Abstract
Background:
Several lines of evidence suggest a dichotomy between immune active and quiescent cancers, with the former associated with a good prognostic phenotype and better responsiveness to immunotherapy. Central to such dichotomy is the master regulator of the acute inflammatory process interferon regulatory factor (IRF)-1. However, it remains unknown whether the responsiveness of IRF-1 to cytokines is able to differentiate cancer immune phenotypes.
Methods:
IRF-1 activation was measured in 15 melanoma cell lines at basal level and after treatment with IFN-γ, TNF-α and a combination of both. Microarray analysis was used to compare transcriptional patterns between cell lines characterised by high or low IRF-1 activation.
Results:
We observed a strong positive correlation between IRF-1 activation at basal level and after IFN-γ and TNF-α treatment. Microarray demonstrated that three cell lines with low and three with high IRF-1 inducible translocation scores differed in the expression of 597 transcripts. Functional interpretation analysis showed mTOR and Wnt/β-cathenin as the top downregulated pathways in the cell lines with low inducible IRF-1 activation, suggesting that a low IRF-1 inducibility recapitulates a cancer phenotype already described in literature characterised by poor prognosis.
Conclusion:
Our findings support the central role of IRF-1 in influencing different tumour phenotypes.
Keywords: IRF-1, IFN-γ, immune phenotype, NF-κB, TNF-α, nuclear translocation
A dichotomy between immunologically active and quiescent cancer phenotypes has been recently demonstrated for several types of cancer (Wang et al, 2002, 2004; Galon et al, 2006; Leffers et al, 2009; Ascierto et al, 2012). Of particular interest, in colorectal cancer the type, density and location of immune cells have been shown to predict patient survival (Galon et al, 2006). Moreover, metastases from cutaneous melanoma have been previously shown to segregate into two different subclasses according to the expression of genes annotated with immune function (Wang et al, 2002; Marincola et al, 2003). The recognition of the importance of the host immune system in controlling tumour progression have prompted several groups around the world to include immunological biomarkers in the conventional classification of tumours (Galon et al, 2012).
Central to the existence of the immune active phenotype in cancer is interferon regulatory factor (IRF)-1, the master regulator of the acute inflammatory process. Recent studies observed that the constitutive activation of the interferon (IFN)-γ/signal transducers and activators of transcription (STAT)-1/IRF-1 axis (Th1 phenotype) bears good prognostic connotation and predicts better responsiveness to anti-cancer therapy (Ascierto et al, 2011). IRF-1 is a nuclear transcription factor crucial to inflammation, immunity, cell proliferation and apoptosis (Kroger et al, 2002). Its synthesis is induced in response to IFN-γ (Taniguchi et al, 2001). The biological effects of IFN-γ are elicited through activation of intracellular molecular signaling networks, of which the best characterised is the JAK (Janus kinase)-STAT pathway. Conventional IFN-γ signaling follows the binding of IFN-γ to its cell surface receptor, which consists of two subunits IFNGRs (interferon gamma receptors)-1 and -2, and each molecule interacts with a member of the JAK family, which are non-receptor protein tyrosine kinases. JAKs phosphorylate STATs receptors and transcriptional co-activators leading to the downstream expression and activation of IRF-1 (Platanias, 2005). Although the relevance of IRF-1 to the immune biology of cancer is emerging, it remains unknown whether the responsiveness of IRF-1 to IFN-γ is able to point out different cancer behaviours. Here, we explored the significance of the cytokines-induced activation of IRF-1 in melanoma cell lines in vitro as a determinant of the immune behaviour of 15 melanoma cell lines. This question is relevant, because it may suggest that intrinsic characteristics of tumour cells that can be defined in vitro may be responsible for the responsiveness of cancer cells to signals produced by other cells, including immune cells, within the tumour microenvironment.
