Abstract
Adenoid cystic carcinomas (ACCs) are among the most enigmatic of human malignancies. These aggressive salivary cancers frequently recur and metastasize despite definitive treatment, with no known effective chemotherapy regimen. Here, we determined the ACC mutational landscape and report the exome or whole genome sequences of 60 ACC tumor/normal pairs. These analyses revealed a low exonic somatic mutation rate (0.31 non-silent events/megabase) and wide mutational diversity. Interestingly, mutations selectively involved chromatin state regulators, such as SMARCA2, CREBBP, and KDM6A, suggesting aberrant epigenetic regulation in ACC oncogenesis. Mutations in genes central to DNA damage and protein kinase A signaling also implicate these processes. We observed MYB-NFIB translocations and somatic mutations in MYB-associated genes, solidifying these aberrations as critical events. Lastly, we identified recurrent mutations in the FGF/IGF/PI3K pathway that may potentially offer new avenues for therapy (30%). Collectively, our observations establish a molecular foundation for understanding and exploring new treatments for ACC.
Adenoid cystic carcinomas (ACCs) are malignancies that cause significant morbidity and mortality. Typically arising from salivary glands, they are characterized by unpredictable growth, extensive perineural invasion, and high rates of metastasis, ultimately resulting in low survival rates. Treatment remains limited to surgery and radiation, and no systemic agent has been found to be effective.1 Although high EGFR and c-kit expression have been identified2,3, drugs against these targets have not led to substantive clinical responses.4 Other initial findings include the identification of MYB-NFIB fusion proteins but their significance remains unclear.5 A deeper grasp of the underlying genomic foundations of ACC is needed to understand its molecular basis and guide the development of effective therapies.
We sequenced the entire exome (n=55) or genome (n=5) of 60 ACC samples with matched normal DNA (Table 1). This approach generated 2,221 gigabases of mapped sequence with 92.4% of the target sequence covered to at least 10x depth. Sequencing generated a mean exome and genome coverage of 106x and 37x, respectively. To ensure the accuracy of our massively parallel sequencing data, we conducted extensive validation of nearly every candidate somatic mutation identified (2,751 variant calls) (Supplementary Fig. 1) using targeted re-sequencing (Supplementary Fig. 2, Supplementary Table 1). In addition, we also performed FISH analysis for the MYB-NFIB translocation (Supplementary Fig. 3, Supplementary Table 2).
Table 1.
ACC whole exome and whole genome sequencing statistics.
| Statistic | Whole exome | Whole Genome |
|---|---|---|
| Tumor/normal pairs sequenced | 55 | 5 |
| Total Gb sequenced | 971.5 | 1249.4 |
| Mean fold tumor target coverage | 100 (23–167) | 38 (34–45) |
| Mean fold normal target coverage | 112 (25–185) | 35 (33–37) |
| Mean somatic mutation rate per Mb | 0.4 (0.06–0.8) | 0.3 (0.2–0.5) |
| Mean number of validated nonsynonymous mutations/patient | 11 (1–36) | 11 (5–17) |
| Mean number of transcribed noncoding mutations/patient (UTRs) | 6 (1–13) | 3 (1–6) |
| Total number of structural rearrangements | N/A | 17 |
| Total number of frame-preserving genic rearrangements | N/A | 1 |
We identified a mean of 22 somatic mutations per sample, corresponding to approximately 0.31 non-silent mutations per MB. This rate is quite low compared to most adult solid tumors such as head and neck squamous cell carcinoma6,7 and colon cancer8 yet similar to hematologic malignancies and neuroblastoma.9–11 The transition/transversion ratio (Ti/Tv) was 1.1, similar to some carcinogen-driven malignancies6,7,12 but unlike most described cancer types.13 The somatic mutation frequency correlated with solid histology (Wilcoxon rank-sum test, p = 4.0 × 10−2), and MYB translocations occurred in 57% of samples (34/60).
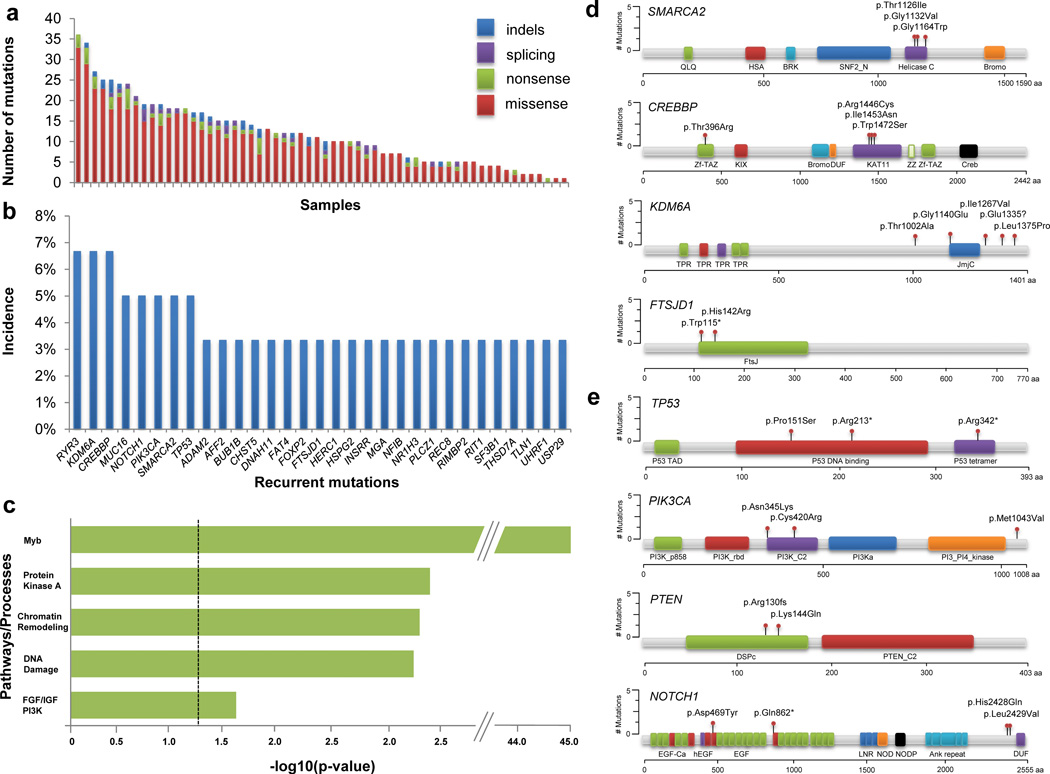
We validated 710 distinct nonsynonymous mutations across 643 genes (1–36 per tumor) (Fig. 1a, Supplementary Table 3, Supplementary Fig. 4). This represents substantial mutational heterogeneity across tumors (Fig. 1b, Fig. 2). We employed CHASM, a widely used approach for distinguishing driver from passenger mutations,14 to identify multiple potential driver mutations, including those in PIK3CA, TP53, PTEN, SMARCA2, KDM6A, and CREBBP (Supplementary Tables 4, 5). Analysis of these driver genes demonstrated marked enrichment in pathways involved in chromatin remodeling, DNA damage, MYB, protein kinase A (PKA) signaling, and PI3K signaling (Fig. 1c–e). For example, despite low overall mutation frequency, 35% of ACC tumors were mutated in chromatin regulators. These data suggest that the ACC mutational landscape is characterized by combinations of MYB pathway alterations and mutations in specific biological processes (Fig. 2). Interestingly, a small subset (n=8) were observed with no CHASM-designated driver mutations. It is possible that some mutations in these tumors are drivers not called by CHASM or that other, non-exonic alterations are important in these tumors.
