Abstract
The systematic and complete characterization of the Saccharomyces cerevisiae genome and proteome has been stalled in some cases by misannotated genes. One such gene is YBR074W, which was initially annotated as two independent open reading frames (ORFs). We now report on Ybr074, a metalloprotease family member that was initially predicted to reside in the endoplasmic reticulum (ER). Therefore, we tested the hypothesis that Ybr074 may be an ER quality control protease. Instead, indirect immunofluorescence images indicate that Ybr074 is a vacuolar protein, and by employing protease protection assays, we demonstrate that a conserved M28 metalloprotease domain is oriented within the lumen. Involvement of Ybr074 in ER protein quality control was ruled out by examining the stabilities of several well-characterized substrates in strains lacking Ybr074. Finally, using a proteomic approach, we show that disrupting Ybr074 function affects the levels of select factors implicated in vacuolar trafficking and osmoregulation. Together, our data indicate that Ybr074 is the only multi-spanning vacuolar membrane protease found in the yeast Saccharomyces cerevisiae.
Introduction
The yeast Saccharomyces cerevisiae has long served as a broadly used model organism because its biology is representative of many organisms and because it benefits from comprehensive genetic and biochemical tools (Foury, 1997; Karathia et al, 2011; Suter et al, 2006). While great progress has been made toward elucidating the function of every gene since the yeast genome was sequenced in 1996, many genes remain uncharacterized, despite original projections that the genome would be “solved” by 2007 (Goffeau et al, 1996; Pena-Castillo & Hughes, 2007). In a number of cases, this delay may be attributed to gene misannotation due to errors in the original genome sequencing (Brachat et al, 2003). Until these sequencing errors were corrected in 2003, misannotated genes were not accurately represented in large scale analyses and strain libraries (Giaever et al, 2002; Huh et al, 2003), thus stalling their complete characterization. One such uncharacterized gene is YBR074W, which encodes a protein with a domain homologous to a putative human metalloprotease, ERMP1.
Metalloproteases are a highly diverse set of proteolytic enzymes distributed across families M1 through M91, each representing a set of homologous proteases (Rawlings et al, 2012). They are commonly identified by a conserved HEXXH active site motif that is required for catalysis and to coordinate metal ions. Metalloprotease activity is mediated by one or two divalent ions, often zinc, which activate water molecules for nucleophilic attack on substrate peptide bonds (Rawlings & Salvesen, 2013). Metalloproteases can be secreted into the extracellular space or reside intracellularly. Secreted matrix metalloproteases in humans are of particular interest because of their prominent role in remodeling of the extracellular matrix, a process associated with both normal physiology and pathology during development, wound healing, and cancer cell metastasis (Klein & Bischoff, 2011; Sbardella et al, 2012). Within the cell, metalloproteases play an important role in the maturation and trafficking of secreted proteins and in the turnover of long-lived, superfluous, or damaged proteins and organelles. The latter function occurs primarily in the lysosome, which is functionally analogous to the yeast vacuole (Li & Kane, 2009).
The yeast, Saccharomyces cerevisiae, encodes 47 metalloproteases, including 7 non-peptidase homologs that are predicted to lack enzymatic activity. YBR074W is predicted to encode a protein that belongs to the M28 family of metalloproteases, which is comprised of both amino- and carboxy-peptidases that require two zinc ions for activity (Rawlings et al, 2012). The rat ortholog of Ybr074, named Felix-ina (FXNA), was reported to be an ER-localized protein involved in ovarian development (Garcia-Rudaz et al, 2007).
Protease activity plays a critical role in vacuole function. The yeast vacuole is also critical for, nutrient and ion storage, osmoregulation, and detoxification (Broach et al, 1991; Li & Kane, 2009). To augment its role in the degradation of macromolecules, the vacuole in Saccharomyces cerevisiae is equipped with seven known proteases, including the soluble metalloproteases aminopeptidase I (Ape1), aminopeptidase Y (Ape3) and carboxypeptdiase S (Cps1).
In this paper we report on the initial characterization of Ybr074, a yeast M28 metalloprotease family member. We present evidence that Ybr074 is a glycosylated transmembrane protein with a lumenal-facing protease domain. We also find evidence to support previous proteomic data suggesting that Ybr074 resides in the yeast vacuole (Wiederhold et al, 2009), thus adding another protease to the repertoire of this organelle.
Materials and Methods
Computational Analysis
The amino acid sequence of Ybr074 was analyzed using the default parameters of the Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) to identify proteins in the non-redundant protein sequences database, which shared local sequence similarity with Ybr074 (Altschul et al, 1997; Altschul et al, 2005). The European Molecular Biology Library-European Bioinformatics Institute (EMBL-EBI), InterPro Scan sequence search tool, and the PANTHER Classification System version 7.2 were also used to examine proteins related to Ybr074 using default parameters (Mi et al, 2010).
The M28 protease domain of Ybr074, and its putative catalytic and zinc-binding residues, were identified using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) (Marchler-Bauer et al, 2011). The M28 protease domain boundaries of Ybr074 were analyzed using Pfam version 26.0 (Punta et al, 2012). Sequence similarity between Ybr074 and FXNA was examined using Lalign under default settings. The C-terminal domains of Ybr074 and FXNA were compared using Lalign global sequence comparison (Huang & Miller, 1991).
The presence and position of transmembrane segments, as well as the topology of Ybr074 were predicted using TmHMM v. 2.0 (Sonnhammer et al, 1998). N-linked glycosylation sites were predicted using the NetNGlyc 1.0 server, under default settings, from the Eukaryotic Linear Motif (ELM) resource for functional sites in proteins (Blom et al, 2004). The sub-cellular localization of Ybr074 was predicted using PSORT, PSORT II, and WoLF PSORT under default settings (Horton et al, 2007).
Strains, plasmids, yeast growth conditions and molecular techniques
Strains used in this study are listed in Table 1. Standard growth conditions and molecular techniques were used (Adams et al, 1998), unless otherwise indicated. Deletion of YBR074 in the BY4742 background was carried out using Polymerase Chain Reaction (PCR)-mediated gene disruption (Brachmann et al, 1998). Briefly, the BY4742 strain was transformed with a ybr074∷KANMX cassette amplified from the pRS400 vector (forward primer: AAA TTA TCT ACA AGG AAA TAA ATT GAT AGG TAA AGT TAA AGA ATC ACG GCA GAT TGT ACT GAG AGT GCA C; reverse primer: CAG TAG GCG AAT TTG AGT TTA TAA AAA TTT ACA TTT AAA ACT AAT TAG AAC TGT GCG GTA TTT CAC ACC G). Transformants were selected on yeast-peptone containing 2% dextrose (YPD) and 250 µg/mL Geneticin (G-418; Invitrogen, Carlsbad, CA). Colonies incorporating the ybr074∷KANMX cassette were verified by PCR using primers flanking the YBR074 open reading frame (ORF) (upstream primer: CAA TGA CGC CAA ATA TGG ACA CT; downstream primer: AAG AGA GCA CCG TAG AAT GGT T). To generate the ybr074∷NATMX strain, the NATMX cassette was PCR amplified from the pFA6a–NAT-MX6 vector using forward primer: CAA GGA AAT AAA TTG ATA GGT AAA GTT AAA GAA TCA CGG CCG GAT CCC CGG GTT AAT TAA and reverse primer: ATT TGA GTT TAT AAA AAT TTA CAT TTA AAA CTA ATT AGA AGA GCT CGT TTA AAC TGG ATG (Vembar et al, 2010). The NATMX cassette was transformed into the BY4742 strain as described (Adams et al, 1998). Cells were selected on YPD medium supplemented with nourseothricin (NAT; clonNAT, Werner BioAgents, Jena, Germany), and integration of the NATMX cassette was verified by PCR using the same primers used to check for integration of the KANMX cassette.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4742 | MAT α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Brachmann C.B. et al. 1998 |
| ybr074Δ | MAT α ybr074∷NATMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
This study |
| KHY1 | MAT α ybr074∷HIS3MX6-PGAL1-GFP-YBR074 his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
This study |
| pep4Δ | MAT α pep4∷KANMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
Invitrogen |
| pdr5Δ | MAT α pdr5∷KANMX his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
Invitrogen |
A chromosomally-integrated PGAL1-GFP-YBR074 strain was generated using the integration vector pFA6a–His3MX6-PGAL1-GFP, as described (Longtine et al, 1998). Briefly, the His3MX6-PGAL1-GFP cassette was amplified using the following primers: forward primer, AAA TTA TCT ACA AGG AAA TAA ATT GAT AGG TAA AGT TAA AGA ATC ACG GCG AAT TCG AGC TCG TTT AAA C; reverse primer, TTT GTT TTC CGG TAC TTT AGA ACT GAT CTG AAT ACA CTT TTT AAT TTC ATT TTG TAT AGT TCA TCC ATG C, and the resulting DNA fragment was transformed into the BY4742 strain. Transformed cells were selected on synthetic medium lacking histidine. Integration of the His3MX6-PGAL1-GFP cassette was confirmed by amplification of a region upstream of YBR074 and internal to the His3MX6-PGAL1-GFP cassette (forward primer: CAA TGA CGC CAA ATA TGG ACA CT; reverse primer: TTT GTA TAG TTC ATC CAT GC) from purified genomic DNA (Adams et al, 1998).