Although IRF-1 over expression in tumour lesions has been associated with better prognosis and likelihood of response to immunotherapy, such association is not absolute, suggesting that other factors are involved in the process. Several years ago, IRF-1 has been demonstrated to synergise with NF-κB and assemble to form a higher-order nucleoprotein complex, the enhanceosome, a structure that modifies the local chromatin architecture and recruits the RNA polymerase II machinery to the promoter of several genes (Escalante et al, 1998; Panne, 2008). There is ample evidence that components of IFN- and NF-κB-dependent signaling cross-interact to promote or repress gene expression by other pathways. Furthermore, it has been shown that TNF-α, a strong stimulator of NF-κB, and IFN-γ are biologically related to each other. However, although TNF-α and IFN-γ have been shown to cooperate, the mechanisms behind this phenomenon are unknown. Here, we assessed whether IRF-1 and NF-κB are concordantly activated in 15 melanoma cell lines, whether their concurrent activation is maintained after stimulation with IFN-γ, TNF-α and IFN-γ plus TNF-α, respectively, and whether it is mediated by their nuclear co-localisation.
Toward this goal, we used the ImageStream Flow Cytometry technique that permitted rapid and objective analysis of individual-cell IRF-1 nuclear translocation.
The present study shows a strong positive correlation between basal status of IRF-1 activation and its IFN-γ-induced activation, suggesting that IFN-γ has a strong effect towards IRF-1 nuclear translocation. This study also shows that the IFN-γ-inducibility of IRF-1 is able to point out different intrinsic properties of cancer cells. Moreover, we demonstrate that IRF-1 activation is additively induced by IFN-γ and TNF-α and that it is concurrently activated together with NF-κB, although such co-activation is not due to a direct physical interaction of these two transcription factors.
Materials and methods
Melanoma cell culture
Melanoma cell lines were derived from metastatic melanoma lesions from patients treated at the Surgery Branch, National Cancer Institute (NCI), Bethesda, MD, USA kindly donated by Dr. Steven A. Rosenberg. The cells received from Surgery Branch were studied after passage 3. Cells were cultured in bulk at 37 °C, in 5% CO2 with RPMI 1640 medium (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (Cellgro, Mediatech, Inc. Manassas, VA, USA), 0.01% L-Glutamine Pen-Strep Solution (GPS 100 × , Gemini Bio-Products, West Sacramento, CA, USA), 0.001% Ciprofloxacin (10 mg ml−1) and 0.01% Fungizone Amphotericin B (250 μg ml−1, Gibco). Confluent adherent cells were washed twice with phosphate-buffered saline 1X (PBS pH 7.4, Gibco) and detached by exposure to 0.2% Trypsin-EDTA (0.5%: 0.53 mℳ Solution, Gemini Bio-Products). The harvested cells were centrifuged to remove cell debris and re-suspended in fresh medium to a final concentration of 1 × 107cells ml−1. Early-passage cultures were used for all experiments, and no clonal subselection was performed.
Aphidicolin-induced cell-cycle synchronisation
A stock solution of 1 mg ml−1 aphidicolin from Nigrospora sphaerica (Sigma-Aldrich Co., St. Louis, MO, USA) was prepared in sterile dimethyl sulphoxide (DMSO, Sigma-Aldrich Co.) and stored in aliquots at −20 °C. Cells were seeded at a concentration of 1 × 105 ml−1 and, once attached, placed into medium containing 2.5 μg ml−1 aphidicolin, which was an optimal reversible dose based on previous experiments (data not shown). After 24 h, cells were placed in a fresh medium.
In order to assess cell-cycle stage, 5-bromo-2-deoxyuridine (BrdU, Sigma-Aldrich Co.) was added to the cells to a final concentration of 10 μℳ. After 30-min incubation, cells were trypsinised and centrifuged at 3000 g for 5 min. Cells were then washed with PBS, fixed in ice-cold 70% ethanol and stored at 4 °C over night. Before staining, a 30-min incubation on ice with 0.1 ℳ HCl containing 0.7% Triton X-100 (Sigma-Aldrich Co.) was performed. Cells were washed with 1 ml of 0.1 ℳ sodium tetraborate (Sigma-Aldrich Co.), pH 8.5, to neutralise the acid, and re-suspended in PBS containing 0.5% Tween 20 (Sigma-Aldrich Co.). Cells were then incubated with anti-BrdU antibody (BD Biosciences, San Jose, CA, USA) (1 : 200) for 30 min, followed by 30 min with a 1 : 500 dilution of fluorescein (FITC)-conjugated secondary antibody. Cell nuclei were counterstained with 10 μg ml−1 propidium iodide (PI, Sigma-Aldrich Co.), BrdU incorporation and total DNA content were then assayed using a FACSVantage SE cell sorter (BD Biosciences). The resulting data were processed and statistically analysed using CellQuest analysis software (BD Biosciences).