Figure 1. Mutational landscape of adenoid cystic carcinoma.
(a). Number of validated nonsynonymous somatic mutations per sample across 60 ACC cases. (b). Representative list of recurrent nonsynonymous somatic mutations. Multiple mutations within a given sample in the same gene were only counted once. (c). Pathways affected by driver mutations identified by CHASM. Bonferroni FDR-corrected p-values for pathway enrichment are shown. (d). Location of ACC mutations in key chromatin remodeling genes. HSA, helicase-SANT-associated; Zf-TAZ, TAZ zinc finger; DUF, domain of unknown function; KAT11, histone acetylation protein; ZZ, ZZ-type zinc finger; SNF2_N, SNF2 family N-terminal domain; Helicase_C, Helicase conserved C-terminal domain; TPR, tetratricopeptide repeat; JmjC, JmjC hydroxylase domain; FtsJ, FtsJ-like methyltransferase. (e). Location of ACC mutations in key established cancer genes. PI3K_p85B, PI3K p85-binding domain; PI3K_rbd, PI3K ras-binding domain; PI3Ka, PI3K accessory domain; EGF_Ca, calcium-binding EGF domain; hEGF, human growth factor-like EGF; Ank repeat, ankyrin repeat; DSPc, dual specificity phosphatase, catalytic domain; P53 TAD, P53 transactivation motif; P53_tetramer, P53 tetramerization motif.
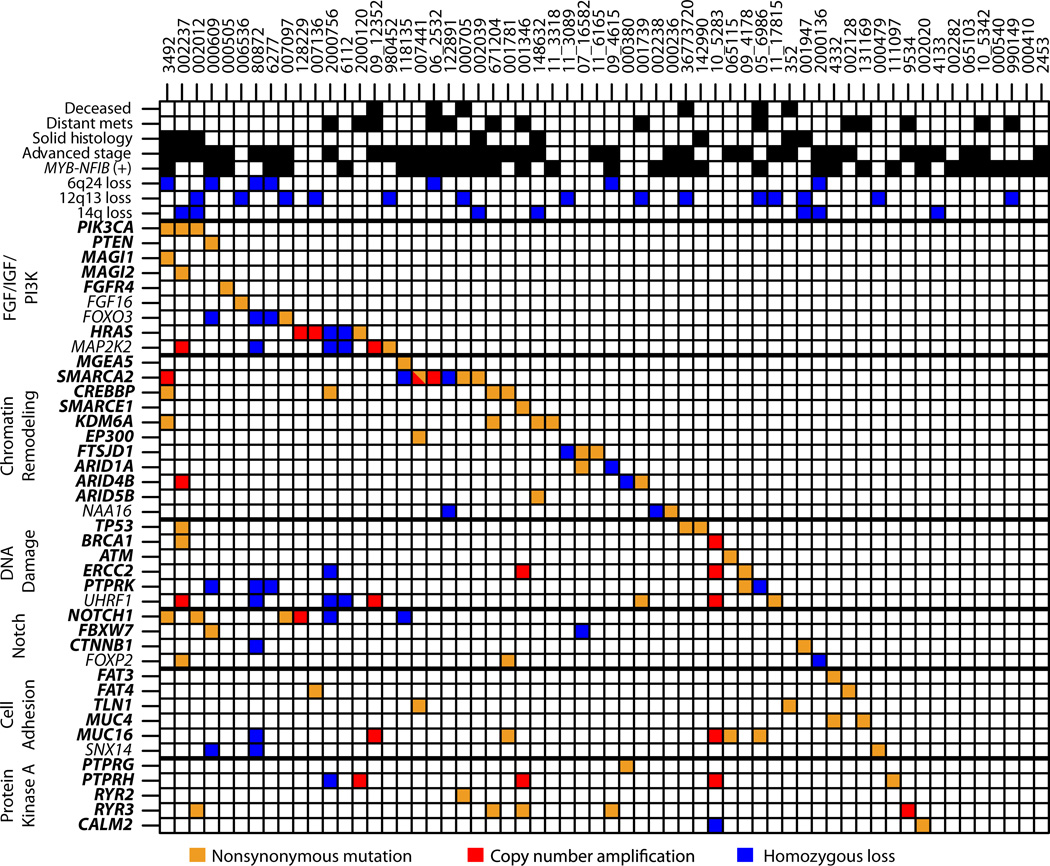
Figure 2. Integrated analysis of adenoid cystic carcinoma genetic alterations.
Clinical data, validated somatic mutations, validated structural variants, and copy number alterations for 60 ACC cases. Genes highlighted in bold contain significant driver mutations identified by CHASM.14 Remaining genes contain selected mutations of interest that did not reach significance by CHASM but which are either altered in other malignancies or are integral components of affected pathways. Orange, nonsynonymous somatic mutation (missense, nonsense, splice site, indel); blue, homozygous copy number deletion; red, high-level copy number amplification.
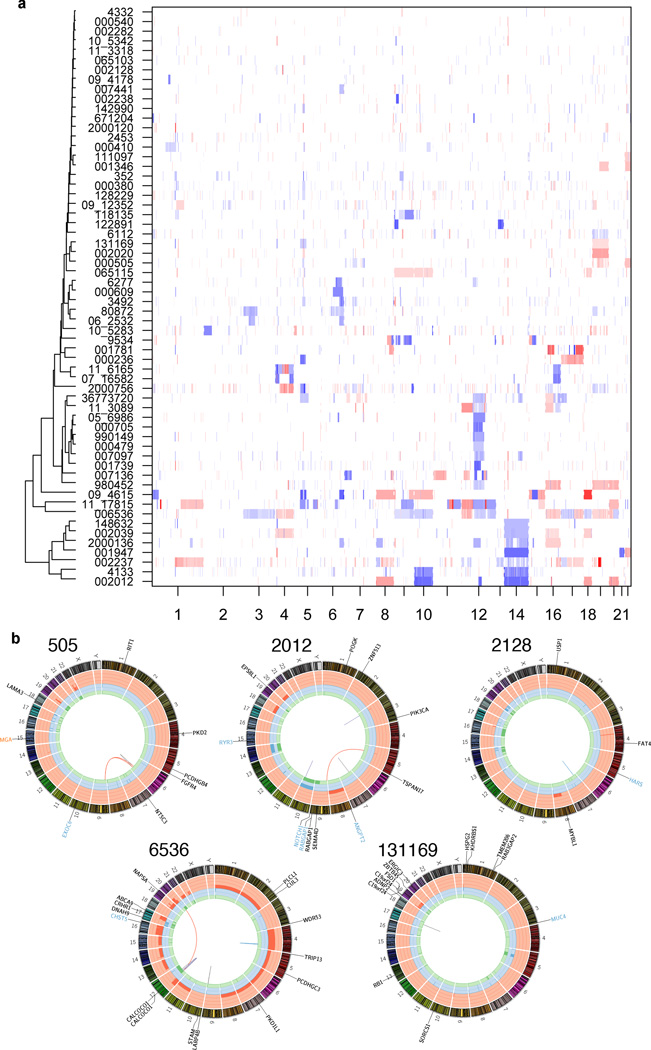
We used exome and genome sequencing data to characterize the copy number landscape of these tumors. We analyzed somatic copy number variations (CNVs) using ExomeCNV15 and found high concordance with a subset (n=12) that underwent array-based analysis. GISTIC2.016 identified recurrent high-level losses in 6q24, 12q13, and 14q (Fig. 3a, Supplementary Table 6).5 Samples with 14q loss were more likely to be of solid histology (Fisher’s exact test, p = 2.0 × 10−4), while samples with 6q24 loss were enriched for advanced stage (p = 4.0 × 10−2). Expression array analysis on 23 ACC tumors found no distinct subgroups (Supplementary Fig. 5). Genes harboring driver mutations were confirmed to be generally expressed in ACC tumors (Supplementary Table 7).
