Abstract
Little is known about oxalate transport in renal epithelia under basal conditions, let alone in hyperoxaluria when the capacity for renal oxalate excretion is increased. Sulfate anion transporter 1 (SAT1, Slc26a1) is considered to be a major basolateral anion-oxalate exchanger in the proximal tubule and we hypothesized its expression may correlate with urinary oxalate excretion. We quantified changes in the renal expression of SAT1 mRNA and protein in two rat models, one with hyperoxaluria (HYP) and one with renal insufficiency (HRF) induced by hyperoxaluria. The hyperoxaluria observed in the HYP group could not simply be ascribed to changes in SAT1 mRNA or protein abundance. However, when hyperoxaluria was accompanied by renal insufficiency, significant reductions in SAT1 mRNA and protein were detected in medullary and papillary tissue. Together, the results indicate that transcriptional modulation of the SAT1 gene is not a significant component of the hyperoxaluria observed in these rat models.
Keywords: Calcium oxalate, Chronic renal failure, Urolithiasis, Kidney stone
Introduction
Hyperoxaluria is a predisposing factor in the etiology of calcium oxalate kidney stone disease with some 30 % of the US stone-forming population exhibiting this clinical condition [1]. Hyperoxaluria may be a consequence of increased hepatic production of oxalate, as in primary hyperoxaluria [2], or from enhanced enteric absorption [3]. Since oxalate is not metabolized by mammals, an increased load presents a continuous challenge to both renal and enteric transport mechanisms involved in its excretion. Microperfusion studies in rats [4] and rabbits [5] indicate that there is a net secretion of oxalate in the proximal tubule under normal conditions, suggestive of active, transepithelial oxalate transport from the blood to the lumen in this nephron segment. In experimental hyperoxaluria in humans [6] and rats [7], the renal clearance of oxalate is enhanced relative to creatinine clearance, which further indicates a significant capacity for tubular oxalate secretion. Given that there is bidirectional tubular transport of oxalate [8–10], elevated excretion of oxalate in experimental hyperoxaluria could result from increased tubular oxalate secretion and/or reduced tubular oxalate reabsorption.
Transcellular secretion of oxalate in the proximal tubule must involve a basolateral oxalate uptake mechanism in series with an oxalate efflux pathway in the apical membrane. While there may be several avenues for apical oxalate exchange [6, 11], there is compelling evidence suggesting that SAT1 (slc26a1), a SO4–oxalate exchanger, is an important component in basolateral oxalate uptake [8, 11–13] that may contribute to the secretory flux of oxalate into the renal tubule. SAT1 protein has been localized to the basolateral membrane in rat proximal tubule [14] and, when heterologously expressed, it exhibits robust SO4–Ox exchanger functionality [14–16]. Furthermore, slc26a1 knockout mice are reported to exhibit hyperoxaluria and hyperoxalemia [13], suggesting that SAT1 plays a role in oxalate homeostasis mediated by renal and/or intestinal epithelia. Based on these observations, we speculated that enhanced renal oxalate excretion in hyperoxaluric rats could be mediated, in part, by increases in the renal abundance of SAT1. To test this hypothesis, we employed two rat models with different degrees of hyperoxaluria promoted by ethylene glycol ingestion [7, 17]: normal rats and nephrectomized (unilateral) rats, together with their respective controls.
SAT1 protein (immunoblots) and mRNA (real-time PCR) abundance were determined in the renal cortex, medulla, and papilla in each group. First, we report here that SAT1 mRNA and protein are present in all regions of the normal rat kidney (cortex > medulla » papilla). Second, the renal cortical, medullary, and papillary expression of SAT1 mRNA and protein in hyperoxaluric rats with two functioning kidneys does not significantly differ from the expression patterns observed in normal untreated rats. Third, when hyperoxaluria induced an impairment in renal function, significant reductions in SAT1 mRNA and protein abundance were apparent in the medullary and papillary regions. These results suggest that increased urinary excretion of oxalate in these animal models is not dependent on the abundance of SAT1 in renal tubular epithelia.
Materials and methods
Animals
Male Sprague–Dawley rats (275–300 g) were utilized in the current studies and all rats had free access to Purina Rat Chow 5001 during the entire course of the study. Experimental protocols were conducted in accordance with the guidelines of the University of Florida Institutional Animal Care and Use Committee and the NIH Guide for the Care and Use of Laboratory Animals. Details regarding the rat models and urinary and serum oxalate and creatinine determinations have been reported previously [7, 17]. Experimental groups for the gene and protein expression studies included control rats (CON); rats provided free access to drinking water containing 0.75 % (vol/vol) ethylene glycol for a period of 4 weeks in order to induce hyperoxaluria (HYP); unilaterally nephrectomized control rats (UNI); and, rats with hyperoxaluria-induced renal failure caused by providing unilateral nephrectomized rats free access to drinking water that contained 0.75 % (vol/ vol) ethylene glycol for a period of 4 weeks (HRF). An additional six untreated control rats of comparable age and weight were used to characterize the expression of SAT1 and GAPDH mRNA in the various regions of the kidney.
RNA isolation
Kidneys were removed, washed in several volumes of icecold saline, decapsulated and stored at 4 °C in 5 volumes of RNA later (Ambion; Austin, TX). Each kidney was then cut longitudinally slightly off-center of the sagittal plane. The papilla was removed and the adjacent medullary tissue was excised with scissors. A central core (≈3–5 m0m3)of medullary tissue and cortical tissue from the outermost 3 mm of the kidney perimeter was utilized for RNA isolation.