Plasmids used in this study are listed in Table 2. To generate a HA-tagged YBR074 constructs, YBR074 was PCR amplified from vector pGP564_1_b11 from the yeast overexpression plasmid library (Jones et al, 2008). PCR amplified YBR074 with a C-terminal HA tag (forward primer: ATG GAG AGC TCG ACA AGT GCG CTG GAT TTA CAA AAG AAA ATG AAT GA; reverse primer: CGA TCT CGA GTT AAG CGT AAT CTG GAA CAT CGT AAG GGT ATA AAA TTA TAG CAT CCT TGA CAA TAA CTA ATC CTT TC) was ligated into the SacI/XhoI sites in the yeast expression vector pKN16 (Table 2). PCR amplified YBR074 with an N-terminal HA-tag (forward primer: AAT CAC GGA TCC AAA TTA AAA AGT GTA TTC AGA TCA GTT CTA AAG; reverse primer: AGA ACT CGA GTT ATA AAA TTA TAG CAT CCT TGA CAA TAA CTA ATC CTT TCT G) was ligated into the BamHI/XhoI sites in the yeast expression vector pKN16 (Table 2). The PGAL1/10 promoter of the resulting plasmid was removed using SacI/SpeI and replaced with the endogenous promoter of YBR074 (forward primer: ATG GAG AGC TCG ACA AGT GCG CTG GAT TTA CAA AAG AAA ATG AAT GA; reverse primer: TCA TAC TAG TGC CGT GAT TCT TTA ACT TTA CCT ATC AAT TTA TTT CCT). This construct was verified by DNA sequence analysis (IDT, Coralville, IA).
Table 2.
Plasmids used in this study
| Plasmid Name | Description | Selectable Marker | Source or reference |
|---|---|---|---|
| pGP564_1_b11 | 2µ overexpression chromosome II: 378129- 393468, including YBR074 |
LEU2 | Jones G.M. et al. 2008 |
| pKN16 | pRS316-PGAL1/10-3HA expression vector |
URA3 | K. Nakatsukasa, Nagoya University |
| pRS426-3HA- YBR074 |
Ybr074 expression | URA3 | This study |
| pRS426-YBR074- 3HA |
Ybr074 expression | URA3 | This study |
| pRS426-YBR074- M28HA |
Ybr074 expression | URA3 | This study |
| GFP-Atg8 | Autophagy reporter | URA3 | D. Klionsky, University of Michigan |
To generate the YBR074-M28HA expression constructs a single HA tag was inserted within the M28 metalloprotease domain by two-stage PCR (Wang & Malcolm, 1999). Secondary structure prediction programs, SSPro version 4.5 (Cheng et al, 2005) and PSIPRED version 3.0 (Buchan et al, 2010), were used to identify a region for insertion of the HA tag without disrupting secondary structures. Specifically, the tag was inserted following residue Pro45 in a predicted coil region between transmembrane segment 1 and the first alpha helix predicted to compose the M28 protease domain (Pollastri et al, 2002). The endogenous promoter and coding sequence of YBR074 were PCR amplified as two separate fragments, PCR1 (forward primer A: ATG GAG AGC TCG ACA AGT GCG CTG GAT TTA CAA AAG AAA ATG AAT GA; reverse primer B: TGC ATA GTC CGG GAC GTC ATA CGG ATA TGG TAG ATT GAG TTT ATA ACG TTC ATG ATC AAA GAT ATA G) and PCR2 (forward primer C: TAT CCG TAT GAC GTC CCG GAC TAT GCA AAA GAG GAT GAG CAC CCT GAA TTC AAT GAC; reverse primer D: AGA ACT CGA GTT ATA AAA TTA TAG CAT CCT TGA CAA TAA CTA ATC CTT TCT G). A third reaction, PCR3 (forward primer A, reverse primer D) was carried out in which a mixture of 5% PCR1 and 5% PCR2 (vol/vol) was used as a template for amplification. The resulting PENDG-YBR074-M28HA cassette was inserted into pRS426 at the SacI/XhoI sites, and the construct was verified by DNA sequence analysis, as above.
Analysis of protein topology
Protease protection assays were performed using microsomes derived from wild type yeast expressing HA-tagged YBR074 from its endogenous promoter on a 2µ plasmid. Microsomes were isolated using the “medium” preparation method (Nakatsukasa & Brodsky, 2010). Reactions were performed in B88 (20mM HEPES, pH 6.8, 150 mM potassium acetate, 5 mM magnesium acetate, 250 mM sorbitol) using 100–500 µg of total microsomal protein, and were incubated in the presence or absence of 100 µg/mL proteinase K (Sigma, St. Louis, MO) supplemented with 1 mM CaCl2 and 1% Triton X-100. Reactions treated with 0.5% SDS were performed in B88 lacking potassium acetate. Reactions were incubated on ice and 25 µL fractions were taken at the indicated times and precipitated in 10% trichloroacetic acid (TCA) for 15 min on ice. Samples taken at 0 min were collected prior to addition of Proteinase K, except in Figure S2, when the 0 min GFP-Ybr074 sample was collected immediately after addition of Proteinase K. Precipitated proteins were pelleted and washed in acetone and then re-suspended in 60 µL TCA sample buffer (80mM Tris-HCl, pH 8, 8mM EDTA, 120 mM DTT, 3.5% SDS, 10% glycerol, 0.08% Tris base, 0.01% Bromophenol Blue). Proteins were resolved by 10% SDS-PAGE, transferred to nitrocellulose, and detected using the following primary antibodies: 1:5000 HRP-conjugated mouse anti-HA (Roche, Indianapolis, IN), 1:1000 rabbit anti-Bos1 (Barlowe lab, Geisel School of Medicine at Dartmouth, Hanover, NH), or 1:5000 rabbit anti-Pdi1 (Denic Lab, Harvard University, Cambridge, MA). The bound antibodies were visualized using Super Signal West Femto kit (Thermo Scientific, Rockford, IL) on a Kodak 440CF Image Station (Eastman Kodak, Rochester, NY).
Determination of glycosylation status and membrane residence
The presence of N-linked glycans on Ybr074 was tested by digestion with Endoglycosidase H (Roche, Indianapolis, IN). Microsomes were prepared as described above from a BY4742 strain expressing HA-tagged YBR074 from its endogenous promoter on a CEN plasmid. Approximately 100 µg of protein were incubated in 50mM citrate buffer, pH 5.5, 1 mM PMSF, 1 µg/mL leupeptin, and 0.5 µg/mL pepstatin A (Sigma, St. Louis, MO) in the presence or absence of 0.005 units of endoglycosidase H overnight at 4°C. Digested protein was resolved by 10% SDS-PAGE and detected by western blot analysis using 1:5000 HRP-conjugated mouse anti-HA antibody (Roche, Indianapolis, IN) as described above. The molecular mass of the resulting protein bands was determined by Rf analysis.
Sodium carbonate extraction was performed using microscomes prepared as described above from a BY4742 strain expressing HA-tagged YBR074 from its endogenous promoter on a CEN plasmid (Sikorski & Hieter, 1989). Approximately 200 µg of microsomal protein was re-suspended in 1 mL B88 or 0.1 M Na2CO3, pH 11.5, in the presence of protease inhibitors (PMSF, leupeptin, pepstatin A; Sigma, St. Louis, MO) at 4°C and incubated for 30 min. Soluble protein was separated from membrane bound protein by ultracentrifugation at 138,000 g (50,000 rpm) for 1 h at 4°C in a Sorvall LC M120-EX Micro Ultracentrifuge using a Sorvall S100AT5 fixed angle rotor. The supernatant was isolated and processed for TCA precipitation as described above. The pellet was washed with B88 containing protease inhibitors, and spun again at 198,000 g (60,000 rpm) for 10 min at 4°C. The supernatant from the second spin was removed and discarded, and the pellet was re-suspended in sample buffer. Samples were resolved by 10% SDS-PAGE and proteins were visualized by western blot analysis as described above.