Figure 3. Structural variations and copy number landscape of adenoid cystic carcinoma.
(a). Clustered copy number profiles of 60 ACC cases. Clustering revealed 4 sub-clusters of tumors with one each defined by loss of 14q, loss of 12q13, a cluster with two samples containing a large number of copy number alterations, and most tumors in a large group with minimal copy number changes. White, normal (diploid) copy number log-ratio; blue, copy number loss; red, copy number amplification. (b). Circos plots73 of genetic alterations in 5 ACC cases. Plots depict validated structural genetic variants, DNA copy number alterations, intra- and inter-chromosomal translocations, and sequence alterations. Loss-of-heterozygosity, green track; amplification, red track; copy number loss, blue track; missense mutation, black gene name; nonsense mutation, blue gene name; splice site mutation, orange gene name; inter-chromosomal translocation, red line; deletion, gray line; intra-chromosomal translocation, blue line; insertion, purple line.
Whole genome paired-end sequencing identified numerous structural variants (SVs) (Fig. 3b, Supplementary Table 8), with the existence of 17 SVs across 5 samples confirmed using PCR (Supplementary Fig. 6, Supplementary Table 9). Combined with FISH data, MYB translocations were the only recurrent SVs detected. However, we cannot exclude the presence of less common recurrent translocations.17 Intriguingly, one sample harbored a tandem duplication within FGFR2. Similar alterations comprised of dimerized, constitutively active variants have been described in hematologic malignancies.18 We did not detect a fusion transcript from this tumor using RT-PCR, but this does not rule out an alternative transcript configuration. Larger numbers will need to be analyzed to further characterize its incidence and impact. The remaining variants were not predicted to be in-frame. Collectively, few CNVs or SVs were found across the cohort, demonstrating the relatively “quiet” nature of the ACC genome. Thus, our data indicate that ACC’s major SVs are MYB translocations and recurrent deletions on 6q24, 12q13, and 14q.
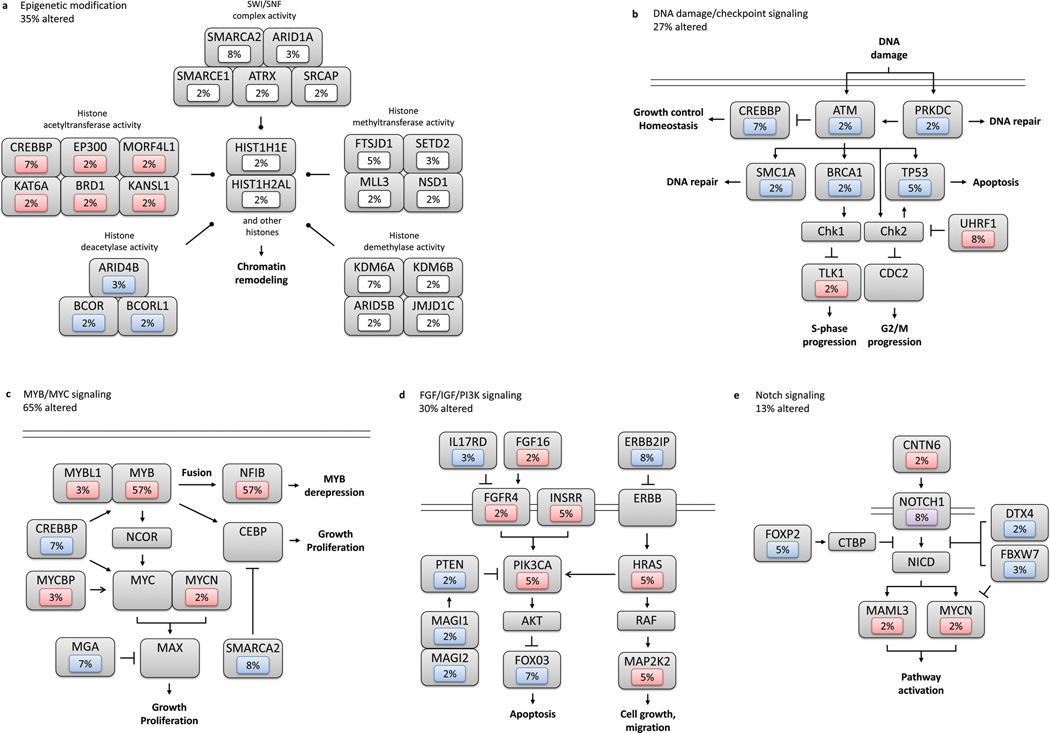
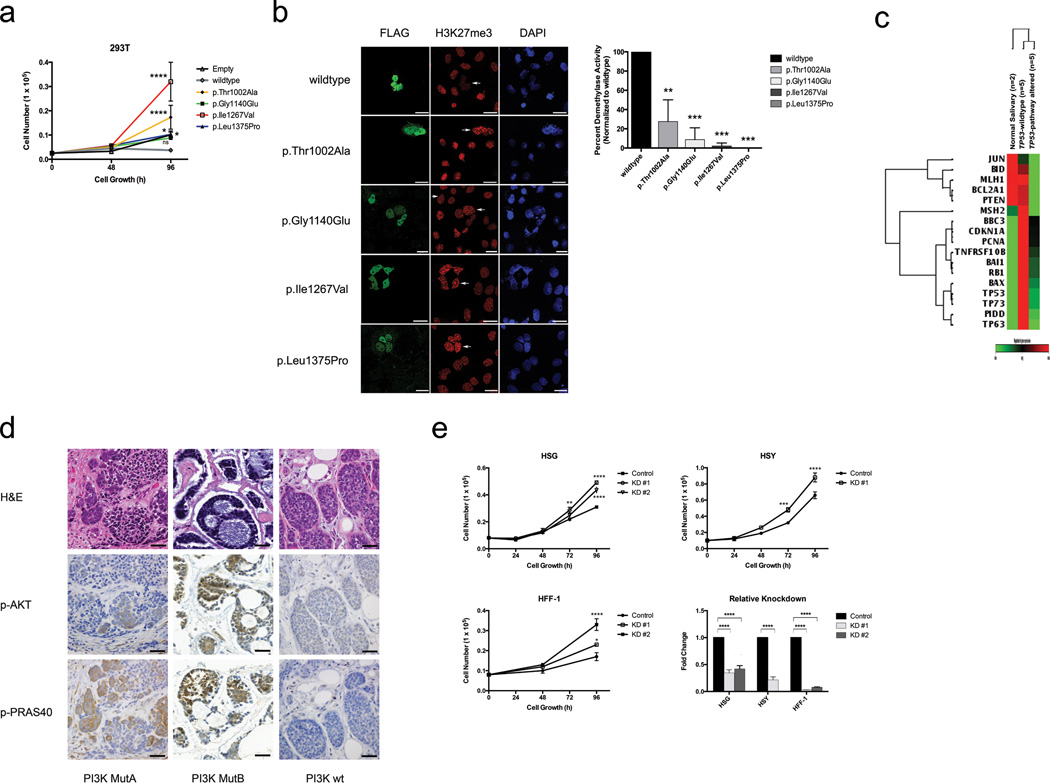
A prominent feature of the ACC mutational landscape is the presence of multiple mutations targeting chromatin remodeling genes (35% incidence) (Fig. 1d, Fig. 4a). Such alterations are increasingly recognized as playing key roles in oncogenesis19,20 and have been reported in various other tumors10,21 but not ACC. Among ACC alterations, chromatin state modifiers were substantially enriched for somatic mutations (q = 4.5 × 10−3). We identified multiple aberrations in the SWI/SNF-related, matrix associated, actin dependent regulator of chromatin (SMARC) family, including SMARCA2 (5%) and single mutations in SMARCE1 (2%), ARID1A (2%), and ATRX (2%). SMARC mutations contribute to the development of both cancers and genetic diseases.21–25 SMARCA2 encodes a core catalytic subunit of the SWI/SNF complex involved in regulating gene transcription.26 All SMARCA2 mutations were clustered within the Helicase C family domain (T1126I, G1132V, G1164W). Notably, mutated helicases have been shown to increase cancer susceptibility, likely by disturbing core repair mechanisms.27 Similarly, we identified likely loss of function in SMARCE1 (2%), another SWI/SNF complex gene with a HMG box domain mutation (Y73C) that likely affects DNA binding.28 Additional recurrent mutations include CREBBP (7%), a histone acetyltransferase that enables transcription across multiple critical signaling pathways.24,25,29 The majority of CREBBP mutations clustered in the critical KAT11 histone acetylation domain (R1446C, I1453N, W1472S).30 We also observed numerous KDM6A missense mutations (7%), a histone demethylase also termed UTX.31 The functional impact of these KDM6A mutations was evaluated via a well-characterized H3K27me3 assay. We observed abrogation of demethylase activity in cells overexpressing mutant but not those overexpressing wildtype KDM6A (Fig. 5a–b). Moreover, whereas wildtype KDM6A suppressed growth, mutants either lost the ability to suppress growth or in some cases, augmented it (dominant phenotype).