Total RNA was isolated from the entire papillary tip (≈35–50 mg) or 100 mg of cortex and medulla were homogenized in 1 ml of TRIzol (Invitrogen, Carlsbad, CA) with three 15-s bursts of a polytron. RNA was extracted with chloroform and separated from the DNA and proteins by centrifugation at 12,000×g for 15 min. The aqueous phase was precipitated with isopropanol and the resulting RNA pellet was washed with 75 % ethanol. Molecular grade glycogen (Ambion; Austin, TX, USA) was added as a carrier prior to precipitation of the papillary RNA to ensure quantitative recovery. The RNA was resuspended in sterile water and quantified by measuring the absorbance at 260 and 280 nm (A260/A280 ratios between 1.8 and 2.0 were considered acceptable). Residual genomic DNA contamination was removed by incubating the RNA with 2 U of TURBO DNase™ (Ambion; Austin, TX, USA) for 1 h at 37 °C. DNase was removed from the RNA samples by addition of 0.1 volume of DNase Inactivation Reagent (Ambion; Austin, TX, USA), incubation at room temperature for 2 min and centrifugation at 10,000× g for 1 min to pellet the DNase inactivation reagent. DNA-free total RNA was quantified as described above and stored at –80 °C.
Oligonucleotide design
Oligonucleotide primers were designed with Primer3 [18] from NCBI reference nucleotide sequences for SAT1 (NM_022287), 18S rRNA (×01117), β-actin (NM_031144), and GAPDH (NM_017008). Each primer was then compared to all available sequences in the GenBank database using the BLAST feature of NCBI to ensure specificity. Resultant sense and antisense primers for SAT1 were 5′-GGCTTTATCAGGTCCTCATGG-3′ and 5′-GATTAG CCTGTCCCACGTTC-3′ corresponding to positions 945–965 and 1,160-1,141, respectively, in the reference sequence, and producing a PCR product of 216 nucleotides. Sense and antisense primers for GAPDH were 5′-TCCCT CAAGATTGTCAGCAA-3′ and 5′-AGATCCACAACGG ATACATT-3′ corresponding to positions 1,270–1,289 and 1,577–1,558, respectively, and producing a PCR product of 308 nucleotides. Sense and antisense primers for 18S rRNA were 5′-CTTTGGTCGCTCGCTCCTC-3′ and 5′-CT GACCGGGTTGGTTTTGAT-3′ corresponding to positions 118–136 and 248–229, respectively, and producing a PCR product of 131 nucleotides. Sense and antisense primers for β-actin were 5′-AGCCATCCAGGCTGTGTT GTCC-3′ and 5′-CTCTCAGCTGTGGTGGTGAA-3′ corresponding to positions 483–504 and 698–679, respectively, and producing a PCR product of 216 nucleotides. All of these primer sets produced amplicons of the expected size, as judged by ethidium bromide staining following agarose gel electrophoresis.
Real-time PCR
Real-time PCR was performed using the QuantiTect™ SYBR®Green RT-PCR kit (Qiagen; Valencia, CA, USA) with the DNA Engine Opticon Continuous Fluorescence Detection System (MJ Research, Inc.; San Francisco, CA, USA). Briefly, total RNA (7.8–500 ng) was added to 25 µl of 2× QuantiTect SYBR Green RT-PCR Master Mix, 0.5 µM of each gene-specific primer and 0.5 µl of Quani-Tect RT mix. The reactions were adjusted to 50 µl total volume with RNase-free water and incubated at 50 °C for 30 min. Reverse transcriptases were deactivated and the HotStarTaq DNA polymerase was activated by incubation at 95 °C for 15 min. Reactions were then subjected to 45 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. Fluorescence data were collected at each extension step. After each PCR run, a melting curve analysis was performed using the Opticon MONITOR software of the DNA Engine Opticon™ System to verify the specificity of the RT-PCR product. In addition, all PCR runs also included the appropriate controls, including no template controls to enable detection of contamination and no reverse transcriptase controls to test for contaminating genomic DNA. The Opticon MONI-TOR™ software was used to calculate the critical threshold value (CT) for each sample and the abundance of each target gene was analyzed using the comparative method [19].
Initially, we compared the expression levels of the three most common reference genes (β-actin, 18S rRNA, and GAPDH) in cortical, medullary, and papillary tissue obtained from control rats using real-time PCR. Expression levels of β-actin and 18S rRNA were higher and lower, respectively, in the medulla compared to the cortex and papilla, while GAPDH levels did not significantly differ among the three sites (data not shown). Consequently, GAPDH was used to normalize the real-time PCR data for the characterization of the spatial distribution of SAT1 in the control rat kidney. GAPDH was further validated to ensure that (1) control gene expression was unaffected by experimental treatment or tissue being examined and (2) that amplification efficiency of the target and reference gene was approximately equal, an assumption for the validity of the comparative method [19].
SAT1 immunoblots
Immunodetection of slc26a1 transport protein on Western blots was performed using a primary antibody to SAT1 (Dr. L. Karniski, University of Iowa). The membranes were incubated for 1 h in 5 % blocking solution (w/v) containing non-fat dry milk and Tris-buffered saline with 0.1 % Tween-20 and then incubated overnight at 4 °C. This was followed by a 30-min incubation with a commercial secondary sheep anti-mouse antibody (Amersham Biosciences; Newark, NJ, USA). Following each incubation period with primary or secondary antibodies, the blots were extensively washed with Tris-buffered saline with 0.1 % Tween-20. For detection, the membranes were reacted for 1 min with chemiluminescence reagent (Amersham, Piscataway, NJ, USA) and exposed to autoradiographic hyperfilm-ECL (Amersham). The intensity of the resulting singular band at ~ 70 kDa was quantified using Image J (NIH). Membranes were re-probed with a monoclonal antibody to GAPDH (Ambion; Austin, TX, USA), to ensure equal loading of the total protein used.
