SUMMARY
Eukaryotic cells sterilize the cytosol by using autophagy to route invading bacterial pathogens to the lysosome. During macrophage infection with Mycobacterium tuberculosis, a vacuolar pathogen, exogenous induction of autophagy can limit replication, but the mechanism of autophagy targeting and its role in natural infection remain unclear. Here we show that phagosomal permeabilization mediated by the bacterial ESX-1 secretion system allows cytosolic components of the ubiquitin-mediated autophagy pathway access to phagosomal M. tuberculosis. Recognition of extracelluar bacterial DNA by the STING-dependent cytosolic pathway is required for marking bacteria with ubiquitin, and delivery of bacilli to autophagosomes requires the ubiquitin-autophagy receptors p62 and NDP52 and the DNA-responsive kinase TBK1. Remarkably, mice with monocytes incapable of delivering bacilli to the autophagy pathway are extremely susceptible to infection. Our results reveal an unexpected link between DNA sensing, innate immunity, and autophagy and indicate a major role for this autophagy pathway in resistance to M. tuberculosis infection.
INTRODUCTION
Autophagy is an evolutionarily conserved process in eukaryotes whereby cytoplasmic components are enveloped and sequestered by membranous structures that subsequently fuse to lysosomes for degradation. In response to nutrient-limiting conditions, general autophagy serves a catabolic function by mediating nonselective consumption of organelles and other cellular components to generate substrates for energy metabolism and protein synthesis (Deretic and Levine, 2009). In contrast, selective autophagy functions to specifically renovate the cell by targeting protein aggregates and specific organelles for removal through the use of ubiquitin-mediated targeting (Youle and Narendra, 2011). For example, damaged mitochondria are designated for destruction by targeting only the nonfunctional organelles to the lysosome via the autophagy pathway, a process termed mitophagy. Ubiquitin-tagged mitochondria are directed into the general autophagy pathway via the action of the adaptor protein p62, which binds both ubiquitin and the autophagosome-associated protein LC3, although other factors are probably required. Once LC3 is targeted to the cargo, other components of the general autophagy pathway, including ATG proteins such as ATG5, function to form autophagosomes and deliver the organelle to the lysosome (Youle and Narendra, 2011).
Autophagy also plays an important role in innate defense against invading intracellular pathogens (Deretic and Levine, 2009). The prevailing view is that autophagy functions to eliminate intracellular microbes that enter into the cytosol by sequestering them into autophagosomes and delivering them to the lysosome. Furthermore, some pathogens employ autophagy evasion mechanisms that are critical for long-term, persistent infection (Kudchodkar and Levine, 2009). Previous studies of Salmonella enterica serovar Typhimurium (S. Typhimurium) and Listeria monocytogenes infection of cultured epithelial cells have shown that bacteria that exit the endosomal pathway and enter into the cytosol are ubiquitinated and delivered to autophagosomes via recognition by the cytosolic autophagy receptors p62 and NDP52 (Thurston et al., 2009; Zheng et al., 2009). Yet how cytosolic bacteria are recognized and targeted for ubiquitination is currently unknown.
Much of the groundbreaking work on the role of autophagy in mycobacterial clearance was performed using Mycobacterium bovis Bacille Calmette-Gue´ rin (BCG), the attenuated vaccine strain (Gutierrez et al., 2004; Singh et al., 2006). Curiously, in these studies, targeting of LC3 to BCG-containing vacuoles required exogenous stimulation of autophagy. Although this vaccine strain has been extremely helpful in modeling many basic functions of M. tuberculosis, it lacks several virulence factors, including the type VII secretion system ESX-1 (Mahairas et al., 1996; Pym et al., 2003). This is a key difference between M. tuberculosis and BCG, as mutants lacking ESX-1 are defective for replication within macrophages, are severely attenuated in animal models of infection, and fail to activate innate immunesignaling responses of macrophages (Guinn et al., 2004; Hsu et al., 2003; Stanley et al., 2003; Wong and Jacobs, 2011). Furthermore, BCG does not undergo selective autophagy and recruitment of LC3 to the phagosomal membrane unless autophagy is experimentally induced (Gutierrez et al., 2004; Singh et al., 2006; Zhao et al., 2008). During infection with Mycobacterium marinum, an ectothermic pathogen related to M. tuberculosis, ESX-1 secretion is required for vacuolar escape (Smith et al., 2008) and for subsequent localization of ubiquitin (Collins et al., 2009) and LC3 (Lerena and Colombo, 2011) to the bacterial surface. However, unlike M. marinum, M. tuberculosis remains membrane bound, although eventual escape has been observed late in infection (van der Wel et al., 2007). Although inducing autophagy exogenously via starvation, treatment with rapamycin, interferon gamma (IFN-γ), or vitamin D3 or genetic depletion of autophagy inhibitors can lead to decreased bacterial replication in macrophages (Gutierrez et al., 2004; Kumar et al., 2010; Singh et al., 2006; Yuk et al., 2009), how M. tuberculosis interfaces with the selective autophagy pathway from within the phagosome, and the contribution of autophagic targeting by macrophages to host resistance, is unknown.
