, 2010). Note, however, that our study has found no evidence that the tumours of PALB2 mutation carriers are more likely to be triple negative, and instead found that if anything they might be less likely.
Acknowledgments
We wish to thank the participants in all the contributing studies, Heather Thorne (OAM), Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded 2001–2009 by National Health and Medical Research Council of Australia (NHMRC) and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. We thank the Breast Cancer Family Registry (BCFR) which is funded by the National Cancer Institute, National Institutes of Health, under RFA CA-06-503 and through cooperative agreements with Cancer Care Ontario (U01 CA69467), the Cancer Prevention Institute of California (U01 CA69417), Columbia University (U01 CA69398), Fox Chase Cancer Center (U01 CA69631), the Huntsman Cancer Institute at the University of Utah (U01 CA69446), and The University of Melbourne (U01 CA69638). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the BCFR, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government or the BCFR. The Australian site of the BCFR was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation, and the Victorian Breast Cancer Research Consortium. We also wish to thank A/Prof Ian Campbell and the VBCRC Cancer Genetics Laboratory, Peter MacCallum Cancer Centre for their assistance with the preparation of the DNA samples (Victorian Breast Cancer Research Consortium). This work was also supported by the NHMRC (Project Grant APP1029974 and APP1023698) and the Victorian Breast Cancer Research Consortium (VBCRC). ZLT was supported by Postgraduate Scholarships provided by the Faculty of Medicine, Dentistry and Health Sciences, The University of Melbourne and the NHMRC (Dora Lush Postgraduate Fellowship). TN-D is the recipient of a postdoctoral fellowship from the Susan G Komen for the Cure Foundation. MCS is an NHMRC Senior Research Fellow and a Group Leader of the VBCRC. GJL is an NHMRC Principal Research Fellow and Group Leader of the VBCRC. JLH is an NHMRC Australia Fellow and a VBCRC Group Leader.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Andrulis IL, Anton-Culver H, Beck J, Bove B, Boyd J, Buys S, Godwin AK, Hopper JL, Li F, Neuhausen SL, Ozcelik H, Peel D, Santella RM, Southey MC, van Orsouw NJ, Venter DJ, Vijg J, Whittemore AS. Comparison of DNA- and RNA-based methods for detection of truncating BRCA1 mutations. Hum Mutat. 2002;20:65–73. doi: 10.1002/humu.10097. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PDP, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjakoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella C, Andrews L, Hodgson SV, Fisher SA, Lewis CM, Solomon E, Tucker K, Friedlander M, Bankier A, Southey MC, Venter DJ, Hopper JL. Log odds of carrying an Ancestral Mutation in BRCA1 or BRCA2 for a defined personal and family history in an Ashkenazi Jewish woman (LAMBDA) Breast Cancer Res. 2003;5:R206–R216. doi: 10.1186/bcr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armes JE, Egan AJ, Southey MC, Dite GS, McCredie MR, Giles GG, Hopper JL, Venter DJ. The histologic phenotypes of breast carcinoma occurring before age 40 years in women with and without BRCA1 or BRCA2 germline mutations—a population-based study. Cancer. 1998;83 (11:2335–2345. [PubMed] [Google Scholar]
- Armes JE, Trute L, White D, Southey MC, Hammet F, Tesoriero A, Hutchins AM, Dite GS, McCredie MRE, Giles GG, Hopper JL, Venter DJ. Distinct molecular pathogeneses of early-onset breast cancers in BRCA1 and BRCA2 mutation carriers: a population-based study. Cancer Res. 1999;59:2011–2017. [PubMed] [Google Scholar]
- Bernstein JL, Teraoka S, Southey MC, Jenkins MA, Andrulis IL, Knight JA, John EM, Lapinski R, Wolitzer AL, Whittemore AS, West D, Seminara D, Olson ER, Spurdle AB, Chenevix-Trench G, Giles GG, Hopper JL, Concannon P. Population-based estimates of breast cancer risks associated with ATM gene variants c.7271T>G and c.1066-6T>G (IVS10-6T>G) from the breast cancer family registry. Hum Mutat. 2006;27:1122–1128. doi: 10.1002/humu.20415. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Sokolenko AP, Iyevleva AG, Abysheva SN, Blaut M, Bremer M, Christiansen H, Rave-Frank M, Dork T, Imyanitov EN. PALB2 mutations in German and Russian patients with bilateral breast cancer. Breast Cancer Res Treat. 