Protein localization
Immunofluorescence microscopy was performed, as described previously, with minor modifications (Adams et al, 1998). Spheroplasted cells were incubated in poly-lysine coated wells for 10 min at room temperature. Cells were then washed three times with 30 µL of sterile filtered blocking solution (PBS, pH 7.4, 0.5% BSA, 0.5% ovalbumin, 0.01% fish skin gelatin; Sigma, St. Louis, MO) supplemented with 0.1% Triton X-100 (Sigma, St. Louis, MO). Cells were then incubated in blocking solution, including Triton X-100, for 30 min in a humidified chamber, followed by an overnight incubation with 1:500 mouse anti-HA (Roche, Indianapolis, IN) and 1:500 rabbit anti-Kar2 (Brodsky et al, 1993), or 1:100 rabbit anti-HA (Cell Signaling Technology, Beverly, MA) and 1:750 mouse anti-Vph1 (Invitrogen/Molecular Probes, Carlsbad, CA) overnight at 4°C in a humidified chamber. The next day, cells were washed four times with 30 µL blocking solution and incubated with 1:250 Alexa Fluor 488 goat anti-mouse IgG and 1:250 Alexa Fluor 568 goat anti-rabbit IgG (Invitrogen, Molecular Probes, Eugene OR). Images were visualized using a Leica TCS SP5 (Leica microsystems, Buffalo Grove, IL).
Live cell imaging of GFP-Ybr074 was examined in the BY4742 strain with chromosomally-integrated GFP-Ybr074 under the control of a galactose promoter, and selected using the HIS3MX gene as described above. Cells were grown in medium lacking histidine and supplemented with 2% galactose to induce expression, or 2% raffinose as a negative control. Strain KHY1 (Table 1) cells were induced overnight at 30°C and harvested at an OD600 of ~0.8. Approximately 0.4 OD600 cell equivalents were harvested by centrifugation at 13,000 g for 30 sec and washed in 1mL PBS, pH 7.4. Next, the yeast were re-suspended in PBS, pH 7.4, to a concentration of 4 OD600/mL and 5 µL of the suspension was mounted on a glass slide for visualization. The cells were visualized using an Olympus BX60 fluorescence microscope and QED InVivo software. GFP-Ybr074 fluorescence signal was absent when cells were grown in media supplemented with raffinose instead of galactose (Figure S4).
The localization of FXNA in COS7 cells was visualized using an N-terminally FLAG-tagged FXNA expression plasmid, which required the acquisition of a C-terminally FLAG-tagged FXNA construct that was a generous gift from Dr. S. Ojeda (Oregon Health and Science University, Portland, OR). Briefly, N-terminally FLAG-tagged FXNA was amplified from the pcDNA3.1Zeo-FXNA-FLAG template using forward primer: ATC GGG ATC CAT GGA CTA CAA AGA CGA TGA CGA CAA GGA GTG GAG CTC GGA GTC GGC GGC CGT and reverse primer: CGA TGA TAT CTT AAA ACA CAA AGA GAC TAT AGG TGG AAA CCC ACG CCG AGG GAA ATG AC. Amplified FLAG-FXNA was inserted into pcDNA3.1Neo vector using the BamHI/EcoRV sites and the construct was confirmed by DNA sequencing analysis. The N- and C-terminally FLAG-tagged FXNA constructs were transformed into COS7 cells using lipofectamine 2000, according to the manufacturer’s instruction (Invitrogen, Carlsbad, CA). Transfected cells were grown on coverslips in a 12-well dish, rinsed with PBS, fixed using 3% paraformaldehyde, and rinsed with PBS supplemented with 10mM glycine (PBS-G). The cells were permeablized at room temperature using 0.5% Triton X-100 in PBS-G followed by another rinse in PBS-G. Non-specific binding sites were blocked using 0.25% ovalbumin in PBS-G at room temperature. Next, the coverslips were incubated at room temperature for 30 min with polyclonal anti-FLAG antibody (1:500 dilution; Cell Signaling Technology, Danvers, MA), monoclonal anti-PDI antibody (1:500 dilution; Enzo Life Sciences, Ann Arbor, MI), and anti-Lamp1 antibody H4A3 (1:500 dilution; Abcam, Cambridge, MA ). The coverslips were washed in blocking buffer and then incubated for 30 min with secondary antibodies Alexa Fluor goat anti-mouse 488 (1:250, Invitrogen, Carlsbad, CA), and Alexa Fluor goat anti-rabbit 568 (1:250, Invitrogen, Carlsbad, CA). Finally, the coverslips were washed in PBS-G and mounted onto slides. Images were captured using an Olympus BX60 fluorescence microscope and QED InVivo software.
Proteomic analysis
To identify putative Ybr074 substrates, and/or proteins whose levels may be regulated by Ybr074, an analysis of a GFP-tagged protein library in the ybr074Δ strain background was conducted as described (Cohen & Schuldiner, 2011). Briefly, a ybr074∷NATMX MATa strain was mated with the cytosolic mCherry-tagged GFP collection (Breker et al, 2013) using Synthetic Genetic Array (SGA) techniques as described (Cohen & Schuldiner, 2011; Tong & Boone, 2006). Haploid cells expressing a GFP-tagged protein in the ybr074Δ background were selected and grown to logarithmic growth phase in synthetic complete medium at 30°C in 384-well growth plates. Cells were then transferred to 384-well microscope plates (Matrical Bioscience, Spokane, WA) and visualized using an automated inverted fluorescence microscope ScanR system (Olympus) using a swap robot (Hamilton). Images were then manually reviewed for localization information and analyzed for fluorescence intensity using the ScanR analysis program and processed by Adobe Photoshop CS3 software.
Results
Ybr074 is a glycosylated transmembrane protein bearing homology to the M28 family of metalloproteases
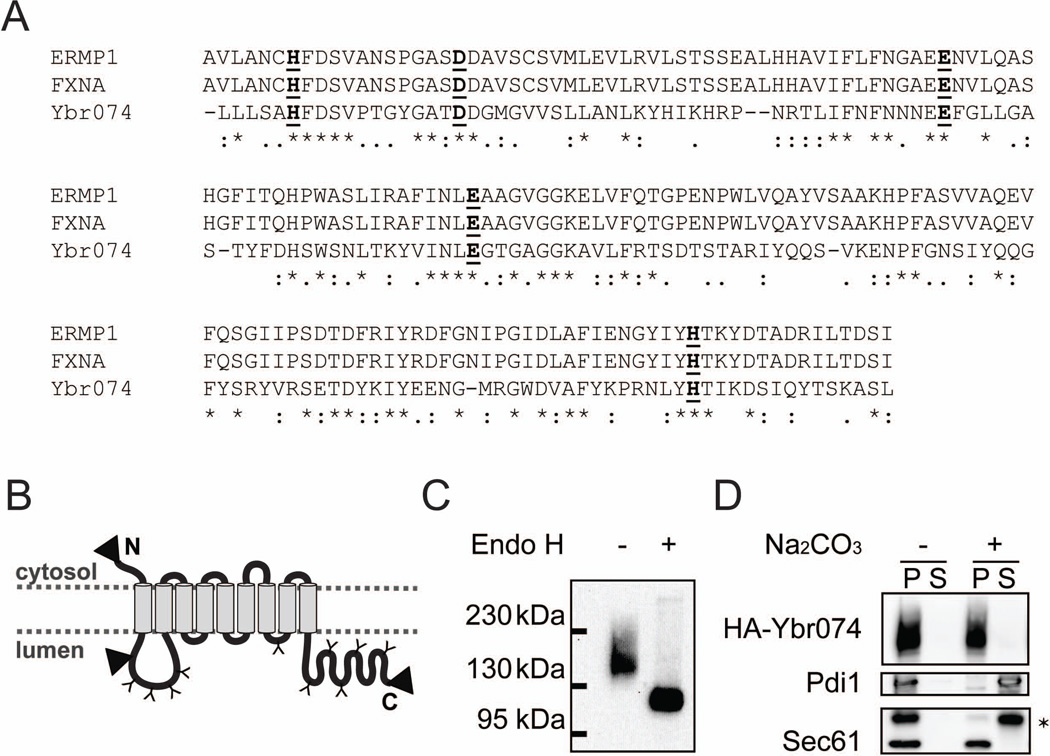
As an initial characterization of Ybr074, we examined the predicted localization and topology of this protein using various computational prediction programs. Ybr074 was found to be a type III membrane protein comprised of 9 predicted helical transmembrane segments (TMs) based on analysis using TMHMM v. 2.0 (Figure S1) (Krogh et al, 2001). PSORT and PSORTII predictions place Ybr074 in the ER membrane with a cytosolic N-terminus and a lumenal C-terminus (Horton et al, 2007; Nakai & Horton, 1999). Analysis of Ybr074 using the Protein family database (Pfam) reveals a conserved M28 metalloprotease domain between TM1 and TM2 (Leu151-Leu314) facing the endomembrane system lumen (Figure 1A). Furthermore, 10 predicted lumenal-facing N-linked glycosylation sites dispersed throughout the protein were identified using NetNGlyc v. 1.0 (Figure 1B).