Here we report that wild-type (WT) M. tuberculosis cells elicit ubiquitin-mediated autophagy targeting in resting macrophages, resulting in the delivery of bacilli to lysosomes. Targeting requires both the bacterial ESX-1 system and the host cytosolic DNA-sensing pathway, revealing a novel link between nucleic acid sensing and selective autophagy of intracellular pathogens. Remarkably, we show that autophagy is a major mechanism of host control during M. tuberculosis infection in vivo.
RESULTS
Autophagic Targeting of M. tuberculosis
To determine whether M. tuberculosis is specifically targeted by selective autophagy, we first examined the dynamics of the autophagosome-specific marker LC3 over the course of WT M. tuberculosis infection of naive macrophages. Primary murine bone marrow-derived macrophages (BMDMs) derived from GFP-LC3 transgenic mice were infected with M. tuberculosis expressing mCherry, and localization of GFP-LC3 was analyzed via microscopy at defined times after infection. Two hours after infection, ~15% of intracellular bacteria colocalized with GFP-LC3, and by 4 hr this increased to ~30% of the bacterial population (Figure 1A, top panels and Figure 1B). Although the number of small GFP-LC3 puncta increased during the infection, targeting of LC3 to larger structures in the cell occurred exclusively at the M. tuberculosis phagosome. Three-dimensional confocal imaging of these cells revealed that GFP-LC3 envelops the entire M. tuberculosis phagosome (Movie S1 available online). Similar results were observed during infection of the macrophage-like cell line RAW 264.7 stably expressing GFP-LC3 (Figures S1A and S1B), as well as BMDMs immunostained with an antibody specific for endogenous LC3 (Figures S1C and S1D). M. tuberculosis also colocalized with another autophagy protein, ATG12, in both BMDMs (Figures 1C and 1D) and RAW 264.7 cells (Figures S1G and S1H). Western blot analysis of endogenous LC3 during M. tuberculosis infection revealed an increase in conversion of LC3-I to LC3-II, consistent with autophagy activation (Figure 1E). In addition, BMDMs from Atg5flox/flox-Lyz-Cre mice (Zhao et al., 2008), which contain a genomic deletion of Atg5 within monocytes/macrophages (hereafter referred to as Atg5− mice), were unable to recruit LC3 to phagosomes containing M. tuberculosis (Figure S2). Thus, WT M. tuberculosis is specifically recognized and targeted by the autophagy pathway in the absence of autophagy inducers.