2011;126:545–550. doi: 10.1007/s10549-010-1290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71:2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench G, Spurdle AB, Gatei M, Kelly H, Marsh A, Chen XQ, Donn K, Cummings M, Nyholt D, Jenkins MA, Scott C, Pupo GM, Dork T, Bendix R, Kirk J, Tucker K, McCredie MRE, Hopper JL, Sambrook J, Mann GJ, Khanna KK. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94:205–215. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, Derlatka P, Cendrowski K, Kupryjanczyk J. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YC, Steele L, Chu LH, Kelley K, Davis H, John EM, Tomlinson GE, Neuhausen SL. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126:227–230. doi: 10.1007/s10549-010-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MRE, Venter DJ, Hopper JL. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–457. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- Dite GS, Makalic E, Schmidt DF, Giles GG, Hopper JL, Southey MC. Tumour morphology of early-onset breast cancers predicts breast cancer risk for first-degree relatives: the Australian Breast Cancer Family Registry. Breast Cancer Res. 2012;14:R122. doi: 10.1186/bcr3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eerola H, Heikkila P, Tamminen A, Aittomaki K, Blomqvist C, Nevanlinna H. Relationship of patients' age to histopathological features of breast tumours in BRCA1 and BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2005;7:R465–R469. doi: 10.1186/bcr1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston CW, Ellis IO, Pinder SE. Pathological prognostic factors in breast cancer. Crit Rev Oncol Hematol. 1999;31:209–223. doi: 10.1016/s1040-8428(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Erkko H, Dowty JG, Nikkila J, Syrjaekoski K, Mannermaa A, Pylkas K, Southey MC, Holli K, Kallioniemi A, Jukkola-Vuorinen A, Kataja V, Kosma VM, Xia B, Livingston DM, Winqvist R, Hopper JL. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clinical Cancer Res. 2008;14:4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Ghadirian P, Akbari MR, Hamel N, Giroux S, Sabbaghian N, Darnel A, Royer R, Poll A, Fafard E, Robidoux A, Martin G, Bismar TA, Tischkowitz M, Rousseau F, Narod SA. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9 (6:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford LG, Easton DF, Reis JS, Sofronis A, Gillett CE, Lakhani SR, Hanby A. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology. 2006;49:22–34. doi: 10.1111/j.1365-2559.2006.02453.x. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Karkkainen H, Aaltonen K, Milne RL, Heikkila P, Aittomaki K, Blomqvist C, Nevanlinna H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15:3214–3222. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- Hellebrand H, Sutter C, Honisch E, Gross E, Wappenschmidt B, Schem C, Deissler H, Ditsch N, Gress V, Kiechle M, Bartram CR, Schmutzler RK, Niederacher D, Arnold N, Meindl A. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum Mutat. 2011;32:E2176–E2188. doi: 10.1002/humu.21478. [DOI] [PubMed] [Google Scholar]
- Hopper JL, Jenkins MA, Dowty JG, Dite GS, Apicella C, Keogh L, Win AK, Young JP, Buchanan D, Walsh MD, Rosty C, Baglietto L, Severi G, Phillips KA, Wong EM, Dobrovic A, Waring P, Winship I, Ramus SJ, Giles GG, Southey MC. Using tumour pathology to identify people at high genetic risk of breast and colorectal cancers. Pathology. 2012;44:89–98. doi: 10.1097/PAT.0b013e32834e8e5b. [DOI] [PubMed] [Google Scholar]
- James PA, Doherty R, Harris M, Mukesh BN, Milner A, Young MA, Scott C. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24:707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O'Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, Farid LM, Venter D, Antoniou A, Storfer-Isser A, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BAJ, Ford D, Peto J, Stoppa-Lyonnet D, Bignon YJ, Struewing JP, Spurr NK, Bishop DT, Klijn JGM, Devilee P, Cornelisse CJ, Lasset C, Lenoir G, Barkardottir RB, Egilsson V, Hamann U, Chang-Claude J, Sobol H, Weber B, Stratton MR, Easton DF. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90:1138–1145. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- Lakhani SR, van de Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- Longacre TA, Ennis M, Quenneville LA, Bane AL, Bleiweiss IJ, Carter BA, Catelano E, Hendrickson MR, Hibshoosh H, Layfield LJ, Memeo L, Wu H, O'Malley FP. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Mod Pathol. 2006;19:195–207. doi: 10.1038/modpathol.3800496. [DOI] [PubMed] [Google Scholar]