Figure 4. Diversity and frequency of ACC genetic changes culminating in aberrant signaling pathways.
Alterations are defined by somatic mutations, homozygous deletions, high-level focal amplifications, and structural variants validated by FISH or PCR. Frequencies are expressed as a percentage of all cases. Red background denotes activating alteration, blue background denotes inactivating alteration, white background denotes numerous alterations, and purple background denotes alteration of unclear significance. Interactions based upon IPA or Pathway Commons.38 (a). Epigenetic modification. (b). DNA damage/checkpoint signaling pathway. (c). MYB/MYC signaling pathway, with fusion denoted by recurrent t(6;9) translocation between MYB and NFIB. (d). FGF/IGF/PI3K signaling pathway. (e). Notch signaling pathway.
Figure 5. Functional consequences of genetic alterations in KDM6A, TP53, PI3K, and FAT4 pathway genes.
(a). Overexpression of KDM6A mutants identified in ACC compared to overexpressed wildtype demonstrates increased growth. Experiments performed in triplicate. (b). KDM6A tumor-specific mutants exhibit abrogated H3K27me3 demethylase activity relative to wildtype KDM6A. Representative immunostaining of cells overexpressing wildtype or mutant KDM6A is shown with corresponding quantitation. Arrows represent cells expressing FLAG-tagged KDM6A construct. Experiments performed in triplicate. Scale bars, 20µm. (c). Heatmap shows differential expression of key TP53 transcriptional targets in TP53-pathway altered ACC samples (TP53-mutant or MDM2-amplified) compared to TP53-wildtype ACC samples and normal salivary tissue. (d). PI3K-mutant ACCs demonstrate marked downstream p-AKT and p-PRAS40 immunohistochemistry staining relative to PI3K-wildtype ACCs. Scale bars, 120µm. (e). FAT4 knockdown demonstrates increased growth in human foreskin fibroblasts (HFF-1), immortalized human salivary cells (HSG), and human salivary adenocarcinoma cells (HSY). Experiments performed in triplicate. KD, knockdown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns, not significant (ANOVA). Error bars, ± 1 S.E.M.
Other related mutations include those involved in histone acetyltransferase/deacetylase activity (EP300, ARID4B, ARID5B, BRD1), histone methyltransferase/demethylase function (FTSDJ1, MLL3), as well as histones themselves (HIST1H2AL, HIST1H1E). EP300 binds CREBBP as a transcriptional co-activator that regulates cell proliferation and differentiation32, and harbored a splice site mutation within its KAT11 histone acetylation domain (G1429_splice). HIST1H2AL encodes a core component of the nucleosome, while HIST1H1E is known to link histones for compaction into higher-order structures. Importantly, histone mutations have been implicated in cancers such as pediatric gliomas.8,33,34 While mutations in our study were observed within known domains, (R18L in core histone H2A/H2B/H3/H4 and K75N in linker histone H1/H5 domains, respectively), further investigation will be necessary to determine their functional impact. Overall, a diversity of chromatin genes was involved, suggesting that disparate means of chromatin dysregulation may promote ACC development through convergent pathways. Given the emerging importance of epigenetic agents on other disorders35,36, similar treatments for ACC cases with chromatin derangements may hold promise.
We observed a second group of mutations in genes involved in DNA damage response (q = 5.6 × 10−3) (Fig. 4b). In TP53 (5% incidence), one missense (P151S) and 2 nonsense mutations (R213*, R342*) in the central binding and tetramer motifs were found (Fig. 1e). Similarly UHRF1, a ubiquitin ligase involved in the p53-dependent DNA damage checkpoint37, was altered in 8% of patients (2 mutations, 3 homozygous deletions). TXNIP, a protein frequently repressed in cancers38,39, was noted to have a frameshift insertion in its arrestin domain (L129fs). We also discovered 2 missense mutations each within individual samples in ATM, BRCA1, and DCLRE1A, a DNA cross-link repair gene. Notably, markedly decreased expression in key TP53 transcriptional targets was specifically identified in tumors with TP53 pathway alterations (binomial test, p = 1.0 × 10−4) (Fig. 5c).