Statistical analyses
Linear regression analysis was performed on the CT values generated from the three serially diluted RNA pools for each gene using SigmaStat (SysStat Software, Point Richmond, CA, USA). Amplification efficiencies for a target gene were compared among the three sites by testing for homogeneity of regression [20]. Amplification efficiencies between target and reference amplicons were compared by calculating the ΔCT (CT,target–CT,reference) for each point of the serial dilution and then fitting the ΔCT versus log mRNA quantity to a linear equation using least-squares linear regression [19]. Efficiencies were considered equivalent when the resultant P value for regression exceeded 0.05. The effect of experimental treatment on housekeeping gene expression was determined by analysis of variance on the rank-transformed CT values. Treatment sums of squares were partitioned into main effects of nephrectomy and hyperoxaluria via orthogonal contrasts using the GLM procedures of the statistical analysis system [21]. Since the main effect of nephrectomy was significant in all sites tested for the reference gene, all subsequent treatment effects were tested against, and expressed relative to, the appropriate control, i.e., CON for HYP (Fig. 2) and UNI for HRF (Fig. 3). Differences in mRNA abundance between CON and HYP or between UNI and HRF groups were tested independently for each site by comparing the CT values after normalizing to the geometric mean of the internal reference gene with Student’s t test [22]. SAT1-to-GAPDH protein ratios were tested for differences between treatments with the Mann–Whitney rank sum test.
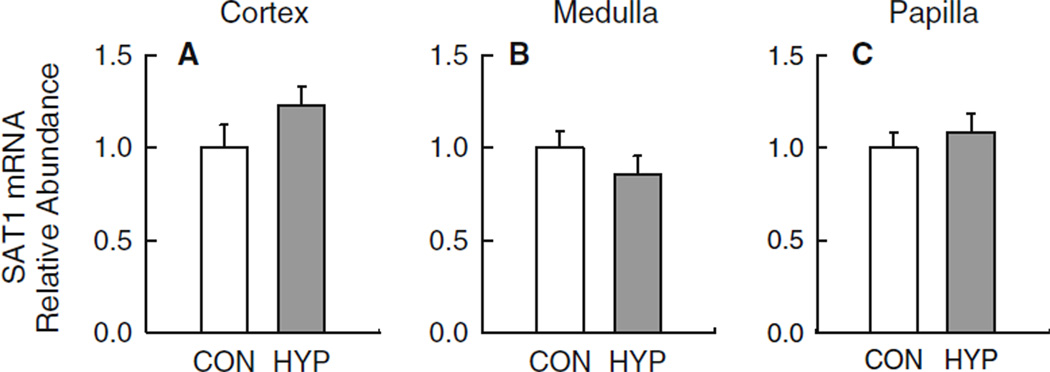
Fig. 2.
Hyperoxaluria alone (HYP) has a minor impact on the mRNA expression levels for Slc26a1 in three regions of the kidney. Experimental groups included normal control rats (CON; n = 11) and hyperoxaluric rats (HYP; n = 11). Quantitative real-time PCR was performed with primers specific to SAT1 using 100 ng total RNA isolated from the renal cortex (a), medulla (b) and papilla (c) of each rat as detailed in ‘‘Materials and methods’’. Critical threshold (CT) values obtained for each sample were normalized to the geometric mean of the internal reference gene GAPDH [22]. Data are presented relative to CON rats. No difference in mRNA abundance was detected for any region examined p > 0.10
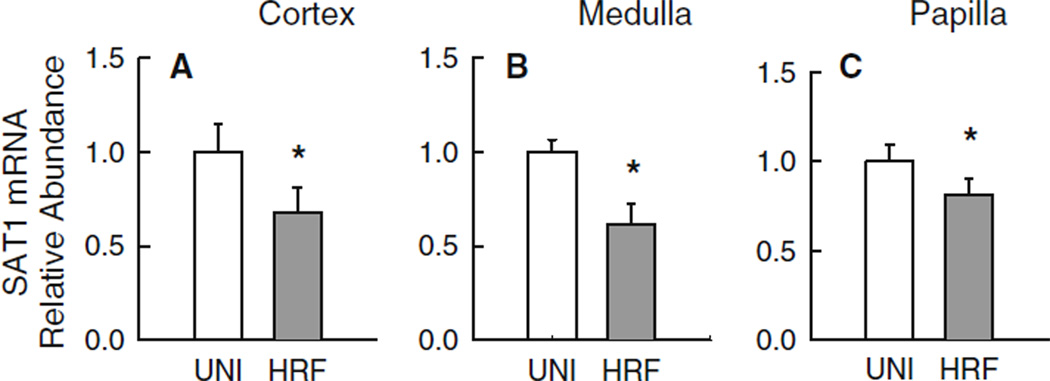
Fig. 3.