Immune stimulation
IRF-1 and NF-κB translocation was activated by 1-h exposure to recombinant human IFN-γ (10 ng ml−1) and recombinant human TNF-α (10 ng ml−1) (R&D Systems, Inc., MN, USA), respectively. Concentration and time of exposure were chosen as optimal to achieve maximum translocation of the two transcription factors, according to preliminary experiments (data not shown). Following activation, cells were fixed in 4% (v/v) paraformaldehyde (PEA, Polysciences Inc, Warrington, PA, USA), permeabilised with 100% methanol and stained as outlined below.
ImageStream Flow Cytometry analysis
IRF-1 and the p65 subunit of NF-κB were visualised by indirect immunofluorescence labeling. The primary mouse monoclonal anti-IRF-1 (clone B1, sc-137061) and rabbit polyclonal anti-NF-κB p65 (ab7970) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and Abcam (Cambridge, MA, USA), respectively. Fixed cells were incubated for 45 min at room temperature with both primary antibodies (1 μg per 106 cells), washed twice with PBS and incubated for 45 min in the dark with both secondary antibodies (1 μg per 106 cells) Alexa Fluor 647-conjugated F(ab')2 fragment donkey anti-mouse IgG and Alexa Fluor 488-conjugated F(ab')2 fragment donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA). After 30-min incubation with the secondary antibodies, 1 μg DAPI per 106 cells was added for the nuclear staining. Cells were finally washed twice with PBS and re-suspended in a PBS solution containing 0.1% sodium azide and 1% pen-strep-neomycin solution (Sigma-Aldrich Co.). Cells were then stored at 4 °C until analysis.
The basal status of IRF-1 activation was assessed first by immunofluorescence staining, which showed a different pattern of IRF-1 nuclear translocation among the cell lines (Supplementary Figure S1) and then by ImageStream analysis. For ImageStream analysis, a total of 2000–5000 events was collected from each sample. Data were acquired using INSPIRE and analysed by IDEAS software (Amnis Corp., Seattle, WA, USA). IRF-1 and NF-κB nuclear translocation was quantitatively measured using similarity analysis on in-focus single cells, and expression levels were analysed as the median fluorescence intensity (George et al, 2006; Zuba-Surma et al, 2007).
Microarray analysis
Total RNA was isolated using miRNeasy minikit (Qiagen, Valencia, CA, USA) according to the manufacture's protocol. RNA quality and quantity was estimated using Nanodrop (Thermo Scientific, Barrington, IL, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). First- and second-strand cDNAs were synthesised from 300 ng of total RNA according to the manufacturer's instructions (Ambion WT Expression Kit, Affymetrix Inc., Santa Clara, CA, USA). cDNAs were fragmented, biotinylated and hybridised to the GeneChip Human Gene 1.0 ST Arrays (Affymetrix WT Terminal Labeling Kit). The arrays were washed and stained on a GeneChip Fluidics Station 450 (Affymetrix); scanning was carried out using the GeneChip Scanner 3000 and image analysis using the Affymetrix GeneChip Command Console Scan Control. Expression data were normalised, background-corrected and summarised using the RMA algorithm, http://www.partek.com/. Data were log-transformed (base 2) for subsequent statistical analysis.
Cluster analysis was performed using BRB-ArrayTools developed by the Biometric Research Branch, National Cancer Institute (http://linus.nci.nih.gov/BRB-ArrayTools.html) (Simon et al, 2007) or Partek Genomics Suite software (St Louis, MO, USA) as appropriate. Functional gene network analysis was performed using the Ingenuity Pathway Analysis system tools 3.0, which transforms large data sets into a group of relevant networks containing direct and indirect relationships among genes based on known interactions in the literature (http://www.ingenuity.com).
Statistical analysis
Two-tailed Spearman correlation tests were applied to compare basal and IFN-γ-, TNF-α- and IFN-γ plus TNF-α-induced IRF-1 translocation and to compare IRF-1 and NF-κB nuclear activation among the cell lines. A P-value ⩽0.05 was considered statistically significant. Tests were performed using the Prism GraphPad software (GraphPad Software, Inc., La Jolla, CA, USA) and were all two-sided.