Figure 1.
Ybr074 is a glycosylated membrane protein with a lumenal-facing putative protease domain. (A) Sequence alignment comparing the M28 metalloprotease domains of Ybr074 with those of the human ortholog ERMP1 and the rat ortholog FXNA. * denotes amino acid identity, : denotes a conserved amino acid substitution, and . denotes a semi-conserved amino acid substitution. Conserved metal-binding amino acids are shown in bold and are underlined. (B) Computationally predicted topology of Ybr074 is shown with putative N-linked glycosylation sites: Asn96, Asn121, Asn189, Asn217, Asn656, Asn768, Asn796, Asn811, Asn866, Asn937 (indicated as a “Y”). The domain homologous to M28 metalloprotease family members is located between transmembrane segments one and two, and spans Leu151-Leu314. Triangles denote sites where an HA-epitope tag was inserted at the N-terminus, between transmembrane segments one and two (Pro45), and at the C-terminus. (C) N-terminally HA-tagged Ybr074 in isolated crude membranes was treated in the presence or absence of endo H. (D) Sodium carbonate extraction of N-terminally HA-tagged Ybr074 from crude membrane extracts is shown. Samples were separated into pellet (P) and soluble (S) fractions. The peripherally associated membrane protein disulphide isomerase (Pdi1) and the intergral membrane protein Sec61 were used as controls. * denotes a non-specific protein recognized by the anti-Sec61 antibody.
To examine whether Ybr074 is modified by N-linked glycans, as predicted, crude membrane preparations were isolated from wild type yeast expressing an N-terminally HA-tagged form of Ybr074 (HA-Ybr074). The membranes were then incubated in the absence or presence of endoglycosidase H (Endo H) to remove N-linked glycans. Treatment with Endo H resulted in a shift in the molecular mass of Ybr074 from ~147 kDa to ~123 kDa (Figure 1C). Assuming that each N-linked glycan contributes ~3 kDa to the total molecular mass of Ybr074, this allows us to estimate the presence of 8 N-linked glycans. Because N-linked glycosylation of secretory proteins occurs in the ER, this result is consistent with the fact that Ybr074 enters through the ER (Aebi et al, 2010; Helenius & Aebi, 2004; Schwarz & Aebi, 2011). However, these data do not inform on the final destination of this endomembrane system protein.
To confirm that Ybr074 is an integral transmembrane protein, crude membrane extracts containing HA-Ybr074 were subjected to sodium carbonate extraction to disrupt electrostatic interactions that mediate peripheral membrane protein associations with the lipid bilayer. As anticipated, the peripheral ER lumenal protein, Pdi1 (Lambert & Freedman, 1985; Mizunaga et al, 1990), was released from the membrane upon treatment with sodium carbonate and partitioned to the soluble fraction (Figure 1D). In contrast, the integral ER membrane protein, Sec61 (Stirling et al, 1992), remained associated with the pellet after treatment with sodium carbonate. Similarly, Ybr074 remained associated with the membrane fraction, indicating that Ybr074 is an integral membrane protein, as predicted.
Ybr074 has a lumenal-facing protease domain
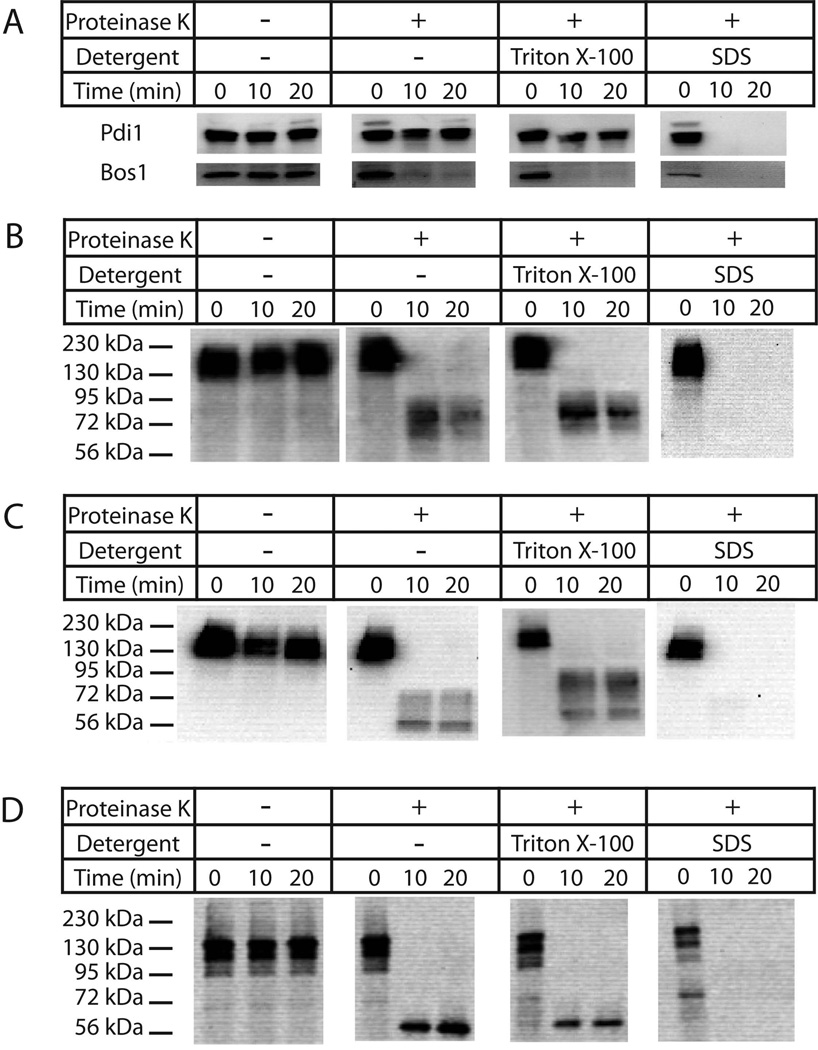
To determine the topology of Ybr074, sensitivity to digestion by proteinase K was examined using Ybr074 constructs in which an HA epitope tag was appended either at the N-terminus, adjacent to the protease domain, or at the C-terminus (see arrowheads, Figure 1B). Crude membrane fractions containing each tagged form of Ybr074 were incubated in the absence or presence of proteinase K, and only lumenal-facing epitope tags were expected to be protected from proteinase K digestion. As a control, Pdi1 was resistant to proteinase K-mediated degradation throughout the 20 min time course. In contrast, the cytosolic-facing tail-anchored protein, Bos1 was proteinase K sensitive (Figure 2A) (Newman et al, 1992). Although the N-terminus of Ybr074 is computationally predicted to be cytosolic, the Ybr074 fragment containing an N-terminal HA tag was protected from proteinase K-mediated degradation (Figure 2B). However, when the larger GFP tag was appended to the N-terminus of Ybr074, it became proteinase-sensitive, suggesting the GFP tag is oriented towards the cytosol (Figure S2). One explanation for this discrepancy is that the smaller HA epitope is protected from proteinase K digestion by its proximity to the membrane or by tertiary interactions of Ybr074.
Figure 2.
Topological analysis of Ybr074. Crude membrane extracts were isolated from yeast expressing HA-tagged Ybr074 protein. Protein samples were incubated in the presence or absence of proteinase K and Triton X-100 or SDS, as indicated. (A) The cytosolic tail-anchored protein Bos1, and the peripherally membrane associated lumenal protein Pdi1 were used as controls. Proteinase K-digested Ybr074 fragments are shown for the (B) HA-Ybr074, (C) Ybr074-M28HA, and (D) Ybr074-HA species.