- Mann GJ, Thorne H, Balleine RL, Butow PN, Clarke CL, Edkins E, Evans GM, Fereday S, Haan E, Gattas M, Giles GG, Goldblatt J, Hopper JL, Kirk J, Leary JA, Lindeman G, Niedermayr E, Phillips KA, Picken S, Pupo GM, Saunders C, Scott CL, Spurdle AB, Suthers G, Tucker K, Chenevix-Trench G. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006;8:R12. doi: 10.1186/bcr1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marginean F, Rakha EA, Ho BC, Ellis IO, Lee AHS. Histological features of medullary carcinoma and prognosis in triple-negative basal-like carcinomas of the breast. Mod Pathol. 2010;23:1357–1363. doi: 10.1038/modpathol.2010.123. [DOI] [PubMed] [Google Scholar]
- Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Spurdle A, Robson M, Sherman M, Mulligan AM, Couch FJ, Engel C, McGuffog L, Healey S, Sinilnikova OM, Southey MC, Terry MB, Goldgar D, O'Malley F, John EM, Janavicius R, Tihomirova L, Hansen TV, Nielsen FC, Osorio A, Stavropoulou A, Benitez J, Manoukian S, Peissel B, Barile M, Volorio S, Pasini B, Dolcetti R, Putignano AL, Ottini L, Radice P, Hamann U, Rashid MU, Hogervorst FB, Kriege M, van der Luijt RB, Peock S, Frost D, Evans DG, Brewer C, Walker L, Rogers MT, Side LE, Houghton C, Weaver J, Godwin AK, Schmutzler RK, Wappenschmidt B, Meindl A, Kast K, Arnold N, Niederacher D, Sutter C, Deissler H, Gadzicki D, Preisler-Adams S, Varon-Mateeva R, Schonbuchner I, Gevensleben H, Stoppa-Lyonnet D, Belotti M, Barjhoux L, Isaacs C, Peshkin BN, Caldes T, de la Hoya M, Canadas C, Heikkinen T, Heikkila P, Aittomaki K, Blanco I, Lazaro C, Brunet J, Agnarsson BA, Arason A, Barkardottir RB, Dumont M, Simard J, Montagna M, Agata S, D'Andrea E, Yan M, Fox S, Rebbeck TR, Rubinstein W, Tung N, Garber JE, Wang X, Fredericksen Z, Pankratz VS, Lindor NM, Szabo C, Offit K, Sakr R, Gaudet MM, Singer CF, Tea MK, Rappaport C, Mai PL, Greene MH, Sokolenko A, Imyanitov E, Toland AE, Senter L, Sweet K, Thomassen M, Gerdes AM, Kruse T, Caligo M, Aretini P, Rantala J, von Wachenfeld A, Henriksson K, Steele L, Neuhausen SL, Nussbaum R, Beattie M, Odunsi K, Sucheston L, Gayther SA, Nathanson K, Gross J, Walsh C, Karlan B, Chenevix-Trench G, Easton DF, Antoniou AC. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2010;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCredie MRE, Dite GS, Southey MC, Venter DJ, Giles GG, Hopper JL. Risk factors for breast cancer in young women by oestrogen receptor and progesterone receptor status. Br J Cancer. 2003;89:1661–1663. doi: 10.1038/sj.bjc.6601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchawar J, Korch C, Byers T, Pitts TM, Li EF, McCredie MRE, Giles GG, Hopper JL, Southey MC. Population-based estimate of the contribution of TP53 mutations to subgroups of early-onset breast cancer: Australian breast cancer family study. Cancer Res. 2010;70:4795–4800. doi: 10.1158/0008-5472.CAN-09-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL, Ozcelik H, Southey MC, John EM, Godwin AK, Chung W, Iriondo-Perez J, Miron A, Santella RM, Whittemore A, Andrulis IL, Buys SS, Daly MB, Hopper JL, Seminara D, Senie RT, Terry MB. BRCA1 and BRCA2 mutation carriers in the Breast Cancer Family Registry: an open resource for collaborative research. Breast Cancer Res Treat. 2009;116:379–386. doi: 10.1007/s10549-008-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DL, Anderson TJ, Sakamoto G. Infiltrating Carcinoma: Major Histological Types. Churchill Livingstone: Edinburgh; 1987. [Google Scholar]
- Papi L, Putignano AL, Congregati C, Piaceri I, Zanna I, Sera F, Morrone D, Genuardi M, Palli D. A PALB2 germline mutation associated with hereditary breast cancer in Italy. Fam Cancer. 2010;9:181–185. doi: 10.1007/s10689-009-9295-z. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish SD, Lach FP, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew CG, D Auerbach A, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genetics. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- Sawyer S, Mitchell G, McKinley J, Chenevix-Trench G, Beesley J, Chen XQ, Bowtell D, Trainer AH, Harris M, Lindeman GJ, James PA. A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol. 2012;30 (35:4330–4336. doi: 10.1200/JCO.2012.41.7469. [DOI] [PubMed] [Google Scholar]