Results
IRF-1 basal activation positively correlates with IFN-γ- and TNF-α-induced activation
In order to assess IRF-1 responsiveness to IFN-γ, we stimulated the 15 melanoma cell lines and evaluated the IRF-1 nuclear translocation by ImageStream IDEAS analysis software (George et al, 2006; Zuba-Surma et al, 2007). The cells were previously synchronised in early S-phase with aphidicolin and the efficiency of the synchronisation was assessed by the BrdU-based cell-cycle assay (Supplementary Figure S2). The basal IRF-1 nuclear translocation scores of the 15 cell lines are displayed in the Supplementary Table 1.
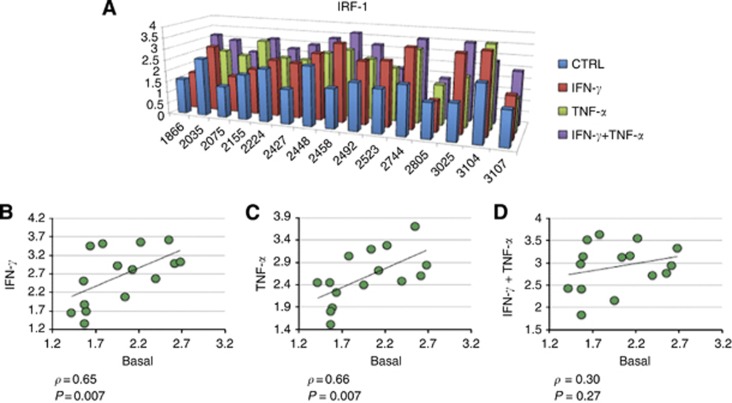
Our data indicate that the cell lines expressed significantly higher IRF-1 level in the nucleus after stimulation with IFN-γ (P=0.001, Figure 1A). Moreover, the correlation of IRF-1 activation between controls and IFN-γ-stimulated cells was statistically significant (P=0.007, ρ=0.65, Figure 1B), suggesting that IFN-γ has a strong effect in inducing IRF-1 activation.
Figure 1.
IRF-1 activation after cytokine stimulation. (A) IRF-1 nuclear translocation scores in controls, IFN-γ-, TNF-α- and IFN-γ plus TNF-α-stimulated cell lines. Correlation of IRF-1 nuclear translocation scores between basal level and after (B) IFN-γ, (C) TNF-α, and (D) IFN-γ plus TNF-α stimulation. ρ: Spearman correlation.
We next assessed whether IRF-1 activation induced by TNF-α and by IFN-γ plus TNF-α was correlated to its basal level as well. The correlation after TNF-α stimulation resulted even slightly stronger than the one induced by IFN-γ alone (P=0.007, ρ=0.66, Figure 1C), confirming the role of TNF-α as a strong inducer of IRF-1 nuclear translocation. However, the correlation of IRF-1 activation between controls and IFN-γ plus TNF-α-stimulated cells was not statistically significant although it maintained a positive trend (P=0.27, ρ=0.30, Figure 1D).
The correlation between basal IRF-1 and the delta of IRF-1 activation after stimulation with IFN-γ, TNF-α and IFN-γ plus TNF-α did not reach statistical significance, suggesting that the IRF-1 activation was mainly dependent on the cytokine stimulation rather than on the basal level of IRF-1 activation (Supplementary Figure S3).