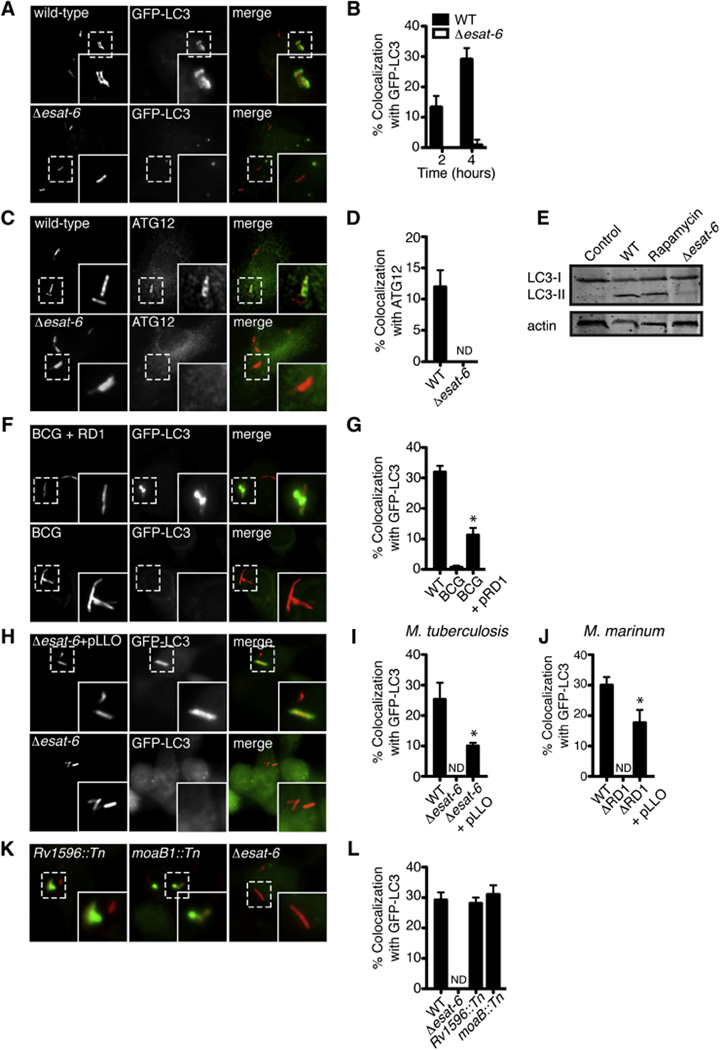
Figure 1. M. tuberculosis Targeting to Autophagosomes Requires Phagosomal Permeabilization via ESX-1.
(A) Fluorescence images of GFP-LC3 BMDMs (green) infected for 4 hr with mCherry-expressing WT or Δesat-6 M. tuberculosis (red).
(B) Quantitative analysis of GFP-LC3 colocalization with WT and Δesat-6 M. tuberculosis at indicated times after infection. Results are the means ± SEM of three independent experiments.
(C) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT or Δesat-6 M. tuberculosis (red) and immunostained with anti-ATG12 (green).
(D) Quantitative analysis of colocalization with ATG12 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(E) Western blot analysis of LC3 and actin (loading control) of BMDMs infected for 4 hr with WT M. tuberculosis.
(F) Fluorescence images of RAW 264.7 cells stably expressing GFP-LC3 (green) infected for 6 hr with either mCherry-expressing BCG or BCG complemented with RD1 (red).
(G) Quantitative analysis of GFP-LC3 colocalization with WT M. tuberculosis, BCG, or BCG complemented with RD1 at 6 hr post-infection. Results are the means ± SEM of three independent experiments (n = 3 per group, *p < 0.005).
(H) Fluorescence images of GFP-LC3 RAW 264.7 cells (green) infected with the Δesat-6 M. tuberculosis (red) or Δesat-6 M. tuberculosis expressing listeriolysin-O (LLO) (red) (n = 3 per group, *p < 0.005).
(I) Quantitative analysis of GFP-LC3 colocalization with WT M. tuberculosis, Δesat-6 M. tuberculosis, and Δesat-6 M. tuberculosis expressing LLO. Results are the means ± SEM of three independent experiments (n = 3 per group, *p < 0.005).
(J) Quantitative analysis of GFP-LC3 colocalization with WT M. marinum, ΔRD1 M. marinum, and ΔRD1 M. marinum expressing LLO. Results are the means ± SEM of three independent experiments (n = 3 per group, *p < 0.005).
(K) Fluorescence images of GFP-LC3 RAW 264.7 cells (green) infected for 6 hr with Rv1596::Tn, moaB::Tn and Δesat-6 M. tuberculosis (red).
(L) Quantitative analysis of GFP-LC3 colocalization with WT, Δesat-6 Rv1596::Tn, and moaB::Tn M. tuberculosis. Results are the means ± SEM of three independent experiments.
See also Figure S1 and Movie S1.
Because BCG lacks genes encoding ESX-1 (Mahairas et al., 1996; Pym et al., 2003) and previous work showed that ESX-1 is required for autophagy targeting of M. marinum (Lerena and Colombo, 2011), we tested whether this secretion system was required for native LC3 targeting to M. tuberculosis during infection. Whereas infection with BCG induced no LC3 recruitment, restoration of ESX-1 secretion via complementation with the RD1 locus of M. tuberculosis led to a partial yet significant increase in targeting during infection of GFP-LC3 RAW 264.7 cells (Figures 1F and 1G). Likewise, an M. tuberculosis mutant defective in ESX-1 secretion, Δesat-6, failed to recruit LC3 (Figures 1A, bottom panels and 1B) and did not induce LC3 processing (Figure 1C). Consistent with previous studies, WT M. marinum also colocalized with GFP-LC3 in an ESX-1-dependent manner (Figures S1E and S1F) (Lerena and Colombo, 2011). These data indicate that the ESX-1 secretion system is required for targeting mycobacteria to autophagosomes during infection.