Hyperoxaluria in the context of renal insufficiency (HRF) is accompanied by significant reductions in SAT1 mRNA abundance in medullary and papillary tissues. Experimental groups included unilateral-nephrectomized rats (UNI; n = 11) and rats in chronic renal failure induced by hyperoxaluria (HRF; n = 11). Real-time PCR was performed with primers specific to SAT1 using 100 ng total RNA isolated from the renal cortex (a), medulla (b) and papilla (c) of each rat as detailed in ‘‘Materials and Methods’’. Critical threshold (CT) values obtained for each sample were normalized to the geometric mean of the internal reference gene GAPDH [22]. Data are presented relative to UNI rats. Differences in abundance are denoted by an asterisk, p < 0.05
Results
Rat models of hyperoxaluria
As shown in Table 1, both HYP and HRF rat models exhibit significant hyperoxalemia and hyperoxaluria compared to CON and UNI rats. The HRF rats have compromised renal function as evidenced by a two-fold increase in serum creatinine and a significant reduction in creatinine clearance compared to HYP, CON, and UNI. The renal clearance of oxalate is markedly elevated in both HYP and HRF models compared to their respective controls CON and UNI. Oxalate clearance is comparatively lower in HRF than HYP because of compromised renal function. The oxalate/creatinine ratios ranged from about 0.3 in CON and UNI to values >1 in HYP and HRF indicating varying degrees of net renal oxalate secretion in the latter. The general findings presented in Table 1 confirm previous characterizations of these rat models [7, 17, 23].
Table 1.
A comparison of serum concentrations and renal handling of oxalate (Ox) and creatinine (Cr) in CON, UNI, HYP and HRF rats
| Group | Serum Ox (µmol/L) | Ox excretion (µmol/24 h) | Ox clearance (ml/min) | Serum Cr (µmol/L) | Cr clearance (ml/min) |
|---|---|---|---|---|---|
| CON | 9.2±0.9 | 8.9 ± 0.4 | 0.69 ± 0.12 | 48.9 ± 1.4 | 2.40 ± 0.09 |
| HYP | 25.9 ± 5.4* | 54.0 ± 8.9* | 2.25 ± 0.25* | 51.5 ± 1.5 | 2.18 ± 0.14 |
| UNI | 12.3±0.8 | 10.8 ± 0.6 | 0.65 ± 0.06 | 39.5 ± 5.4 | 2.90 ± 0.13 |
| HRF | 45.7 ± 4.9*,† | 90.9 ± 6.1*,† | 1.42 ± 0.19*† | 109.0 ± 5.9*,† | 1.00 ± 0.15*,† |
Results are presented as the mean ± SEM for n = 8–11 rats per treatment group
Significant difference from CON, p ≤ 0.05
Significant difference between HRF and HYP p ≤ 0.05
Validation of real-time PCR amplification efficiencies
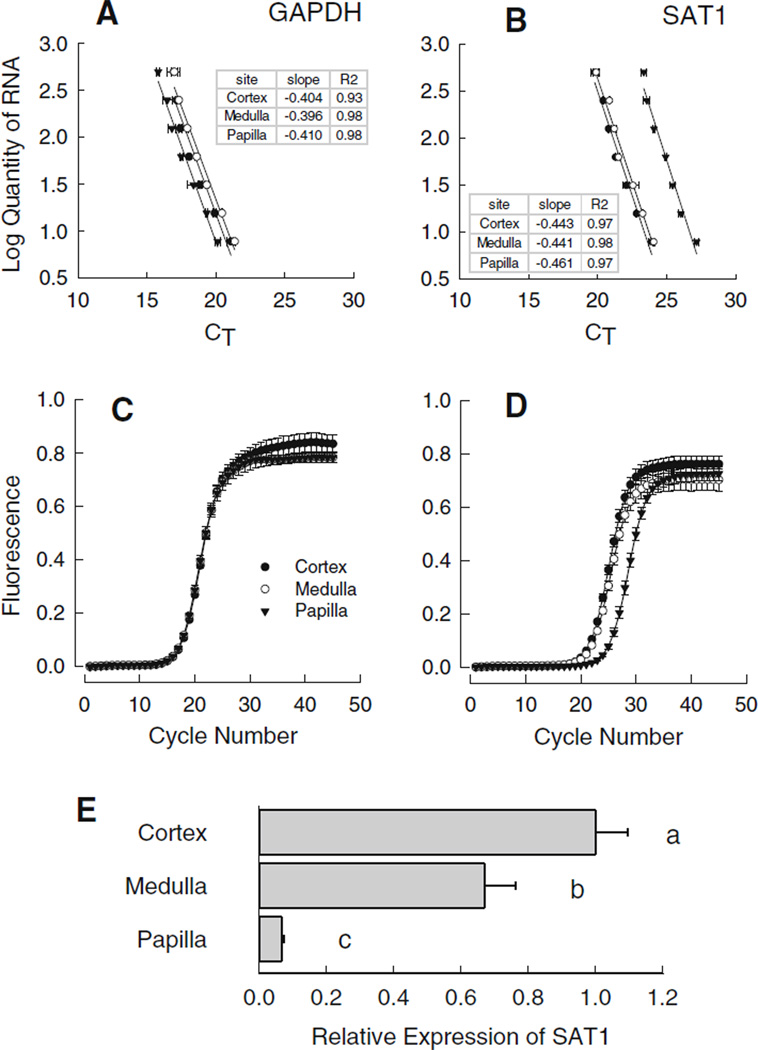
As we were interested in comparing gene expression in three different regions of the rat kidney, we first tested whether the amplification efficiencies for target and reference gene transcripts were similar among template RNAs isolated from the cortex, medulla and papilla. Two-fold serial dilutions (500 ng–7.8 ng) of each renal RNA pool were used to generate standard curves for the reference gene, GAPDH (Fig. 1a) and the target gene, SAT1 (Fig. 1b). Plots were fit to a linear equation by regression analysis and slopes were tested for homogeneity [20]. Slopes and coefficients of determination obtained for the cortex, medulla and papilla for each gene are provided in the inset of the appropriate panel. No significant differences among the slopes for the three sites were detected for SAT1 or GAPDH; hence amplification efficiencies are comparable among the three renal sites.