- Smith LD, Tesoriero AA, Ramus SJ, Dite G, Royce SG, Giles GG, McCredie MRE, Hopper JL, Southey MC. BRCA1 promoter deletions in young women with breast cancer and a strong family history: a population-based study. Eur J Cancer. 2007;43:823–827. doi: 10.1016/j.ejca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey M, Teo Z, Dowty J, Odefrey F, Park D, Tischkowitz M, Sabbaghian N, Apicella C, Byrnes G, Winship I, Baglietto L, Giles G, Goldgar D, Foulkes W, Hopper J, kConFab for the Breast Cancer Family Registry A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12:R109. doi: 10.1186/bcr2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, Tesoriero AA, Smith LD, Jennings K, Grubb G, Royce SG, Walsh MD, Barker MA, Young JP, Jass JR, St, John DJB, Macrae FA, Giles GG, Hopper JL. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J Clin Oncol. 2005;23:6524–6532. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- Southey MC, Ramus SJ, Dowty JG, Smith LD, Tesoriero AA, Wong EEM, Dite GS, Jenkins MA, Byrnes GB, Winship I, Phillips KA, Giles GG, Hopper JL. Morphological predictors of BRCA1 germline mutations in young women with breast cancer. Br J Cancer. 2011;104:903–909. doi: 10.1038/bjc.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey MC, Tesoriero AA, Andersen CR, Jennings KM, Brown SM, Dite GS, Jenkins MA, Osborne RH, Maskiell JA, Porter L, Giles GG, McCredie MRE, Hopper JL, Venter DJ. BRCA1 mutations and other sequence variants in a population based sample of Australian women with breast cancer. Br J Cancer. 1999;79:34–39. doi: 10.1038/sj.bjc.6690008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp 2009Stata Statistical Software:Release 11. StataCorp LP: College Station, TX [Google Scholar]
- Sy SMH, Huen MSY, Chen JJ. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo ZL, Park DJ, Provenzano E, Chatfield CA, Odefrey FA, Nguyen-Dumont T, Dowty JG, Hopper JL, Winship I, Goldgar DE, Southey MC. Prevalence of PALB2 mutations in Australasian multiple-case breast cancer families. Breast Cancer Res. 2013a;15:R17. doi: 10.1186/bcr3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo ZL, Sawyer SD, James PA, Mitchell G, Trainer AH, Lindeman GJ, Shackleton K, Cicciarelli L, Southey MC.2013bThe incidence of PALB2 c.3113G>A in women with a strong family history of breast and ovarian cancer attending familial cancer centres in Australia Fam Cancer;e-pub ahead of print 8 March 2013. [DOI] [PubMed]
- Tischkowitz M, Capanu M, Sabbaghian N, Li L, Liang X, Vallée MP, Tavtigian SV, Concannon P, Foulkes WD, Bernstein L, WECARE Study Collaborative Group. Bernstein JL, Begg CB. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33:674–680. doi: 10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz M, Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70:7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz M, Xia B, Sabbaghian N, Reis-Filho JS, Hamel N, Li GL, van Beers EH, Li LL, Khalil T, Quenneville LA, Omeroglu A, Poll A, Lepage P, Wong N, Nederlof PM, Ashworth A, Tonin PN, Narod SA, Livingston DM, Foulkes WD. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Eynden GG, Colpaert CG, Couvelard A, Pezzella F, Dirix LY, Vermeulen PB, Van Marck EA, Hasebe T. A fibrotic focus is a prognostic factor and a surrogate marker for hypoxia and (lymph)angiogenesis in breast cancer: review of the literature and proposal on the criteria of evaluation. Histopathology. 2007;51:440–451. doi: 10.1111/j.1365-2559.2007.02761.x. [DOI] [PubMed] [Google Scholar]
- Van den Eynden GG, Smid M, Van Laere SJ, Colpaert CG, Van der Auwera I, Bich TX, van Dam P, den Bakker MA, Dirix LY, Van Marck EA, Vermeulen PB, Foekens JA. Gene expression profiles associated with the presence of a fibrotic focus and the growth pattern in lymph node-negative breast cancer. Clin Cancer Res. 2008;14:2944–2952. doi: 10.1158/1078-0432.CCR-07-4397. [DOI] [PubMed] [Google Scholar]
- Win AK, Young JP, Lindor NM, Tucker KM, Ahnen DJ, Young GP, Buchanan DD, Clendenning M, Giles GG, Winship I, Macrae FA, Goldblatt J, Southey MC, Arnold J, Thibodeau SN, Gunawardena SR, Bapat B, Baron JA, Casey G, Gallinger S, Le Marchand L, Newcomb PA, Haile RW, Hopper JL, Jenkins MA. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol. 2012;30:958–964. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu JM, Christ N, Liu XG, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma JL, Wu JX, Ye L, Cai H, Xia B, Yu XC. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]