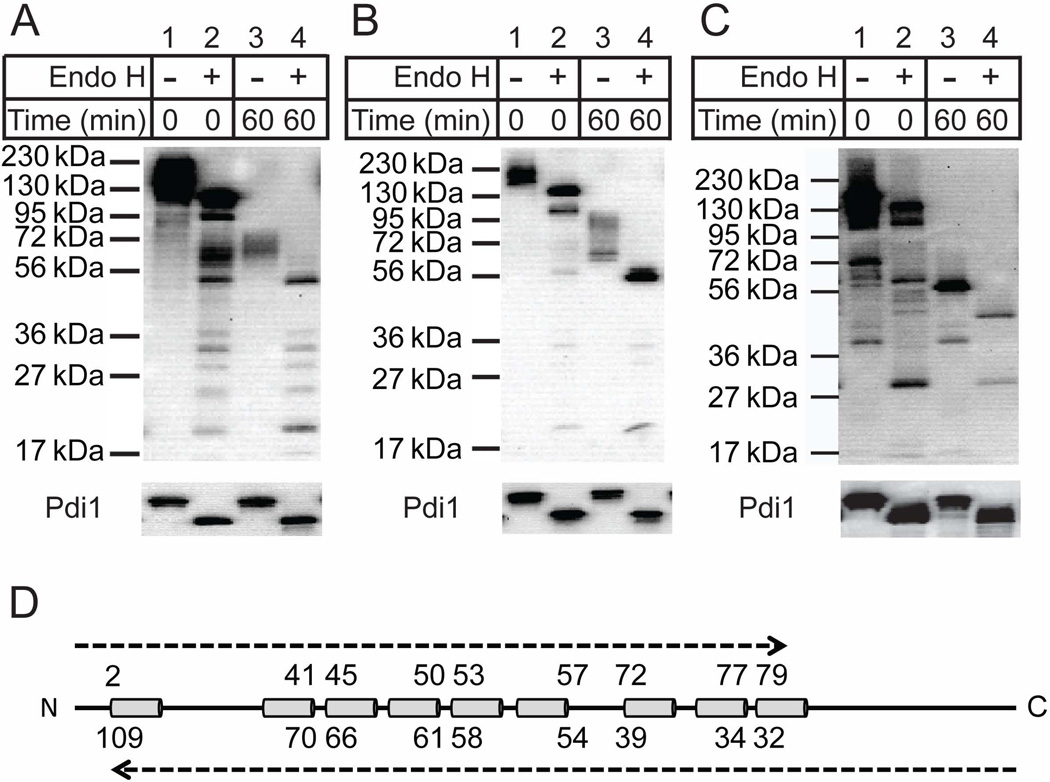
Both the HA tag adjacent to the protease domain and that appended to the C-terminus were protected from proteinase K digestion (Figure 2C and 2D), suggesting they are oriented within the lumen. Please note that the Ybr074 fragment shown in Figure 2D ran at ~56 kDa in contrast to higher molecular mass species present in Figures 2B and 2C. However, these protease-protected Ybr074 fragments remained insensitive to digestion even when, in principle, the membrane was disrupted by addition of Triton X-100. Degradation was only achieved when SDS was substituted as the detergent. The simplest explanation for this phenomenon, and one that is consistent with the heavily glycosylated protease and C-terminal domains of Ybr074, is that these HA epitopes may stably fold and/or aggregate during protease digestion and are proteinase K resistant unless they are denatured by SDS. To differentiate between these possibilities, proteinase K digested Ybr074 fragments were treated in the absence or presence of Endo H to remove N-linked glycans, which can only be appended to lumenal-facing regions of Ybr074. The protease domain of Ybr074 is predicted to be modified by four N-linked glycans, while the C-terminal domain is predicted to be modified by five N-linked glycans. In both cases, treatment with Endo H resulted in a shift in molecular mass of the HA-containing peptides (Figure 3A,B,C; note shifts in molecular mass across lanes 3 and 4). These collective results strongly suggest that the protease domain and C-terminal domain are lumenal, as predicted.
Figure 3.
Proteinase K digested Ybr074 fragments were treated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of Endo H. (A) HA-Ybr074, (B) Ybr074-M28HA, and (C) Ybr074-HA forms are shown. (D) Representative image of Ybr074 with transmembrane helices depicted as cylinders. The predicted molecular masses of Ybr074 fragments are shown in kDa units. The top set of molecular masses corresponds to fragments spanning from the N-terminus to the predicted cytosolic boundary of each transmembrane helix. The bottom set of molecular weight masses corresponds to fragments spanning from the C-terminus to the predicted boundary of each transmembrane helix.
Ybr074 resides in the vacuole
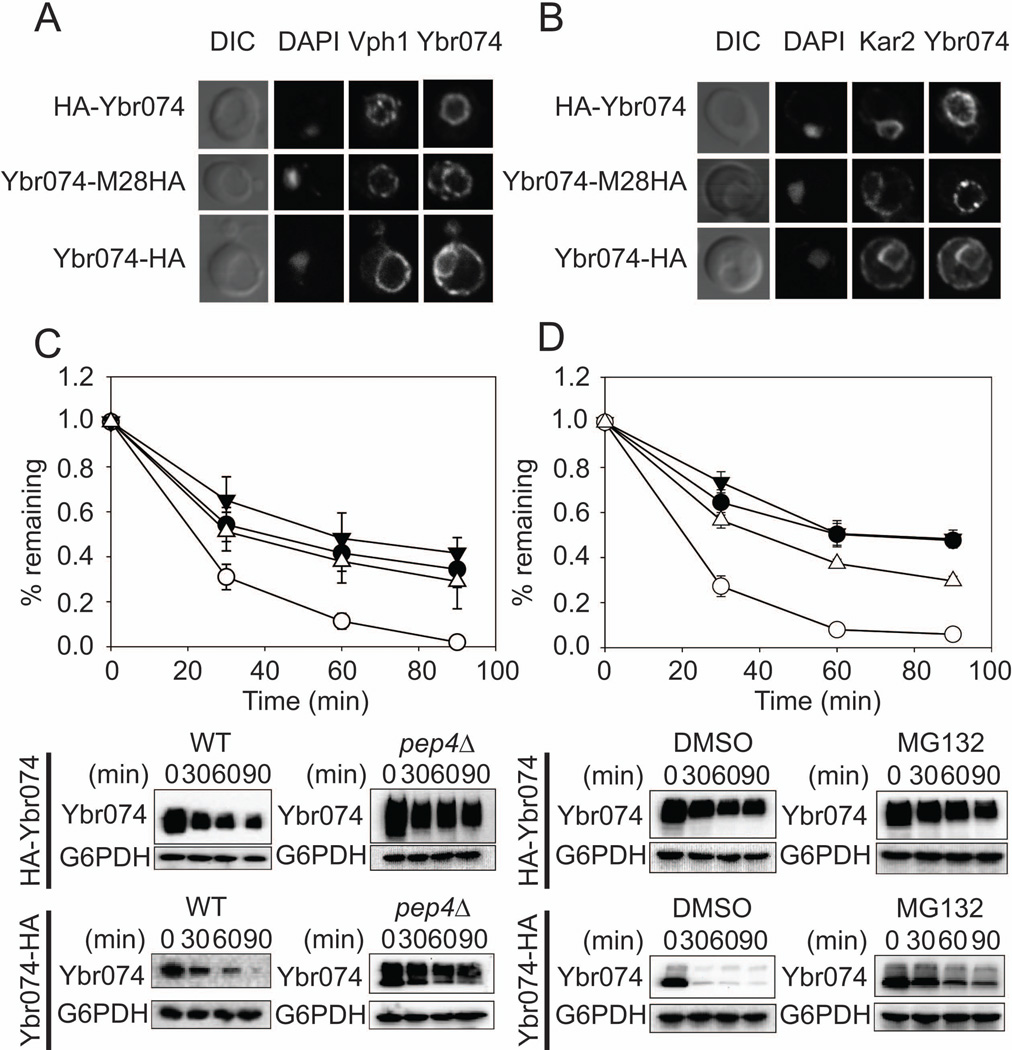
To determine the subcellular localization of Ybr074, the three HA-tagged forms of Ybr074 were subjected to indirect immunofluorescence microscopy. HA-Ybr074 and Ybr074 tagged at the M28 protease domain (Ybr074-M28HA) both strongly co-localized with the vacuolar protein, Vph1 (Kawasaki-Nishi et al, 2001) (Figure 4A). In contrast, the C-terminally HA-tagged Ybr074 construct (Ybr074-HA) co-localized with the ER molecular chaperone, Kar2 (Rose et al, 1989) (Figure 4B). The vacuolar localization of HA-Ybr074 and Ybr074-M28HA was unexpected in light of both the computational predictions and the fact that a C-terminally FLAG-tagged rat ortholog, FXNA, overexpressed in COS7 cells appeared to be ER-localized (Garcia-Rudaz et al 2007). However, it is worth noting that the overexpression of heterologous membrane proteins may cause them to aberrantly accumulate in the ER. Nevertheless, we considered the possibility that the C-terminal placement of the epitope tag on both Ybr074 and FXNA may directly affect their localization. However, indirect immunofluorescence of both N-terminally and C-terminally FLAG-tagged FXNA constructs showed that FXNA co-localizes with the ER marker PDI in COS7 cells, regardless of where the FLAG epitope was appended (Figure S3). Therefore, vacuolar localization appears to be unique to Ybr074. Consistent with this view, live cell imaging of N-terminally GFP-tagged Ybr074 in yeast indicated vacuolar localization (Figure S4), suggesting that vacuolar localization of Ybr074 is not specific to the HA epitope tag. Notably, GFP-Ybr074 fluorescence was present as a vacuolar ring and did not occupy the vacuolar lumen. This is consistent with protease protection data indicating the N-terminal GFP tag is cytosolic (Figure S2). In further agreement with vacuolar localization of Ybr074 observed herein, a previous proteomic study demonstrated that native untagged Ybr074 can be found in highly enriched vacuolar membranes (Wiederhold et al, 2009).