Fig. 1.
Analysis of slc26a1 gene expression in rat kidney. a, b Validate the comparative method used in the analysis of gene expression with real-time PCR for slc26a1. Equal masses of total RNA isolated from the renal cortex (filled circles), medulla (open circles), and papilla (filled triangle) of six normal control rats were combined to produce RNA pools for each segment. Real-time PCR amplifications were performed with gene-specific primers and standard curves were constructed from twofold serial dilutions (500 ng–7.8 ng) of each renal RNA pool. Each point represents the mean ± SEM of duplicate wells from two independent real-time assays. Plots were fit to a linear equation by regression analysis and the resultant slopes (insets) were tested for homogeneity [20] to ensure the amplification efficiencies were similar. No significant differences (p > 0.05) among the slopes for the three sites were detected for either GAPDH (a) or SAT1 (b). Panels c (GAPDH) and d (SAT1) represent the mean amplification profiles ± SEM generated by real-time PCR analysis for cortex (filled circles), medulla (open circles), and papilla (filled triangle) obtained from normal control rats (n = 6). In panel e, the abundance SAT1 gene expressed is presented relative to the cortex as computed from the CT value obtained for each individual sample and corrected for GAPDH by the comparative method [19]. Bars without a common superscript differ p < 0.05
Regional distribution of slc26A1 in the normal rat kidney
As an initial step in characterizing the renal expression of SAT1, the regional distribution in terms of mRNA abundance in three defined regions of the normal rat kidney was quantified by real-time PCR. Figure 1c and d, respectively, depict the mean amplification profiles ± SEM for GAPDH and SAT1 generated by real-time PCR analysis for cortex, medulla, and papillary tissues from normal control rats. From these data, we calculated the expression of SAT1 relative to GAPDH as shown in Fig. 1e. SAT1 mRNA abundance was highest in the renal cortex with levels approximately tenfold higher than in the papilla, while medullary expression of SAT1 mRNA was intermediate relative to the other two regions.
SAT1 mRNA and protein in hyperoxaluria
Patterns of mRNA expression for SAT1 in HYP rats did not differ from CON rats irrespective of the renal site examined (Fig. 2a–c). These results would suggest that a significant modulation of the slc26a1 gene does not cause hyperoxaluria or result from chronic hyperoxaluria. In contrast, however, SAT1 does appear to be significantly modulated in the various regions of the rat kidney when the hyperoxaluria is associated with renal insufficiency. Thus, renal expression of SAT1 mRNA in HRF rats was significantly lower than in UNI rats in cortex (Fig. 3a), medulla (Fig. 3b) and papilla (Fig. 3c).
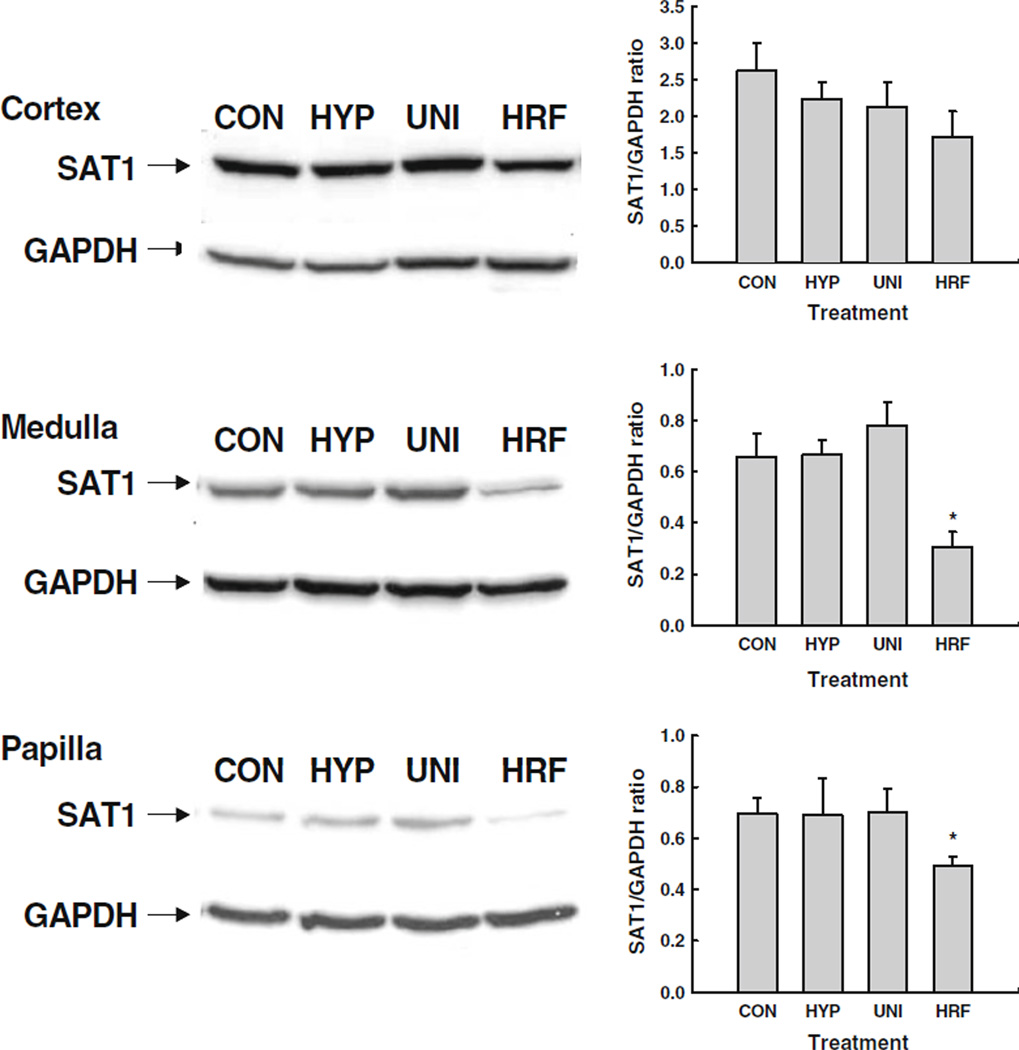
To establish whether the SAT1 protein expression profiles paralleled those observed for the mRNA, Western blot analysis was performed using protein isolated from each renal segment for a subset (n = 4) of the rats in each experimental group. Levels of SAT1 protein were similar among CON, HYP and UNI rats in the renal cortex (Fig. 4a), medulla (Fig. 4b) and papilla (Fig. 4c). In contrast, SAT1 protein abundance was significantly reduced in the medulla and papilla of HRF rats. A similar trend was observed for cortical expression of SAT1 in HRF rats, but this reduction did not achieve statistical significance. Nevertheless, the concomitant reduction in SAT1 protein levels does ascribe functional significance to the somewhat modest decreases (20–40 %) in mRNA abundance depicted in Fig. 3.