IRF-1-inducible activation predicts differential transcriptome behaviour in melanoma cell lines
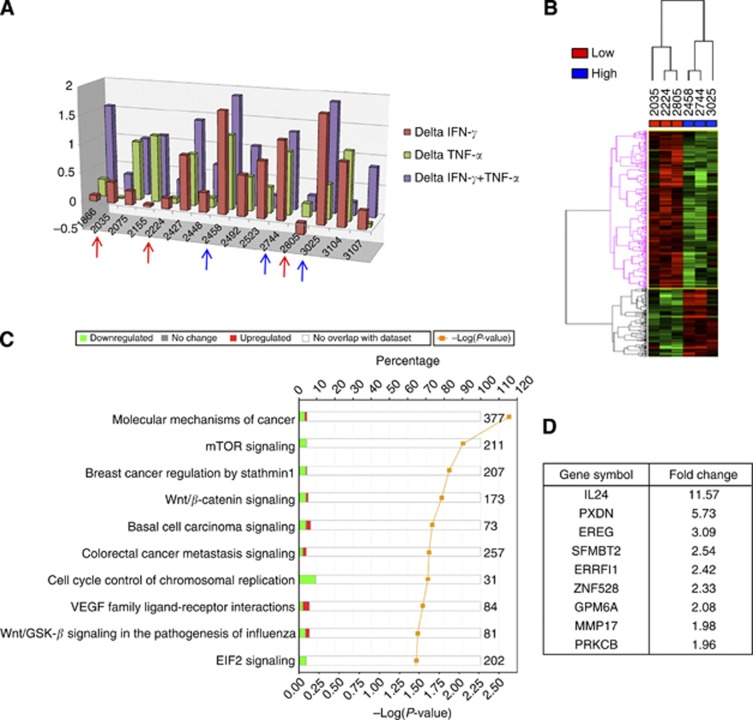
To assess whether the responsiveness of IRF-1 activation to cytokine stimulation was able to distinguish different phenotypes in melanoma cell lines, we selected three cell lines with an overall low and three with an overall high inducible IRF-1 activation among the three experimental points (IFN-γ, TNF-α and IFN-γ plus TNF-α stimulation) and performed microarray analysis (Figure 2A). When comparing the three cell lines with low vs the three with high IRF-1-inducible translocation scores, we found 597 annotated transcripts whose expression was significantly different among the two groups (P=0.05, Supplementary Data). A self-organising map based on such transcripts completely discriminated the three cell lines with low from the three with high IRF-1-inducible activation (Figure 2B). Interestingly, when using these transcripts for functional interpretation analysis, we found mTOR and Wnt/β-cathenin as the top downregulated pathways in the cell lines with low inducible IRF-1 activation (Figures 2C and D, Supplementary Figures S4 and S5). Noteworthy, such pathways have been already shown to be associated with a poor phenotype in melanoma (Bittner et al, 2000; Spivey et al, 2012). These findings suggest that differential IRF-1 responsiveness to IFN-γ and TNF-α reflects a differential behaviour of cell lines in terms of tumour aggressiveness and poor phenotype.
Figure 2.
Delta IRF-1 activation after cytokine stimulation. (A) Bar graph showing the delta of IRF-1 activation scores in the 15 melanoma cell lines. The three cell lines with an overall low and the three with an overall high inducible IRF-1 activation are highlighted by the red and blue arrows, respectively. (B) Self-organising map of three low and three high inducible IRF-1 expressing cell lines. (C) Functional interpretation analysis of the transcripts differentially expressed between the two groups of cell lines. (D) Top transcripts differentially expressed between high and low IRF-1 cell lines.
Quite interestingly, IL-24 was upregulated by up to 11-fold in cell lines with high inducible IRF-1. This is particularly intriguing as IL-24 has been shown to induce apoptosis in melanoma cell lines (Ekmekcioglu et al, 2008), thus its upregulation might reflect its potential synergism with IRF-1.
IRF-1 and NF-κB are additively activated
We next evaluated whether IRF-1 and NF-κB were mutually exclusively or concurrently activated in the 15 melanoma cell lines at basal level and after IFN-γ, TNF-α and IFN-γ plus TNF-α stimulations. Toward this goal, we used ImageStream Flow Cytometry analysis. Cells displaying different IRF-1 and NF-κB subcellular localisation were first singularly visualised by ImageStream INSPIRE and then analysed with ImageStream IDEAS software, which allowed the assessment of population statistics of each single cell line (Figures 3A and B).
Figure 3.
NF-kB and IRF-1 activation. (A) Single-cell images showing different NF-κB and IRF-1 subcellular distribution. From the top to the bottom: NF-κB and IRF-1 predominantly located in the cytoplasm; NF-κB predominantly translocated to the nucleus and IRF-1 predominantly localised in the cytoplasm; NF-κB predominantly localised in the cytoplasm and IRF-1 predominantly translocated to the nucleus; and both NF-κB and IRF-1 predominantly translocated to the nucleus. (B) Population statistics of the IRF-1 and NF-κB nuclear translocation of the cell line # 2155. (C) Correlation between NF-κB and IRF-1 nuclear translocation scores in controls and (D) in IFN-γ plus TNF-α-stimulated cell lines. ρ: Spearman correlation.