Mounting evidence strongly suggests that ESAT-6, the major ESX-1-secreted substrate, has membrane-damaging activity (de Jonge et al., 2007; Hsu et al., 2003; Smith et al., 2008) that functions to permeabilize the phagosomal membrane, allowing access of the bacterium to the cytosol (Stamm et al., 2003; van der Wel et al., 2007). Because pore-forming toxins such as listeriolysin-O (LLO) are required for cytosolic access and to induce autophagy during L. monocytogenes infection (Birmingham et al., 2007; Py et al., 2007), we hypothesized that ESX-1-mediated autophagy was due to ESAT-6 pore formation. To test this, we created an ESX-1 mutant strain that expressed and secreted an autoactivated form of the heterologous poreforming toxin LLO (Singh et al., 2008). Expression of LLO in either M. tuberculosis or M. marinum partially restored targeting of GFP-LC3 to ESX-1 mutants (Figures 1H–1J), suggesting that the requirement of ESX-1 secretion in autophagy targeting is pore formation of the phagosomal membrane.
During macrophage infection, the majority of M. tuberculosis cells block phagosomal maturation into lysosomes (Armstrong and Hart, 1971), a process that requires a functional ESX-1 secretion system (MacGurn and Cox, 2007; Tan et al., 2006), and the bacilli reside and grow in an endosome-like compartment (Sturgill-Koszycki et al., 1996). To test whether bacteria colocalized with LC3 due to this block in intracellular trafficking, we infected GFP-LC3 RAW macrophages with M. tuberculosis mutants that have functional ESX-1 secretion systems but fail to inhibit phagolysosome fusion (Brodin et al., 2010; MacGurn and Cox, 2007). We observed that both Rv1506c::Tn and moeB1::Tn mutant cells, deficient in molybdopterin and acytrehalose-containing glycolipids, respectively, colocalized with GFP-LC3 to a similar extent as WT M. tuberculosis (Figures 1K and 1L). Triple-labeling experiments demonstrated that these GFP-LC3+ bacteria also colocalized with ubiquitin (Figure S1I). Collectively, these results further demonstrate that the ESX-1 type VII secretion system serves to expose phagosome-bound M. tuberculosis to the cytosol, where the bacilli are recognized and targeted to autophagic compartments.
Role of LC3 and Ubiquitin Adaptors in M. tuberculosis Targeting to the Autophagy Pathway
To understand the mechanism by which LC3 is recruited to ESX-1+ M. tuberculosis, we tested whether p62 and NDP52 are required for targeting bacilli to autophagosomes. As shown in Figures 2A–2D, ~20%–25% of the M. tuberculosis population recruited both p62 and NDP52 to phagosomes in an ESX-1-dependent fashion. Triple-labeling experiments showed that ~70% of the LC3+ population also localized with both p62 and NDP52 (Figures 2E and 2F). Knockdown of either p62 or NDP52 expression in GFP-LC3 RAW 264.7 cells (Figure S3) resulted in a dramatic decrease in LC3 colocalization (Figure 2G). Thus, as with S. Typhimurium, both adaptors are required for autophagy targeting of M. tuberculosis (Cemma et al., 2011).