Fig. 4.
SAT1 protein abundance in cortical (a), medullary (b) and papillary (c) regions of the kidney in control rats (CON), hyperoxaluric rats (HYP), unilateral-nephrectomized rats (UNI) and rats in chronic renal failure induced by hyperoxaluria (HRF). On the left of each panel is a representative Western blot for renal tissue collected from a rat in each experimental group. On the right is the average SAT1 protein abundance after GAPDH correction for n = 4 rats per experimental group. Note that SAT1 abundance in the medulla and papilla was significantly reduced in HRF rats compared to CON and UNI rats. *p<0.05
Discussion
Sodium-independent, stilbene-sensitive, sulfate–oxalate exchange has been observed in proximal tubule basolateral membrane vesicle preparations from the mammalian kidney [8, 24]. It has been argued that this basolateral sulfate–oxalate activity provides for the final cellular exit step in tubular sulfate reabsorption and the initial cellular uptake step in transepithelial oxalate secretion [14, 16, 24]. The sulfate–oxalate exchange process (or part thereof) has been shown to be mediated by a renal SAT1 protein [14] which is targeted to the basolateral membrane in rat proximal tubule [14] and in renal cell lines (OK and LLC-PK1) [25]. When SAT1 mRNA is expressed heterologously, many of the resultant transport characteristics resemble the sulfate–oxalate exchange process observed in basolateral membrane vesicles [14, 16].
Given its potential role in tubular oxalate handling, we hypothesized that SAT1 expression may be altered in hyperoxaluric conditions to affect an increase in tubular oxalate secretion; an adaptive response similar to that reported for NaSi (SLC13a1) in rat renal cortex following dietary sulfate changes [26, 27]. That a single SLC26 gene product can influence oxalate homeostasis has been clearly demonstrated in slc26a6 null mice wherein intestinal oxalate absorption is greatly enhanced in the absence of this oxalate exchanger [28]. Using two rat models with differing severity of hyperoxaluria,we observed that (1) hyperoxaluria, per se, is not associated with altered renal expression of the SAT1 anion transporter; (2) increased urinary excretion of oxalate in rats with renal insufficiency induced by hyperoxaluria is accompanied by significant reduction in the expression of the SAT1 mRNA and its protein product in the medulla and papilla; and (3) there is a significant distribution of SAT1 in extra-cortical regions of the kidney.
SAT1 in hyperoxaluria
Although SAT1 may have a constitutive role in basolateral oxalate exchange, and hence, in renal tubular oxalate secretion, hyperoxaluria alone (HYP) was not associated with any significant changes in SAT1 mRNA or protein abundance in any of the renal segments examined. Hyperoxaluria in the context of renal insufficiency (HRF) was, however, associated with a significant decrease in both SAT1 mRNA and protein in medullary and papillary tissues. While these findings do not preclude a role for SAT1 in the urinary excretion of oxalate, they clearly demonstrate that the potential role of SAT1 in contributing to hyperoxaluria in rats is more complex than simple changes in mRNA and protein abundance. Although little is known about the regulation of SAT1 and oxalate transport in the kidney, it was recently reported that an ethylene glycol metabolite and oxalate precursor, glyoxylate, can induce an up-regulation of SAT1 mRNA in HepG2 cells [29]. Glyoxylate was also identified as a substrate of SAT1 in the same study. While we did not measure urinary glyoxylate excretion in the present study, we presume the excretion of this ethylene glycol metabolite was elevated and apparently did not correlate with an upregulation of SAT1 mRNA in renal tissue.
The decrease in SAT1 message and protein abundance observed in the medullary and papillary tissues of HYP rats is in agreement with earlier findings in a different rat model of chronic renal failure (CRF) produced by 4/5 nephrectomy [30]. This observation suggests that factors other than hyperoxaluria/hyperoxalemia may underlie the reduction in SAT1 observed in the HRF rats used in the present study. In this regard, results from another study we conducted [31] indicated that experimental hyperoxaluria induced by ethylene glycol causes enzymuria suggesting a degree of renal injury. Thus, it is possible that the reduced SAT1 expression in HRF is a consequence of renal damage. We have also previously reported [17] that there is a nascent metabolic acidosis in the HRF model which is also apparent in CRF [30] hence it is also possible that the changes in SAT1 abundance correlate with the acid–base status of the animal. In Xenopus oocytes, the rat liver isoform of SAT1 has been shown to mediate robust SO4–HCO3 exchange [11, 32] in addition to its SO4–Ox exchanger activity. A variety of other transport mechanisms, independent of SAT1 abundance, may also be involved in enhanced urinary oxalate excretion in hyperoxaluria. These include increases in SAT1 functional activity not revealed by total mRNA and protein abundance determined here; a reduction in tubular oxalate reabsorption, (since there is bidirectional transport in the proximal tubule [8, 24]); or by changes in tubular secretion mediated by other oxalate exchangers. These possibilities need to be examined in future studies.