TNF-α alone and in combination with IFN-γ significantly increased the NF-κB translocation score compared with the controls (Supplementary Figure S6).
When assessing the concurrent activation of IRF-1 and NF-κB, we found that IRF-1 and NF-κB nuclear translocation scores positively correlated in all the experimental conditions. However, such correlation reached a statistical significance only at basal level (P=0.03, ρ=0.53, Figure 3C) and after IFN-γ plus TNF-α stimulation (P=0.02, ρ=0.60, Figure 3D).
In addition, given the hypothesis that if IRF-1 and NF-κB interact with each other they should both co-localise to the nucleus, we tested their nuclear co-localisation score as well. Contrary to our hypothesis, IRF-1 and NF-κB did not significantly co-localise in any experimental condition. Thus, although their activation resulted in an additive effect, it was not mediated by their physical interaction, at least under our experimental conditions.
Discussion
Several recent studies have shown that a neoplastic phenotype enriched in immune infiltrates correlates with good prognosis and enhanced likelihood to respond to immunotherapy (Galon et al, 2006; Ascierto et al, 2011, 2012). Our group observed that IFN-γ has a central role in the immunologically active cancer phenotype characterised by enhanced expression of IRF-1. This, in turn, correlates with the activation of transcripts associated with improved survival in cancer (Galon et al, 2006; Ascierto et al, 2012; Spivey et al, 2012). However, the mechanisms leading to the activation of IRF-1 in cancer are still unknown. Assessing the cytokine-mediated IRF-1 activation may help in understanding the intrinsic mechanisms underlying differential immune behaviour of cancer cells.
IFN-γ is a pleiotropic cytokine that induces gene expression through activation by tyrosine and serine phosphorylation of components of the JAK-STAT pathway (Darnell et al, 1994; Darnell, 1997; Stark et al, 1998), by binding to gamma-activated sequence (Stark et al, 1998; Shultz et al, 2009) or to IFN-stimulated response elements in the promoters of the IFN-stimulated genes. In this report, we assessed IRF-1 activation by using the ImageStream Flow Cytometry approach. Knowledge of the cellular localisation of a given transcription factor can provide insights about its activation; indeed, when the transcription factor is primarily sequestered in the cytosol, its ability to regulate gene transcription could be inhibited. We used the ImageStream nuclear localisation score to assess IRF-1 activation (George et al, 2006).
After treatment with IFN-γ and TNF-α, we observed a significant increase of IRF-1 nuclear translocation. The induction of IRF-1 activation after cytokine stimulation was a primary response that did not require protein synthesis, as it occurred within 1 h upon stimulation. Thus, even though several reports have shown that the effect of IFN-γ and TNF-α occur at the transcription and protein level (Lamb et al, 1995; Pine, 1997; Lee et al, 2000), our data suggest that the IRF-1 activation may be mediated by mechanisms, which do not require de novo protein synthesis.
Given that different cell lines have different amplitude of response to cytokine stimulation, we asked whether the inducibility of IRF-1 by IFN-γ and TNF-α was able to distinguish a parallel different behaviour of cancer cells. Toward this aim, we measured the difference of nuclear translocation score between controls and stimulated cells and further selected three cell lines with low and three cell lines with high delta of IRF-1 nuclear translocation among the three experimental points (IFN-γ, TNF-α and IFN-γ plus TNF-α stimulation). When comparing the basal transcriptome of these cell lines, we found 597 annotated transcripts significantly different between the two groups. Most interestingly, functional interpretation of such transcripts revealed mTOR and Wnt/β-signaling among the mostly downregulated pathways in cell lines with high IRF-1 responsiveness to IFN-γ and TNF-α. This is in perfect concordance to previous studies, which showed the existence of at least two immune phenotypes in melanoma: a Th17 phenotype associated with overexpression of components of Wnt and mTOR pathways, enhanced cellular motility, a more undifferentiated status and poor prognosis, and a Th1 immune phenotype associated with the expression of melanocytic lineage-specific transcripts, a more differentiated status and better prognosis (Wang et al, 2002; Spivey et al, 2012). The latter has been shown to centre around the activation of IRF-1 and to be characterised by a classic IFN-signature. Thus, the differential IRF-1 inducibility reflects a different cancer behaviour that has already been described in literature.