Figure 4.
Ybr074 localization and stability depends on the placement of the HA epitope tag. (A) N-terminally HA-tagged Ybr074 (HA-Ybr074) and M28 metalloprotease domain HA-tagged Ybr074 (Ybr074-M28HA) co-localize with the vacuolar marker Vph1. The vacuole is visualized by Differential Interference Contrast (DIC) imaging, and the nucleus is evident by DAPI staining. (B) The C-terminally HA-tagged form of Ybr074 (Ybr074-HA) co-localizes with the ER chaperone, Kar2. DIC images depict the vacuole. The nucleus is marked with DAPI staining. (C) C-terminally-tagged Ybr074 (Ybr074-HA) is unstable in comparison to the N-terminally tagged form of the protein (HA-Ybr074). Cycloheximide chase analyses were performed as described in Materials and Methods in a wild type (WT) strain expressing HA-Ybr074 (●), or Ybr074-HA (○), and in a pep4Δ strain expressing HA-Ybr074(▼), or Ybr074-HA (Δ), and (D) in a pdr5Δ strain expressing HA-Ybr074 treated with the 26S proteasome inhibitor MG132 (▼) or DMSO (●) or expressing Ybr074-HA treated with MG132 (Δ) or DMSO (○). Quantified data represents the means of independent experiments using three to eight colonies, ± SEM. Where error bars are not visible, the SEM is less than the size of the symbol. Bottom panels are representative images from the chase analyses.
Vacuolar Ybr074 is more stable than ER-localized Ybr074
If the C-terminally tagged form of Ybr074 was illegitimately localized to the ER, we hypothesized that this ER-localized from of Ybr074 may exhibit decreased stability. Proteins that accumulate in the ER and are not ER residents may be degraded by the ER associated degradation (ERAD) pathway in a proteasome-dependent manner (McCracken & Brodsky, 1996; Ward et al, 1995). To test this hypothesis, we conducted cycloheximide chase analyses comparing the stabilities of HA-Ybr074 and Ybr074-HA. In a wild type strain, ER-localized Ybr074-HA was significantly less stable than vacuolar HA-Ybr074 (Figure 4C), indicating that the ER-localized Ybr074-HA is targeted for degradation. To examine whether this degradation is dependent on vacuolar function, cycloheximide chase analyses were conducted in a pep4Δ strain, in which ~90% of the proteolytic activity of the vacuole is compromised (Hemmings et al, 1980). We found that Ybr074-HA was now degraded to the same extent as observed for HA-Ybr074 (Figure 4C). Similarly, inhibition of 26S proteasome activity using MG132 in a drug efflux pump mutant strain, pdr5Δ, resulted in stabilization of Ybr074-HA (Figure 4D). These combined results indicate the aberrantly ER-localized C-terminally HA-tagged form of Ybr074 is unstable and that its degradation is dependent on both vacuolar proteases and the proteasome. The addition of a C-terminal epitope tag to Ybr074 may be contributing to ER retention by altering the conformation of Ybr074 in a way that targets it for degradation.
Ybr074 is required neither for autophagy nor for ER associated degradation
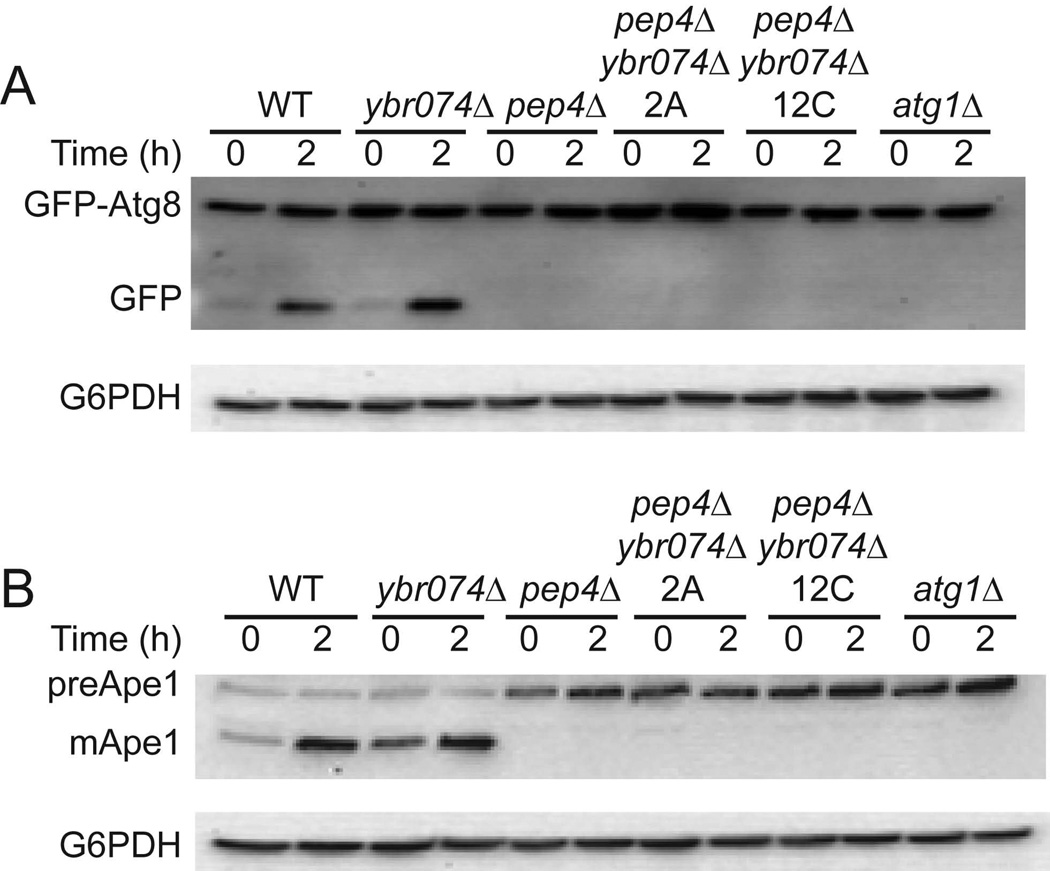
In order to elucidate the biological function of Ybr074, an ybr074Δ strain was challenged using conditions that require vacuolar-mediated osmoregulation (i.e., media supplemented with calcofluor white, congo red, or 1M NaCl) (Nass & Rao, 1999; Ram & Klis, 2006) and conditions known to induce the cytoplasmic and ER protein quality control machineries (i.e., heat stress and media supplemented with DTT or tunicamycin) (Elbein, 1984; Jamsa et al, 1994; Morimoto, 1998). No growth phenotypes were identified under these conditions, suggesting that Ybr074 may be acting redundantly with other yeast proteases. To examine this possibility, candidate genes were disrupted in conjunction with YBR074W, including genes encoding the vacuolar serine protease, Pep4, and another vacuolar M28 metalloprotease, Ape3 (Biagini & Puigserver, 2001; Rawlings et al, 2012). Although growth phenotypes were absent in the double mutant strains, we employed these protease double mutant strains to examine more specifically whether there was an effect on autophagy, particularly since Ybr074 appeared to reside in the yeast vacuole. To this end, we used the GFP-Atg8 reporter to monitor the fusion between autophagosomes and the vacuole under conditions of nitrogen starvation, which triggers a Pep4-dependent clipping of GFP from the GFP-Atg8 fusion protein (Nair et al, 2011). As shown in Figure 5, we found that this event was proficient in each of the strains, except when PEP4 and ATG1 were deleted as controls. We also examined the maturation of the vacuolar protease preApe1 under these same conditions, and found that the Pep4-dependent proteolytic processing of preApe1 was also unaffected by Ybr074 disruption.