Previous work has implicated MYB as an ACC fusion oncogene with NFIB via a t(6;9) translocation40 (Fig. 4c). Indeed, we verified 57% of our cohort as MYB-NFIB(+) by FISH analysis. We extend this initial finding and observe further MYB pathway dysregulation with alterations found in an additional 8% of cases. Specifically, splice site and coding mutations involving exon 10 of MYB were seen, potentially disrupting its leucine-rich negative regulatory domain40 and triggering constitutive activation. Five additional homozygous deletions were identified on CNV analysis (4/5 were MYB-NFIB(+)), though these may be secondary alterations around the translocation. Other mutations identified in the MYB pathway included MYCBP2, MGA, and MCM4. In NFIB, we observed two truncating mutations in the CTF/NFI transcription modulation domains (Y249*, P390fs), in addition to 4 homozygous deletions (all were MYB-NFIB(+)). Our data highlight MYB in ACC as an active oncogenic partner in fusion transcripts but also suggest a separate role for NFIB given the presence of specific mutations in this gene.
Multiple genes involved in the PKA pathway were found to be mutated (27% incidence, q = 4.2 × 10−3). We identified recurrent mutations in RYR3 (7%) and RYR2 (2%), both tetrameric, intracellular calcium channels implicated in breast cancer progression.41,42 Mutations were also observed in PKA pathway-associated genes that are known or potential tumor suppressors, including tyrosine phosphatases (PTPRG, PTPRH, PTPRJ, PTPRK). Nonsense (PTPRG, PTPRH) and frameshift mutations (PTPRK) abrogate their phosphatase domains (E736*, W602*, L457fs, respectively), while an additional 4 samples had PTPRK homozygous deletions. PKA-related genes are known to facilitate CREBBP recruitment via calcium influx43 and CREB phosphorylation, regulating multiple critical developmental processes involved in tumorigenesis.24,44
Mutations in genes important in the FGF/IGF/PI3K pathway were also identified in 30% of our cohort (q = 2.4 × 10−2) (Fig. 4d). Three tumors harbored separate missense mutations in PIK3CA (Fig. 1e). Another tumor had two mutations in the catalytic domain of PTEN (R130fs, K144Q). All five lesions occurred in hotspots (COSMIC v61).45 FOXO346 was also altered in 7% of patients. We further identified mutations in receptor kinase/PI3K-associated genes, including FGF16, FGFR4, IGFBP2, and ILR17RD. Fibroblast growth factors and insulin-like growth factors are potent oncogenic PI3K activators47,48, while IL17RD appears to be a PI3K inhibitor through its interaction with FGFR.49 The samples with PI3K-associated insults all demonstrated solid histology, the most aggressive ACC variant (Fisher’s exact test, p < 1.6 × 10−3). We further evaluated PI3K-altered samples using immunohistochemistry (p-AKT and p-PRAS40). Functional activation of the PI3K pathway was observed in all ACC tumors harboring PIK3CA or PTEN mutations, but not in wild-type tumors (Fig. 5d, Supplementary Fig. 7). This is supported by the presence of significantly enriched PI3K signatures in PI3K-mutant ACC tumors via GSEA (Gene Set Enrichment Analysis) (p < 1.0 × 10−3) (Supplementary Fig. 8a). Together, these findings delineate a previously undescribed subset of ACC patients which we hypothesize may benefit from agents targeting this pathway.50
Despite not meeting significance by CHASM analysis, we observed alterations in the Notch signaling pathway in 13% of samples (Fig 4e). In addition to recurrent NOTCH1 (5% incidence, including 3 missense and one nonsense) and FOXP2 (3%) mutations, predicted functional mutations were found in DTX4, FBXW7, and CNTN6. FBXW7 (R465H) is a tumor suppressor, targeting c-myc and NOTCH1 for degradation.51 Depending on cellular context, NOTCH1 has a dual role in cancer as an oncogene or tumor suppressor.6,7,52–55 Analysis of NOTCH1-altered ACC samples using GSEA56 showed a trend toward Notch signaling enrichment (Supplementary Fig. 8b). These alterations require further characterization, but may have therapeutic implications.57
Intriguingly, we observed mutations in various genes involved in cell metabolism processes. Of particular interest are recurrent alterations in HSPG2 or perlecan (7%), a basement membrane-based proteoglycan known to modulate tumorigenic growth factors, including FGF. FGF signaling promotes mitogenesis and angiogenesis58, while deletion of perlecan heparan sulfate domains has been shown to impair tumorigenesis.59–61 Importantly, ACC produces high perlecan levels within its pseudocystic cribiform structures62,63, and maximum perlecan synthesis occurs during the tumor’s proliferative phase.64 Additionally, we identified a hotspot IDH1 mutation in its catalytic domain (R132H).33,65,66,67
ACC is unique in its propensity for perineural invasion and distant metastasis. We noted numerous mutations in cell adhesion proteins (29% incidence). We found mutations in the active domains of genes responsible for neuronal axon guidance cues, including NTNG1, SEMA3G, and SEMA5A. Netrins and semaphorins are implicated in neuronal invasion68 and metastasis.69 Additionally, mutations in protocadherins were observed, including truncating mutations in FAT3 and FAT4. FAT4 has been hypothesized as a tumor suppressor in gastric adenocarcinoma.70 We examined FAT4 through RNAi knockdown in human salivary cells (HSG, HSY, HTB-41) as well as fibroblasts (HFF-1) (Fig. 5e, Supplementary Fig. 9). Significant increases in growth occurred in all cells tested following FAT4 knockdown but not in controls.
In summary, we report a large-scale ACC analysis and highlight potential driver mutations in novel genes and pathways. Common alterations across ACC include pathways involving chromatin remodeling, MYB/MYC, DNA damage, and tyrosine kinase signaling (Fig. 4). Our discovery of genomic alterations in targetable pathways suggests potential avenues for novel therapies for a typically chemoresistant malignancy. Verified ACC cell lines71,72 are needed to further substantiate the clinical utility of mutations identified here. In total, our data provide insights into the genetic framework underlying ACC oncogenesis and establish a foundation for new therapeutic strategies.
URLs. MSKCC cBioPortal for Cancer Genomics, http://www.cbioportal.org/; dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; ESP5400, http://evs.gs.washington.edu/EVS/; 1000 Genomes Project, http://www.1000genomes.org/; IPA, http://www.ingenuity.com/; Pathway Commons, http://www.pathwaycommons.org/.
ONLINE METHODS
Tumor samples
Primary tumor samples and matched normal specimens (peripheral blood or tissue) were obtained with written informed consent per approved institutional review board protocols (Supplementary Note). Specimens were snap-frozen in liquid nitrogen immediately upon surgical resection and stored at −80°C. Hematoxylin eosin-stained sections were prepared and diagnosis confirmed by a dedicated head and neck pathologist. Microdissection was performed to ensure >70% tumor purity. DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen) and quantified via Quant-iTPicogreen dsDNA assay (Invitrogen). RNA was extracted using the RNeasy Mini kit (Qiagen) and quantified using a Nanodrop spectrophotometer (Thermo Fisher). The identities of tumor and normal DNA samples were reconfirmed by mass spectrometric fingerprint genotyping (Sequenom) as previously described.74
Exome capture and sequencing
Whole exome capture libraries were constructed via Agilent Sureselect 51MB target enrichment (Agilent). Approximately 2–3µg of genomic DNA from each tumor and normal specimen were sheared and ligated to barcoded sequencing adaptors. Enriched exome libraries were sequenced on a HiSeq 2000 platform (Illumina) for generation of paired-end reads (2×76bp) (Supplementary Table 1).