SAT1 in medullary and papillary tissue
Our finding of significant expression of SAT1 mRNA and protein in the medulla and slight expression in the papilla of control and hyperoxaluric rats is novel, in that previous studies have localized SAT1 protein to the renal cortex [14, 33]. Such differences could be related to differences in the sensitivities of the methods employed or inclusion of non-tubular cells harvested in the present study. Indeed, the latter may well be the case in the papilla where expression levels were <10 % of that found in the renal cortex. However, it would be difficult to attribute the relative abundance of SAT1 message and protein in the medullary region (65 % of that measured in cortical tissue) as a non-tubular contaminant. Further studies are required to resolve these issues.
While it has been generally assumed that the bulk of oxalate secretion occurs along the early portions of the nephron [9, 34], significant expression of SAT1 mRNA and protein in the medulla, and to a lesser degree in the papilla, is a reason to reconsider this assumption. Further indication that renal handling of oxalate does not reside solely in the renal cortex comes from studies of dispersed renal papillary cells which exhibited enhanced oxalate uptake in stone-forming rats [35] and studies employing isolated sheets of renal papillary epithelium which were also shown to support net oxalate absorption [36].
In summary, hyperoxaluria, per se, does not appear to be a result of altered expression patterns of the slc26 transporter, SAT1. Moreover, in the context of chronic renal failure, a significant reduction in both SAT1 mRNA and protein levels is observed, despite a ~10-fold increase in urinary oxalate excretion. Together, these data indicate that the increased excretion of oxalate in these rodent models of hyperoxaluria is not modulated by changes in the transcriptional activity of the SAT1 gene.
Acknowledgments
The technical assistance of Michael Green, Ph. D., Candi Morris, and Bonnie Murphey is greatly appreciated. This work was supported by grants from the National Institutes of Health (DK60544, DK56245).
References
- 1.Hatch M. Oxalate status in stone-formers. Two distinct hyperoxaluric entities. Urol Res. 1993;21:55–59. doi: 10.1007/BF00295194. [DOI] [PubMed] [Google Scholar]
- 2.Danpure CJ. Molecular etiology of primary hyperoxaluria type 1: new directions for treatment. Am J Nephrol. 2005;25:303–310. doi: 10.1159/000086362. [DOI] [PubMed] [Google Scholar]
- 3.Hatch M, Freel RW. Intestinal transport of an obdurate anion: oxalate. Urol Res. 2005;33:1–16. doi: 10.1007/s00240-004-0445-3. [DOI] [PubMed] [Google Scholar]
- 4.Knight TF, Sansom SC, Senekjian HO, Weinman EJ. Oxalate secretion in the rat proximal tubule. Am J Physiol. 1981;240:F295–F298. doi: 10.1152/ajprenal.1981.240.4.F295. [DOI] [PubMed] [Google Scholar]
- 5.Senekjian HO, Weinman EJ. Oxalate transport by proximal tubule of the rabbit kidney. Am J Physiol. 1982;243:F271–F275. doi: 10.1152/ajprenal.1982.243.3.F271. [DOI] [PubMed] [Google Scholar]
- 6.Hatch M, Freel RW. The roles and mechanisms of intestinal oxalate transport in oxalate homeostasis. Semin Nephrol. 2008;28:143–151. doi: 10.1016/j.semnephrol.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatch M, Freel RW. Renal and intestinal handling of oxalate following oxalate loading in rats. Am J Nephrol. 2003;23:18–26. doi: 10.1159/000066300. [DOI] [PubMed] [Google Scholar]
- 8.Kuo SM, Aronson PS. Oxalate transport via the sulfate/ HCO3 exchanger in rabbit renal basolateral membrane vesicles. J Biol Chem. 1988;263:9710–9717. [PubMed] [Google Scholar]
- 9.Greger R, Lang F, Oberleithner H, Deetjen P. Handling of oxalate by the rat kidney. Pflugers Arch. 1978;374:243–248. doi: 10.1007/BF00585601. [DOI] [PubMed] [Google Scholar]
- 10.Weinman EJ, Frankfurt SJ, Ince A, Sansom S. Renal tubular transport of organic acids. Studies with oxalate and para-aminohippurate in the rat. J Clin Invest. 1978;61:801–806. doi: 10.1172/JCI108994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krick W, Schnedler N, Burckhardt G, Burckhardt BC. Ability of SAT-1 to transport sulfate, bicarbonate, or oxalate under physiological conditions. Am J Physiol Renal Physiol. 2009;297:F145–F154. doi: 10.1152/ajprenal.90401.2008. [DOI] [PubMed] [Google Scholar]
- 12.Brandle E, Bernt U, Hautmann RE. In situ characterization of oxalate transport across the basolateral membrane of the proximal tubule. Pflugers Arch. 1998;435:840–849. doi: 10.1007/s004240050592. [DOI] [PubMed] [Google Scholar]