When assessing the activation of both IRF-1 and NF-κB, we found the two transcription factors to be concurrently activated. Previous studies on human cell lines have demonstrated that TNF-α and IFNs co-regulate the expression of inflammation-associated genes such as MHC class I, intracellular cell adhesion molecule 1 and vascular cell adhesion molecule-1 in a synergistic manner (Doukas & Pober, 1990; Blanchet et al, 1991; Jahnke & Johnson, 1994; Johnson & Pober, 1994; Drew et al, 1995). However, when testing the co-localisation of the two transcription factors, we were not able to demonstrate their physical interaction, not even after cytokine stimulation. This suggests that the activation of the two transcription factors may determine the same effects; moreover, the fact that the co-localisation was never significant in all the experimental conditions suggests a mutual exclusively activation of IRF-1 and NF-κB at a cell level. Thus, it is likely that IRF-1 and NF-κB are activated in an additive rather than in a synergistic manner under our experimental system. Testing this idea with a variety of experimental models of melanoma is warranted to determine whether IRF-1 and NF-κB interact with each other or act independently both at basal level and after IFN-γ and TNF-α stimulation. Moreover, we cannot exclude the possibility that NF-κB p50 and/or NF-κB p52 expression are activated and may compensate for any loss of NF-κB activity (Cogswell et al, 2000).
In conclusion, our findings suggest that the responsiveness of IRF-1 to the immune cytokines IFN-γ and TNF-α is able to point out two different cancer phenotypes and support the central role of such transcription factor in influencing different tumour behaviours.
Acknowledgments
This study was financed by the National Institutes of Health; all NIH affiliated authors performed this work as part of their official duties. DB's scholarship was supported by the Conquer Cancer Foundation of the American Society of Clinical Oncology (2011 Young Investigator Award). DM's scholarship was supported by the Master and Back Program of Regione Autonoma della Sardegna, Italy.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Ascierto ML, De Giorgi V, Liu Q, Bedognetti D, Spivey TL, Murtas D, Uccellini L, Ayotte BD, Stroncek DF, Chouchane L, Manjili MH, Wang E, Marincola FM. An immunologic portrait of cancer. J Transl Med. 2011;9:146. doi: 10.1186/1479-5876-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, Marincola FM, Manjili MH. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast cancer Res Treat. 2012;131 (3:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406 (6795:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Blanchet O, Bourge JF, Zinszner H, Tatari Z, Degos L, Paul P. DNA binding of regulatory factors interacting with MHC-class-I gene enhancer correlates with MHC-class-I transcriptional level in class-I-defective cell lines. Int J Cancer Suppl. 1991;6:138–145. doi: 10.1002/ijc.2910470725. [DOI] [PubMed] [Google Scholar]
- Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19 (9:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277 (5332:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264 (5164:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Doukas J, Pober JS. IFN-gamma enhances endothelial activation induced by tumor necrosis factor but not IL-1. J Immunol. 1990;145 (6:1727–1733. [PubMed] [Google Scholar]
- Drew PD, Franzoso G, Becker KG, Bours V, Carlson LM, Siebenlist U, Ozato K. NF kappa B and interferon regulatory factor 1 physically interact and synergistically induce major histocompatibility class I gene expression. J Interferon Cytokine Res. 1995;15 (12:1037–1045. doi: 10.1089/jir.1995.15.1037. [DOI] [PubMed] [Google Scholar]