Analysis pipeline and mutation annotation
Reads were aligned to the hg19 (GRCh37) build using the Burrows-Wheeler Aligner (BWA)75 (Supplementary Fig. 1). Further indel realignment, base quality score recalibration, and duplicate read removal were performed using Genome Analysis Toolkit (GATK) v2.2.76 Somatic Sniper v1.0.077 and GATK Somatic Indel Detector76 were used to generate single nucleotide variations (SNVs) and indel calls, respectively, via standard default parameters. MuTect78 was used to confirm SNV calls with conditions described previously.79 Baseline filters (depth 3x coverage in both tumor and normal specimens, >97% normal allelic fraction, >10% tumor allelic fraction) were chosen. We excluded germline variants found in the 1000 Genomes Project80, ESP5400 (NHLBI GO Exome Sequencing Project), and dbSNP132.81 Further filtering was based upon mapping quality score, average read length, and strand bias as previously described.77 Resulting mutations were annotated based on RefSeq (Release 55) using Annovar.82 All candidates were manually inspected via Integrative Genomics Viewer (IGV) v2.183 (Supplementary Fig. 4). Mean somatic mutations per sample (16 nonsynonymous, 6 synonymous) are comprised of exonic SNVs and indels. The non-silent mutations per megabase (MB) rate (0.31) refers to nonsynonymous mutations found within the exon capture target regions (51MB).
Validation of mutations
Nearly all SNVs or indels were resequenced via SOLiD (Life Technologies) or MiSeq (Illumina) platforms to validate putative mutations. Regions encompassing putative candidates were amplified with PCR using Kapa Fast HotStartTaq Polymerase (KapaBiosystems) with WGA genomic DNA generated via Repli-G kits (Qiagen). For SOLiD preparation, 3µg of DNA was sheared, with 200–220 bp fragments selected by agarose gel electrophoresis. DNA was purified and ligated to P1 and P2 adaptors per manufacturer’s instructions. Following PCR amplification, DNA was quantified and size confirmed by Bioanalyzer DNA 1000 assay (Agilent Systems). Approximately 300–500 ng per sample was used for 24hr hybridization with custom SureSelect baits (Agilent Technologies) per manufacturer instructions. Captured DNA was separated from noncaptured DNA per SureSelect protocol. Following emulsion PCR and bead preparation, each sample was loaded into one octet of a SOLiD plate for paired-end sequencing. This was followed by BWA75 alignment, GATK v2.276 variant detection, and manual inspection on IGV 2.1.
For MiSeq sequencing, amplified templates were purified using AMPure (Beckman Coulter). The purified PCR reactions were sequenced bidirectionally with M13 forward and reverse primer and Big Dye Terminator Kit v.3.1 (Applied Biosystems). Dye terminators were removed using the CleanSEQ kit (Beckman Coulter), and the resulting amplicons were barcoded and prepared for sequencing using the Nextera DNASample Prep Kit (Illumina). Libraries were sequenced using the paired-end 150-cycle protocol, followed by BWA75 alignment, GATK v2.276 variant detection, and manual inspection on IGV 2.1 (Supplementary Table 3).
Whole genome capture and sequencing
Library construction and whole genome sequencing were performed as previously described.84 Briefly, 5µg of native DNA from tumor and matched normal sample were separately sheared, standard paired-end adaptors were ligated, and products run on a gel and excised. Bands were purified via mini-elute columns (Qiagen) and enriched by PCR amplification (10 cycles). Paired-end sequencing (2×101bp) was performed on the HiSeq 2000 platform (Illumina) (Supplementary Table 8). The HumanOmni 2.5–8v1 BeadChip Kit (Illumina) was used for genotyping.
Structural variant and copy number analysis
Structural variants (SVs) were identified using CREST (Clipping REveals STructure)85, which employs soft-clipped reads to directly map the breakpoints of SVs. Samples were analyzed using the paired analysis module. Primers were designed for the 1000bp flanking regions surrounding the predicted SV breakpoint using Primer386 (Supplementary Table 9). Using a standard touchdown PCR protocol, products were run on a gel, excised, purified using a NucleoSpin Gel and PCR Clean-up kit (Clontech), and sequenced. Candidates were considered germline if found in both the tumor and normal sample (Supplementary Fig. 6).
Copy number variations (CNVs) were calculated with ExomeCNV15, a statistical method to detect somatic copy number variation using depth-of-coverage from mapped short sequence reads. A subset was also run on AffymetrixGenomeWide SNP 6.0 arrays (Affymetrix) as confirmation. Analysis was performed with the aroma.affymetrix package87 and segmentation was performed using the CBS algorithm in the DNAcopy R package.88 For quality control, the subset of samples genotyped on HumanOmni 2.5–8v1 BeadChips (as part of whole genome sequencing) were also analyzed for CNVs by using tQN89 for normalization, followed by OncoSNP v1.2 for characterization of CNVs and loss-of-heterozygosity (LOH) events.90 Copy number clustering was performed on segmented copy number data. A unified breakpoint profile (region by sample matrix) was derived by combining breakpoints across all samples and determining the minimal common regions of change. Unified breakpoint profiles were computed using the Bioconductor package CNTools.91 Hierarchical clustering was done using the R function hclust, with manhattan distance and Ward’s agglomeration method. Gene level copy number changes across samples were identified using GISTIC2.0 (Genomic Identification of Significant Targets in Cancer)16 (Supplementary Table 6).
Gene expression profiling
Approximately 300ng of RNA was extracted from fresh frozen tissue, and RNA integrity was assessed using the Eukaryote Total RNA Nano Assay of the Agilent 2100 Bioanalyzer (Agilent Systems). Expression profiling of viable RNA was performed using the Human HT-12 Expression BeadChip array (Illumina). Analysis was performed with Partek Genomics Suite v6.5 (Partek).
Fluorescence In Situ Hybridization (FISH)
Tissue microarrays were constructed for tumors with available formalin-fixed, paraffin-embedded (FFPE) tissue. Unstained frozen slides were generated for all other tumors. FISH was performed using a 3-color probe mix consisting of BAC clones for 5’ MYB (RP11–614H6, RP11–104D9) (Green), 3’ MYB (RP11–323N12, RP11–1060C14) (Orange), and 3’ NFIB (RP11–413D24, RP11–589C16) (Red) (BACPAC Resources and Wellcome Trust Sanger Institute92) (Supplementary Fig. 3). DNA was prepared by standard alkaline lysis, and labeled by nick translation with fluorochrome-conjugated dUTPs (Enzo Life Sciences). Paraffin and frozen slide sections were processed as previously described.93
Driver/Passenger and pathway analysis
Driver/Passenger analysis was performed with CHASM v1.0.5 (Cancer-specific High-throughput Annotation of Somatic Mutations), a random-forest-based classification algorithm that predicts whether tumor-derived somatic missense mutations are important contributors to cancer cell fitness.14 The default set of 52 features was used with an FDR cutoff of 0.35 (Supplementary Table 4).