- 13.Dawson PA, Russell CS, Lee S, McLeay SC, van Dongen JM, Cowley DM, Clarke LA, Markovich D. Urolithiasis and hepatotoxicity are linked to the anion transporter SAT1 in mice. J Clin Invest. 2010;120:706–712. doi: 10.1172/JCI31474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karniski LP, Lotscher M, Fucentese M, Hilfiker H, Biber J, Murer H. Immunolocalization of SAT-1 sulfate/oxalate/ bicarbonate anion exchanger in the rat kidney. Am J Physiol. 1998;275:F79–F87. doi: 10.1152/ajprenal.1998.275.1.F79. [DOI] [PubMed] [Google Scholar]
- 15.Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- 16.Xie Q, Welch R, Mercado A, Romero MF, Mount DB. Molecular characterization of the murine Slc26a6 anion exchanger: functional comparison with Slc26a1. Am J Physiol Renal Physiol. 2002;283:F826–F838. doi: 10.1152/ajprenal.00079.2002. [DOI] [PubMed] [Google Scholar]
- 17.Green ML, Hatch M, Freel RW. Ethylene glycol induces hyperoxaluria without metabolic acidosis in rats. Am J Physiol Renal Physiol. 2005;289:F536–F543. doi: 10.1152/ajprenal.00025.2005. [DOI] [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Steel RG, Torrie JH. Principles and procedures of statistics. 2nd edn. New York: McGraw-Hill Book Company; 1980. [Google Scholar]
- 21.SAS. SAS User’s Guide: Basics, Version. 5th edn. Carey: SAS Institute Inc; 1985. [Google Scholar]
- 22.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatch M, Freel RW. Angiotensin II involvement in adaptive enteric oxalate excretion in rats with chronic renal failure induced by hyperoxaluria. Urol Res. 2003;31:426–432. doi: 10.1007/s00240-003-0367-5. [DOI] [PubMed] [Google Scholar]
- 24.Aronson PS. The renal proximal tubule: a model for diversity of anion exchangers and stilbene-sensitive anion transporters. Annu Rev Physiol. 1989;51:419–441. doi: 10.1146/annurev.ph.51.030189.002223. [DOI] [PubMed] [Google Scholar]
- 25.Regeer RR, Lee A, Markovich D. Characterization of the human sulfate anion transporter (hsat-1) protein and gene (SAT1; SLC26A1) DNA Cell Biol. 2003;22:107–117. doi: 10.1089/104454903321515913. [DOI] [PubMed] [Google Scholar]
- 26.Markovich D, Murer H, Biber J, Sakhaee K, Pak C, Levi M. Dietary sulfate regulates the expression of the renal brush border Na/Si cotransporter NaSi-1. J Am Soc Nephrol. 1998;9:1568–1573. doi: 10.1681/ASN.V991568. [DOI] [PubMed] [Google Scholar]
- 27.Sagawa K, DuBois DC, Almon RR, Murer H, Morris ME. Cellular mechanisms of renal adaptation of sodium dependent sulfate cotransport to altered dietary sulfate in rats. J Pharmacol Exp Ther. 1998;287:1056–1062. [PubMed] [Google Scholar]
- 28.Freel RW, Hatch M, Green M, Soleimani M. Ileal oxalate absorption and urinary oxalate excretion are enhanced in Slc26a6-null mice. Am J Physiol (Gastrointest Liver Physiol) 2006;290:G719–G728. doi: 10.1152/ajpgi.00481.2005. [DOI] [PubMed] [Google Scholar]
- 29.Schnedler N, Burckhardt G, Burckhardt BC. Glyoxylate is a substrate of the sulfate–oxalate exchanger, SAT-1, and increases its expression in HepG2 cells. J Hepatol. 2011;54:513–520. doi: 10.1016/j.jhep.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes I, Laouari D, Tutt P, Hampson G, Friedlander G, Silve C. Sulfate homeostasis, NaSi-1 cotransporter, and SAT-1 exchanger expression in chronic renal failure in rats. Kidney Int. 2001;59:210–221. doi: 10.1046/j.1523-1755.2001.00481.x. [DOI] [PubMed] [Google Scholar]
- 31.Green ML, Freel RW, Hatch M. Lipid peroxidation is not the underlying cause of renal injury in hyperoxaluric rats. Kidney Int. 2005;68:1–10. doi: 10.1111/j.1523-1755.2005.00735.x. [DOI] [PubMed] [Google Scholar]
- 32.Quondamatteo F, Krick W, Hagos Y, Kruger MH, Neubauer-Saile K, Herken R, Ramadori G, Burckhardt G, Burckhardt BC. Localization of the sulfate/anion exchanger in the rat liver. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1075–G1081. doi: 10.1152/ajpgi.00492.2005. [DOI] [PubMed] [Google Scholar]
- 33.Markovich D. Physiological roles and regulation of mammalian sulfate transporters. Physiol Rev. 2001;81:1499–1533. doi: 10.1152/physrev.2001.81.4.1499. [DOI] [PubMed] [Google Scholar]
- 34.Greger R. In: Renal transport of organic substances. Greger R, Lang F, Silbernagel S, editors. Berlin: Springer-Verlag; 1981. p. 224. [Google Scholar]
- 35.Sigmon D, Kumar S, Carpenter B, Miller T, Menon M, Scheid C. Oxalate transport in renal tubular cells from normal and stone-forming animals. Am J Kidney. Dis. 1991;17:376–380. doi: 10.1016/s0272-6386(12)80626-3. [DOI] [PubMed] [Google Scholar]
- 36.Chandhoke PS, Fan J. Transport of oxalate across the rabbit papillary surface epithelium. J Urol. 2000;164:1724–1728. [PubMed] [Google Scholar]