- Ekmekcioglu S, Mumm JB, Udtha M, Chada S, Grimm EA. Killing of human melanoma cells induced by activation of class I interferon-regulated signaling pathways via MDA-7/IL-24. Cytokine. 2008;43 (1:34–44. doi: 10.1016/j.cyto.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391 (6662:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313 (5795:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Galon J, Franck P, Marincola FM, Angell HK, Thurin M, Lugli A, Zlobec I, Berger A, Bifulco C, Botti G, Tatangelo F, Britten CM, Kreiter S, Chouchane L, Delrio P, Hartmann A, Asslaber M, Maio M, Masucci GV, Mihm M, Vidal-Vanaclocha F, Allison JP, Gnjatic S, Hakansson L, Huber C, Singh-Jasuja H, Ottensmeier C, Zwierzina H, Laghi L, Grizzi F, Ohashi PS, Shaw PA, Clarke BA, Wouters BG, Kawakami Y, Hazama S, Okuno K, Wang E, O'Donnell-Tormey J, Lagorce C, Pawelec G, Nishimura MI, Hawkins R, Lapointe R, Lundqvist A, Khleif SN, Ogino S, Gibbs P, Waring P, Sato N, Torigoe T, Itoh K, Patel PS, Shukla SN, Palmqvist R, Nagtegaal ID, Wang Y, D'Arrigo C, Kopetz S, Sinicrope FA, Trinchieri G, Gajewski TF, Ascierto PA, Fox BA. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med. 2012;10 (1:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311 (1-2:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Jahnke A, Johnson JP. Synergistic activation of intercellular adhesion molecule 1 (ICAM-1) by TNF-alpha and IFN-gamma is mediated by p65/p50 and p65/c-Rel and interferon-responsive factor Stat1 alpha (p91) that can be activated by both IFN-gamma and IFN-alpha. FEBS Lett. 1994;354 (2:220–226. doi: 10.1016/0014-5793(94)01130-3. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Pober JS. HLA class I heavy-chain gene promoter elements mediating synergy between tumor necrosis factor and interferons. Mol Cell Biol. 1994;14 (2:1322–1332. doi: 10.1128/mcb.14.2.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22 (1:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- Lamb P, Seidel HM, Haslam J, Milocco L, Kessler LV, Stein RB, Rosen J. STAT protein complexes activated by interferon-gamma and gp130 signaling molecules differ in their sequence preferences and transcriptional induction properties. Nucleic Acids Res. 1995;23 (16:3283–3289. doi: 10.1093/nar/23.16.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Hong JH, Seo YS. Tumour necrosis factor-alpha and interferon-gamma synergistically activate the RANTES promoter through nuclear factor kappaB and interferon regulatory factor 1 (IRF-1) transcription factors. Biochem J. 2000;350 (Pt 1:131–138. [PMC free article] [PubMed] [Google Scholar]
- Leffers N, Gooden MJ, de Jong RA, Hoogeboom BN, ten Hoor KA, Hollema H, Boezen HM, van der Zee AG, Daemen T, Nijman HW. Prognostic significance of tumor-infiltrating T-lymphocytes in primary and metastatic lesions of advanced stage ovarian cancer. Cancer Immunol Immunother. 2009;58 (3:449–459. doi: 10.1007/s00262-008-0583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marincola FM, Wang E, Herlyn M, Seliger B, Ferrone S. Tumors as elusive targets of T-cell-based active immunotherapy. Trends Immunol. 2003;24 (6:335–342. doi: 10.1016/s1471-4906(03)00116-9. [DOI] [PubMed] [Google Scholar]
- Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18 (2:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Pine R. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 1997;25 (21:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5 (5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Shultz DB, Rani MR, Fuller JD, Ransohoff RM, Stark GR. Roles of IKK-beta, IRF1, and p65 in the activation of chemokine genes by interferon-gamma. J Interferon Cytokine Res. 2009;29 (12:817–824. doi: 10.1089/jir.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Spivey TL, De Giorgi V, Zhao Y, Bedognetti D, Pos Z, Liu Q, Tomei S, Ascierto ML, Uccellini L, Reinboth J, Chouchane L, Stroncek DF, Wang E, Marincola FM. The stable traits of melanoma genetics: an alternate approach to target discovery. BMC Genomics. 2012;13:156. doi: 10.1186/1471-2164-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Mocellin S, Perez-Diez A, Petersen D, Zhao Y, Simon R, Powell JI, Asaki E, Alexander HR, Duray PH, Herlyn M, Restifo NP, Liu ET, Rosenberg SA, Marincola FM. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2002;62 (13:3581–3586. [PMC free article] [PubMed] [Google Scholar]
- Wang E, Panelli MC, Zavaglia K, Mandruzzato S, Hu N, Taylor PR, Seliger B, Zanovello P, Freedman RS, Marincola FM. Melanoma-restricted genes. J Transl Med. 2004;2 (1:34. doi: 10.1186/1479-5876-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuba-Surma EK, Kucia M, Abdel-Latif A, Lillard JW, Jr, Ratajczak MZ. The ImageStream System: a key step to a new era in imaging. Folia Histochem Cytobiol. 2007;45 (4:279–290. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.