Pathway analyses for enrichment were performed using IPA (Ingenuity Pathway Analysis) (Winter Release 2012, Ingenuity Systems), DAVID v6.794 (Database for Annotation, Visualization and Integrated Discovery), and hypergeometric distribution, with potential CHASM drivers used as input. Enriched genesets reflecting Gene Ontology95 biological processes, Kyoto Encyclopedia of Genes and Genomes (Release 64.0), and Biocarta pathways were corrected for multiple hypothesis testing using the Bonferroni method. Aberrant signaling pathways (Fig. 4) were constructed referencing IPA and Pathway Commons.38 Expression data was analyzed using Gene Set Enrichment Analysis (GSEA) v2.0.10.56 Functional consequences were also evaluated using the MSKCC cBioPortal for Cancer Genomics.96,97
Cell culture
Cell lines were cultured with the following: 293T and COS7 cell lines, DMEM 10% FBS; HSG, DMEM 5% FBS; HSY, DMEM:F12 10% FBS; HTB-41, McCoy 10% FBS; HFF-1, DMEM 15% FBS. All cell lines were obtained from ATCC except for HSG (O. Baker) and HSY cells (R. Wong).
FAT4 knockdown, transfection, and viral transduction
FAT4 knockdown was performed using siRNA or shRNA (Dharmacon). siRNAs were transfected into cells in antibiotic-free medium using Lipofectamine RNAiMAX (Invitrogen), medium was changed after 6h and cells were harvested after 48h. shRNAs were transfected into 293T cells, along with packaging and envelope plasmids. Virus was harvested from cell culture supernatant at 48, 72, and 96h. Cells were transduced with 8 µg/mL polybrene. Two independent RNAi sequences were used for knockdown. Relative knockdown fold change was normalized to respective controls.
KDM6A mutagenesis and overexpression
Flag-tagged KDM6A plasmid was obtained (AddGene, #17438) and site-directed mutagenesis performed with QuikChange II XL (Stratagene). 293T and COS7 cells were transfected with FuGene HD (Promega).
Immunofluorescence
COS7 cells were plated on poly-d-lysine cover slips (BD Biosciences) and transfected with wildtype or mutant FLAG-tagged KDM6A. After a 36h incubation, cells were fixed in 4% paraformaldehyde in PBS, permeabilized in 0.5% Triton X-100 in PBS, and blocked in 10% FBS. Primary antibodies were anti-FLAG (Sigma, F3165; 1:50 dilution) and anti-H3K27me3 (Millipore, 07–449; 1:50 dilution). Secondary antibodies were Alexa Fluor 488 anti-mouse IgG and Alex Fluor 568 anti-rabbit IgG (Molecular Probes; 1:200 dilution). Images were acquired using a SP5 confocal microscope (Leica).
Growth curves
Growth curve assays were performed in triplicate and quantified using the Vi-Cell XR Cell Viability Analyzer (Beckman Coulter) or in real time in quadruplicate with the xCELLigence System (Roche). xCELLigence plates were seeded with 5,000 or 20,000 cells per well, and growth is reported as the cell index, a measure of impedance reflecting viable, adherent cells.
TP53 Signaling Pathway PCR Array
Approximately 200 ng of RNA extracted from fresh frozen tissue was used to make cDNA using SuperScript III (Invitrogen). Gene expression was quantified using the RT2 Profiler PCR Array Human p53 Signaling Pathway (SABioscience). The reactions were performed using a Mastercycler™ ep Realplex 4s and Realplex software (Eppendorf). The 2-(ΔΔCt) method was used to calculate the ΔΔCt values. In all tissue-specific comparisons, individual gene expression levels in tumor specimens were compared with that in normal salivary tissue. Comparisons were made within tissue between TP53-pathway wildtype and TP53-pathway altered tissue. All 84 genes were normalized against the average of two most stably expressed reference genes (GAPDH, HPRT1). Gene expression was considered below the limit of detection if the Ct value was 35.0 or greater.
PI3K immunohistochemistry staining
5µm FFPE tissue sections were stained for pAKT (Cell Signaling Technology; 1:100 dilution) on Benchmark Ultra (Ventana) using the OmniMap DAB anti-Rabbit Detection Kit (Ventana, 760-149) and multimer detection system. 5µm FFPE tissue sections were similarly stained for pPRAS40 (1:40 dilution) using biotinylated secondary antibody (Vector Labs, no. BA-1000; 1:300 dilution) and straptividin detection system (Ventana).
Supplementary Material
Editorial Summary.
Timothy Chan and colleagues report exome and genome sequencing of 60 adenoid cystic carcinoma (ACC) tumor/normal pairs. They identify multiple pathways recurrently disrupted in ACC and provide evidence that KDM6A and PIK3CA are functionally relevant ACC candidate driver genes.
ACKNOWLEDGEMENTS
We thank A. Kayserian and S. Thomas for technical assistance. We are indebted to R.J. Wong, F. Kaye, P.K. Brindle, and O.J. Baker for generously providing cell lines and constructs. We are additionally grateful to J.O. Boyle, S.G. Patel, A.R. Shaha, R.J. Wong, M.C. Mariano, and L. Lopez for facilitating tissue procurement. This grant was supported by the National Institutes of Health (RO1CA154767 (T.A.C.), R21DE023229 (T.A.C.), 5T32CA009685 (A.S.H.)), the Geoffrey Beene Foundation (T.A.C.), the STARR Cancer Consortium (T.A.C.), and the Louis Gerstner Foundation (T.A.C.). A.S.H. was supported by the American Head and Neck Society/American Academy of Otolaryngology-Head and Neck Surgery Foundation Young Investigator Combined Award. D.M.R. was supported by the Howard Hughes Medical Institute Medical Research Fellows Program. T.A.C. would like to thank Jeff and Marnie Kaufman and the Adenoid Cystic Carcinoma Research Foundation for support of the work and inspiration to see it through for the benefit of patients.
Footnotes
ACCESSION NUMBERS
Sequencing and genotype data have been deposited into the NCBI database of Genotypes and Phenotypes (dbGaP) under accession number phs000612.v1.p1.