Figure 5.
Autophagy is not compromised in a ybr074Δ strain. (A) Cells expressing GFP-Atg8 were subjected to nitrogen starvation conditions for 2 h at 30°C and the amount of released GFP was determined by western blot analysis. Glucose 6-phosphate dehydrogenase (G6PDH) was used as a loading control. Clipping of the GFP-Atg8 construct is shown in wild type and ybr074Δ yeast, but is absent in pep4Δ ybr074Δ pep4Δ strains derived from independent matings (“2A” and “12C”), and an atg1Δ (autophagy deficient) strain. (B) Extracts from the same cells from part (A) were examined for maturation of the vacuolar protease preApe1 by western blot analysis, and G6PDH was used as a loading control.
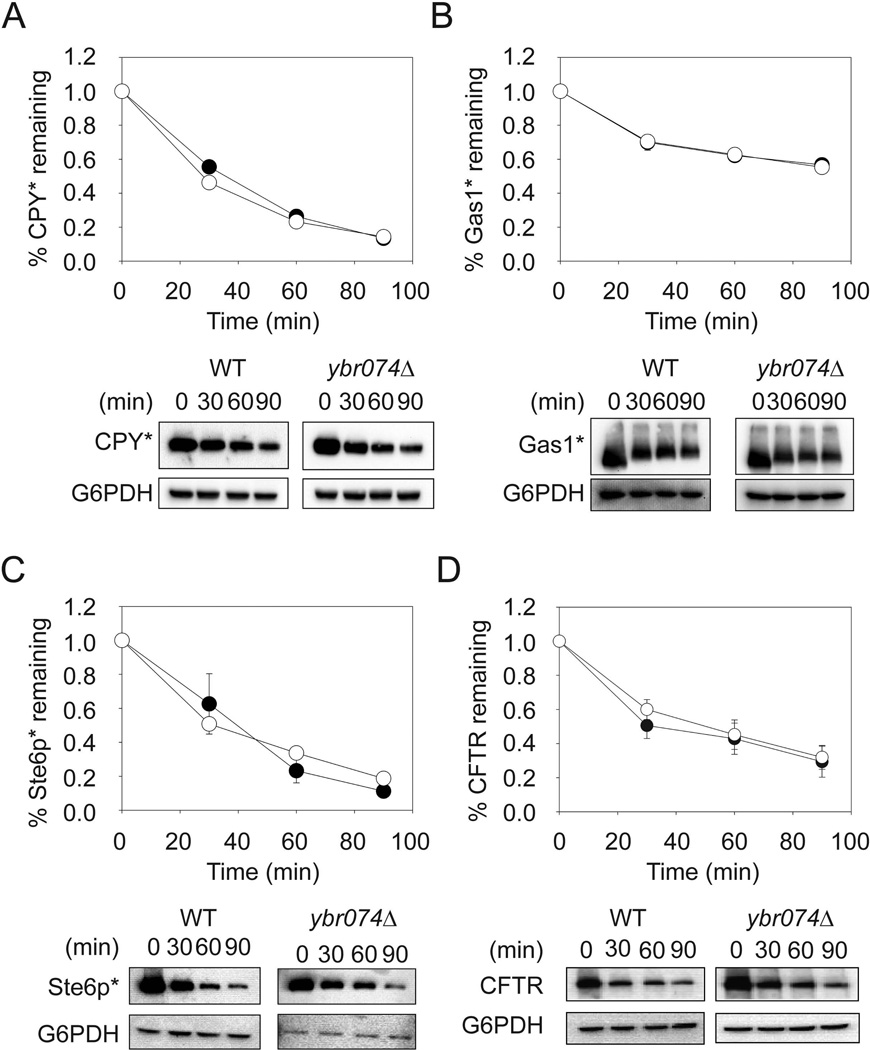
Because Ybr074 harbors a lumenal protease domain and passes through the early secretory pathway, we next assessed the role of Ybr074 in ER protein quality control. To this end, we examined the degradation of four well-characterized ERAD substrates in wild type and ybr074Δ strains. However, there was no significant stabilization of any of these ERAD substrates, whether they were soluble, membrane-anchored, or integral membrane proteins (Figure 6A–D). Consistent with these data, loss of YBR074W failed to activate an unfolded protein response (data not shown). Together, our results suggest that Ybr074 functions in the vacuole to degrade specific substrates and/or acts in a complementary manner with other vacuolar proteases to facilitate protein turnover in this organelle under as yet undefined conditions.
Figure 6.
Ybr074 is not required for the ERAD of diverse substrates. Wild type (●) and ybr074Δ (○) cells expressing (A) CPY*, (B) Gas1*, (C) Ste6p*, or (D) CFTR were subjected to cycloheximide chase analysis. Glucose 6-phosphate dehydrogenase (G6PDH) was used as a loading control. Data represent the means of independent experiments using three to seven colonies, ± SEM. Where error bars are not visible, the SEM is less than the size of the symbol. Bottom panels are representative images for each chase analysis.
Proteomic analysis hints at links between Ybr074 and vacuolar processes
In order to identify factors whose levels are controlled by Ybr074 activity, we conducted a proteomic analysis comparing GFP-tagged protein levels in a wild type and ybr074Δ strain. Proteins that accumulated or showed reduced levels in response to YBR074 deletion are shown in Table S1 and Table S2, respectively. Of particular interest was accumulation of the hydrophillin protein, Gre1, which is upregulated in response to osmotic stress (Garay-Arroyo & Covarrubias, 1999). Since vacuolar functions, including membrane fusion and ion transport, are important in maintaining osmolarity in yeast, accumulation of Gre1 may occur as a result of impaired osmoregulatory function resulting from loss of Ybr074 activity (Hohmann, 2002; Nass & Rao, 1999). Intriguingly, levels of GTPase Activating Protein (GAP), Gyp1, were diminished in response to YBR074W deletion. Gyp1 associates with Rab GTPases Sec4, Ypt1, Ypt7, and Ypt51 (Vps21), and has been shown to inhibit the activity of both Ypt1 (in vivo) and Ypt51 (in vitro) (Brett & Merz, 2008; Du & Novick, 2001). Intriguingly, yeast respond to osmotic stress by inhibiting early Rab-dependent docking and pre-docking events (Brett & Merz, 2008). Furthermore, Ypt51, which is required for sorting of vacuolar proteases, was identified by the Boone lab in a negative genetic interaction with Ybr074 (personal communication) (Robinson et al, 1988; Tong & Boone, 2006). Relieving Gyp1-mediated inhibition of Ypt51 may facilitate trafficking of vacuolar proteases to compensate for loss of Ybr074 activity. Overall, these proteomic data provide further support for a role of Ybr074 in vacuole physiology in the yeast Saccharomyces cerevisiae.
Discussion
Through the collective efforts of yeast researchers, ~ 80% of the yeast genome was characterized as of 2007 (Castillo 2007). However, the endeavor to elucidate the remainder of the yeast genome has been hindered in some cases by gene misannotation. This paper represents the initial characterization of a putative metalloprotease encoded by one such gene, known as YBR074W. As designated by the PANTHER protein classification system, Ybr074 is a member of the FXNA-related Family of Proteins (PTHR12147) (Thomas et al, 2003) and we propose to name this gene Pff1 (Protease in FXNA-related Family 1). The work presented here demonstrates that Pff1 is an N-glycosylated integral membrane protein that resides in the vacuole. We also found that the putative protease domain in Pff1 faces the lumen. As a prelude to future investigations, we then conducted a proteomic analysis that revealed intriguing links between Pff1, vacuolar sorting, and osmoregulation.