AUTHOR CONTRIBUTIONS
T.A.C., A.S.H., K.K., and W.L. designed the experiments. D.M.R., A.S.H., S.E., A.V., A.H., K.H., D.R., L.A.W., J.T.H., J.Z., N.G.I., Y.G., M.A.L., and C.E.R. performed the experiments. A.S.H., W.L., T.A.C., K.K., D.M.R., L.G.T.M., and S.T. analyzed the data. N.K. and R.R.S. provided histopathologic confirmation of sample purity. A.S.H., T.A.C., and W.L. wrote the manuscript. A.S.H., I.G., L.G.T.M., M.D., J.P.S., B.S., N.G.I., C.R.L., E.B., R.L.F., R.R.S., S.C., and S.K. contributed new reagents and provided tissues for analysis. W.L., K.K., A.S.H., T.A.C., L.P., B.J.R., I.D., Y.L., R.S., N.S., B.E.G., and C.S. conducted the bioinformatics analysis.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Adelstein DJ, Koyfman SA, El-Naggar AK, Hanna EY. Biology and management of salivary gland cancers. Semin Radiat Oncol. 2012;22:245–253. doi: 10.1016/j.semradonc.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Agulnik M, et al. Phase II study of lapatinib in recurrent or metastatic epidermal growth factor receptor and/or erbB2 expressing adenoid cystic carcinoma and non adenoid cystic carcinoma malignant tumors of the salivary glands. J Clin Oncol. 2007;25:3978–3984. doi: 10.1200/JCO.2007.11.8612. [DOI] [PubMed] [Google Scholar]
- 3.Holst VA, Marshall CE, Moskaluk CA, Frierson HF., Jr KIT protein expression and analysis of c-kit gene mutation in adenoid cystic carcinoma. Mod Pathol. 1999;12:956–960. [PubMed] [Google Scholar]
- 4.Laurie SA, Ho AL, Fury MG, Sherman E, Pfister DG. Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: a systematic review. Lancet Oncol. 2011;12:815–824. doi: 10.1016/S1470-2045(10)70245-X. [DOI] [PubMed] [Google Scholar]
- 5.Persson M, et al. Clinically significant copy number alterations and complex rearrangements of MYB and NFIB in head and neck adenoid cystic carcinoma. Genes Chromosomes Cancer. 2012;51:805–817. doi: 10.1002/gcc.21965. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research, N. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sathirapongsasuti JF, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vekony H, et al. DNA copy number gains at loci of growth factors and their receptors in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2007;13:3133–3139. doi: 10.1158/1078-0432.CCR-06-2555. [DOI] [PubMed] [Google Scholar]
- 18.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe AP. Chromatin remodeling: why it is important in cancer. Oncogene. 2001;20:2988–2990. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 20.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Houdt JK, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–449. doi: 10.1038/ng.1105. S1. [DOI] [PubMed] [Google Scholar]
- 23.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 24.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masse JE, et al. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 29.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spangle JM, Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol. 2010;84:9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer NG, et al. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc Natl Acad Sci U S A. 2004;101:7386–7391. doi: 10.1073/pnas.0401002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 36.Fenaux P, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tien AL, et al. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435:175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egloff AM, Grandis JR. Molecular pathways: context-dependent approaches to notch targeting as cancer therapy. Clin Cancer Res. 2012;18:5188–5195. doi: 10.1158/1078-0432.CCR-11-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, et al. Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci U S A. 2011;108:13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdul M, Ramlal S, Hoosein N. Ryanodine receptor expression correlates with tumor grade in breast cancer. Pathol Oncol Res. 2008;14:157–160. doi: 10.1007/s12253-008-9045-9. [DOI] [PubMed] [Google Scholar]
- 43.Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- 44.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 45.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Essers MA, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dey JH, et al. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 48.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang RB, et al. A novel interleukin-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J Biol Chem. 2003;278:33232–33238. doi: 10.1074/jbc.M305022200. [DOI] [PubMed] [Google Scholar]
- 50.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 51.Tan Y, Sangfelt O, Spruck C. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett. 2008;271:1–12. doi: 10.1016/j.canlet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 52.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 53.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 2011;71:6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarmento LM, Barata JT. Therapeutic potential of Notch inhibition in T-cell acute lymphoblastic leukemia: rationale, caveats and promises. Expert Rev Anticancer Ther. 2011;11:1403–1415. doi: 10.1586/era.11.73. [DOI] [PubMed] [Google Scholar]
- 58.Iozzo RV, Murdoch AD. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 1996;10:598–614. [PubMed] [Google Scholar]
- 59.Tran PK, et al. Increased intimal hyperplasia and smooth muscle cell proliferation in transgenic mice with heparan sulfate-deficient perlecan. Circ Res. 2004;94:550–558. doi: 10.1161/01.RES.0000117772.86853.34. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Z, et al. Impaired angiogenesis, delayed wound healing and retarded tumor growth in perlecan heparan sulfate-deficient mice. Cancer Res. 2004;64:4699–4702. doi: 10.1158/0008-5472.CAN-04-0810. [DOI] [PubMed] [Google Scholar]
- 61.Savore C, et al. Perlecan knockdown in metastatic prostate cancer cells reduces heparin-binding growth factor responses in vitro and tumor growth in vivo. Clin Exp Metastasis. 2005;22:377–390. doi: 10.1007/s10585-005-2339-3. [DOI] [PubMed] [Google Scholar]
- 62.Munakata R, Irie T, Cheng J, Nakajima T, Saku T. Pseudocyst formation by adenoid cystic carcinoma cells in collagen gel culture and in SCID mice. J Oral Pathol Med. 1996;25:441–448. doi: 10.1111/j.1600-0714.1996.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 63.Cheng J, Saku T, Okabe H, Furthmayr H. Basement membranes in adenoid cystic carcinoma. An immunohistochemical study. Cancer. 1992;69:2631–2640. doi: 10.1002/1097-0142(19920601)69:11<2631::aid-cncr2820691103>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 64.Kimura S, et al. Perlecan (heparan sulfate proteoglycan) gene expression reflected in the characteristic histological architecture of salivary adenoid cystic carcinoma. Virchows Arch. 2000;437:122–128. doi: 10.1007/s004280000209. [DOI] [PubMed] [Google Scholar]
- 65.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Binmadi NO, et al. Plexin-B1 and semaphorin 4D cooperate to promote perineural invasion in a RhoA/ROK-dependent manner. Am J Pathol. 2012;180:1232–1242. doi: 10.1016/j.ajpath.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Rodrigues S, De Wever O, Bruyneel E, Rooney RJ, Gespach C. Opposing roles of netrin-1 and the dependence receptor DCC in cancer cell invasion, tumor growth and metastasis. Oncogene. 2007;26:5615–5625. doi: 10.1038/sj.onc.1210347. [DOI] [PubMed] [Google Scholar]
- 70.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 71.Phuchareon J, Ohta Y, Woo JM, Eisele DW, Tetsu O. Genetic profiling reveals cross-contamination and misidentification of 6 adenoid cystic carcinoma cell lines: ACC2, ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One. 2009;4:e6040. doi: 10.1371/journal.pone.0006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Retraction notice to "Nef from SIVmac239 decreases proliferation and migration of adenoid-cystic carcinoma cells and inhibits angiogenesis" [OO 47 (2011) 847–854] Oral Oncol. 2012;48:95. doi: 10.1016/j.oraloncology.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 73.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013 doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Genomes Project C. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rozen SS HJi. Primer3 on the WWW for general users and for biologist programmers. Totowa, NJ: Humana Press; 2000. [DOI] [PubMed] [Google Scholar]
- 87.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 88.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 89.Staaf J, et al. Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios. BMC Bioinformatics. 2008;9:409. doi: 10.1186/1471-2105-9-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yau C, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol. 2010;11:R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fiegler H, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 93.Gopalan A, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gao J, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004088. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.