Although Pff1 was initially predicted to be an ER-localized protein, we have presented compelling evidence demonstrating vacuolar localization. First, we showed that Pff1, HA-tagged at the N-terminus or adjacent to its protease domain, co-localized with the vacuolar marker Vph1. Second, we showed that this localization was not unique to the HA-epitope tagged form of Pff1, as N-terminally GFP-tagged Pff1 also exhibited vacuolar localization. Third, we showed that C-terminally HA-tagged Pff1, which accumulated in the ER, was highly unstable and degraded. Finally, a previously published proteomic analysis found untagged Pff1 was enriched in a vacuolar membrane preparation (Wiederhold et al, 2009). In contrast to the situation in yeast, we also found that both N- and C-terminally FLAG-tagged forms of FXNA were ER-localized in COS7 cells, consistent with previously published work (Garcia-Rudaz, 2007). In light of this discrepancy between the localization of the yeast and rat orthologs, we noted that while Pff1 was expressed from its endogenous promoter, FXNA was highly overexpressed from the CMV promoter (Chen et al, 2011; Garcia-Rudaz et al, 2007). Since overexpression of FXNA may contribute to its ER retention, re-examining the localization of FXNA expressed from its endogenous promoter may clarify this question. On the other hand, this variation in sub-cellular localization may reflect a species-specific difference in protein targeting. Notably, Pff1 shares 32.5% sequence identity with FXNA across the M28 protease domain, and 29.8% sequence identity across the entire sequence, but Pff1 has an N-terminal transmembrane helix that is absent in FXNA (Figure S1). Furthermore, the C-terminal domains of Pff1 and FXNA are more divergent, sharing only 13% sequence identity (Myers & Miller, 1989). It is possible that these variations may contribute to the differential targeting of Pff1 and FXNA by some mechanism yet to be defined.
Based on our studies, we propose to add Pff1 to the repertoire of the seven known vacuolar proteases. This raises the question of why an eighth protease, such as Pff1, might be expressed in this compartment. To answer this question we considered one characteristic that makes Pff1 unique from other vacuolar proteases. Namely, Pff1 is the only multi-pass transmembrane protease identified in the vacuole. This feature may enable Pff1 to interact with, and potentially modulate the activity of other proteins found in the vacuolar membrane, including vacuolar sorting factors, fusion proteins, and transporters. In fact, the results of our proteomic analysis hint at the possible involvement of Pff1 in vacuolar-mediated osmoregulation. In this context, loss of Pff1 activity might be expected to induce the high osmolarity glycerol pathway or alter vacuolar fusion events under conditions of osmotic stress. Therefore, future work examining these hypotheses may reveal an effect of Pff1 on yeast osmoregulation or vacuolar biology under specific conditions.
Supplementary Material
Acknowledgements
We would like to thank the following people for reagents, advice and helpful discussion: Dr. Charlie Boone, Dr. Vlad Denic, Dr. Charles Barlowe, Dr. Daniel J. Klionsky, Dr. Jeffrey Hildebrand, Dr. Allison O’Donnell, Dr. Christopher J. Guerrero, and Dr. Annette Ahner. This work was supported by the National Institutes of Health Grant GM75061 awarded to J.L.B. Furthermore,V.A.W. acknowledges support from a Howard Hughes Medical Institute Undergraduate Research Fellowship. T.A. and M.S. acknowledge support from the Minerva Foundation and an EMBO Young Investigators Award.
References
- Adams A, Kaiser C. Methods in yeast genetics : a Cold Spring Harbor Laboratory course manual. 1997 edn. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1998. Cold Spring Harbor Laboratory. [Google Scholar]
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends in biochemical sciences. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. The FEBS journal. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini A, Puigserver A. Sequence analysis of the aminoacylase-1 family. A new proposed signature for metalloexopeptidases. Comparative biochemistry and physiology Part B, Biochemistry & molecular biology. 2001;128:469–481. doi: 10.1016/s1096-4959(00)00341-9. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Brachat S, Dietrich FS, Voegeli S, Zhang Z, Stuart L, Lerch A, Gates K, Gaffney T, Philippsen P. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome biology. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. The Journal of cell biology. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Merz AJ. Osmotic regulation of Rab-mediated organelle docking. Current biology : CB. 2008;18:1072–1077. doi: 10.1016/j.cub.2008.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR, Pringle JR, Jones EW. The Molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cole Spring Harbor Laboratory Press; 1991. [Google Scholar]
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. The Journal of cell biology. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic acids research. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Krohn J, Bhattacharya S, Davies B. A comparison of exogenous promoter activity at the ROSA26 locus using a PhiiC31 integrase mediated cassette exchange approach in mouse ES cells. PloS one. 2011;6:e23376. doi: 10.1371/journal.pone.0023376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic acids research. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–159. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- Du LL, Novick P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Molecular biology of the cell. 2001;12:1215–1226. doi: 10.1091/mbc.12.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD. Inhibitors of the biosynthesis and processing of N-linked oligosaccharides. CRC critical reviews in biochemistry. 1984;16:21–49. doi: 10.3109/10409238409102805. [DOI] [PubMed] [Google Scholar]
- Foury F. Human genetic diseases: a cross-talk between man and yeast. Gene. 1997;195:1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Covarrubias AA. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast. 1999;15:879–892. doi: 10.1002/(SICI)1097-0061(199907)15:10A<879::AID-YEA428>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Garcia-Rudaz C, Luna F, Tapia V, Kerr B, Colgin L, Galimi F, Dissen GA, Rawlings ND, Ojeda SR. Fxna, a novel gene differentially expressed in the rat ovary at the time of folliculogenesis, is required for normal ovarian histogenesis. Development. 2007;134:945–957. doi: 10.1242/dev.02795. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. 563–547. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annual review of biochemistry. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hemmings BA, Zubenko GS, Jones EW. Proteolytic inactivation of the NADP-dependent glutamate dehydrogenase in proteinase-deficient mutants of Saccharomyces cerevisiae. Archives of biochemistry and biophysics. 1980;202:657–660. doi: 10.1016/0003-9861(80)90475-0. [DOI] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiology and molecular biology reviews : MMBR. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic acids research. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Advances in Applied Mathematics. 1991;12:337–357. [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jamsa E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nature methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- Karathia H, Vilaprinyo E, Sorribas A, Alves R. Saccharomyces cerevisiae as a model organism: a comparative study. PloS one. 2011;6:e16015. doi: 10.1371/journal.pone.0016015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. The Journal of biological chemistry. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lambert N, Freedman RB. The latency of rat liver microsomal protein disulphide-isomerase. The Biochemical journal. 1985;228:635–645. doi: 10.1042/bj2280635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochimica et biophysica acta. 2009;1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic acids research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. The Journal of cell biology. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic acids research. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunaga T, Katakura Y, Miura T, Maruyama Y. Purification and characterization of yeast protein disulfide isomerase. Journal of biochemistry. 1990;108:846–851. doi: 10.1093/oxfordjournals.jbchem.a123291. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes & development. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Approximate matching of regular expressions. Bulletin of mathematical biology. 1989;51:5–37. doi: 10.1007/BF02458834. [DOI] [PubMed] [Google Scholar]
- Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 2011;7:1546–1550. doi: 10.4161/auto.7.12.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in biochemical sciences. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K, Brodsky JL. in vitro reconstitution of the selection, ubiquitination, and membrane extraction of a polytopic ERAD substrate. Methods Mol Biol. 2010;619:365–376. doi: 10.1007/978-1-60327-412-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass R, Rao R. The yeast endosomal Na+/H+ exchanger, Nhx1, confers osmotolerance following acute hypertonic shock. Microbiology. 1999;145(Pt 11):3221–3228. doi: 10.1099/00221287-145-11-3221. [DOI] [PubMed] [Google Scholar]
- Newman AP, Groesch ME, Ferro-Novick S. Bos1p, a membrane protein required for ER to Golgi transport in yeast, co-purifies with the carrier vesicles and with Bet1p and the ER membrane. The EMBO journal. 1992;11:3609–3617. doi: 10.1002/j.1460-2075.1992.tb05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic acids research. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nature protocols. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic acids research. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Salvesen G. Handbook of proteolytic enzymes [electronic resource] 3rd edn. [S.l.]. Academic Press; 2013. [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and cellular biology. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Sbardella D, Fasciglione GF, Gioia M, Ciaccio C, Tundo GR, Marini S, Coletta M. Human matrix metalloproteinases: an ubiquitarian class of enzymes involved in several pathological processes. Molecular aspects of medicine. 2012;33:119–208. doi: 10.1016/j.mam.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Current opinion in structural biology. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings / International Conference on Intelligent Systems for Molecular Biology ; ISMB International Conference on Intelligent Systems for Molecular Biology. 1998;6:175–182. [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Molecular biology of the cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Auerbach D, Stagljar I. Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond. BioTechniques. 2006;40:625–644. doi: 10.2144/000112151. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic acids research. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL. J domain co-chaperone specificity defines the role of BiP during protein translocation. The Journal of biological chemistry. 2010;285:22484–22494. doi: 10.1074/jbc.M110.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wiederhold E, Gandhi T, Permentier HP, Breitling R, Poolman B, Slotboom DJ. The yeast vacuolar membrane proteome. Molecular & cellular proteomics : MCP. 2009;8:380–392. doi: 10.1074/mcp.M800372-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.