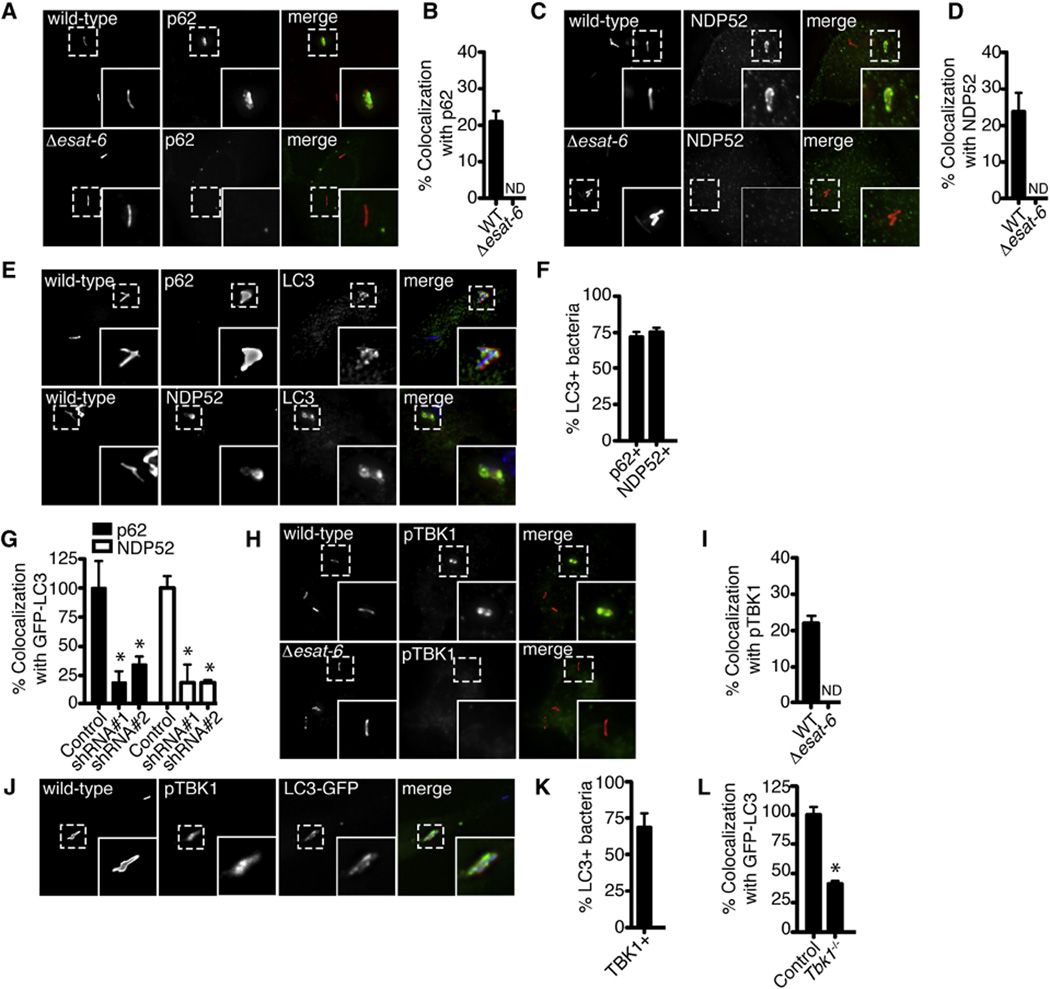
Figure 2. The Autophagy Receptors p62 and NDP52 and the Kinase TBK1 Are Required for Efficient Delivery of M. tuberculosis to Autophagosomes.
(A) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT or Δesat-6 M. tuberculosis (red) and immunostained with anti-p62 (green).
(B) Quantitative analysis of colocalization with p62 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(C) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT or Δesat-6 M. tuberculosis (red) and immunostained with anti-NDP52 (green).
(D) Quantitative analysis of colocalization with NDP52 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(E) Fluorescence images of BMDMs infected with WT M. tuberculosis (blue) for 4 hr and immunostained with anti-LC3 (green), anti-p62 (red), or anti-NDP52 (red).
(F) Quantitative analysis of M. tuberculosis colocalization with p62, NDP52 and LC3 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(G) Quantitative analysis of GFP-LC3 colocalization with WT M. tuberculosis after shRNA knockdown of p62 or NDP52 in GFP-LC3 RAW 264.7 cells. Results are the means ± SEM of three independent experiments. Data are expressed as a percentage relative to control (scrambled shRNA) knockdown cells (n = 3 per group, *p < 0.006). See also Figure S3.
(H) Fluorescence images of BMDMs infected with either mCherry-expressing WT or Δesat-6 M. tuberculosis (red) for 4 hr and immunostained with anti-phospho- TBK1 (red).
(I) Quantitative analysis of M. tuberculosis colocalization with phospho-TBK1 and LC3 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(J) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT M. tuberculosis (blue) and immunostained with anti-LC3 (green) and antiphospho-TBK1 (red) (n = 3 per group, *p < 0.01).
(K) Quantitative analysis of M. tuberculosis colocalization with TBK1 and LC3 at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
(L) Quantitative analysis of BMDMs from Tbk1+/+ and Tbk1−/− mice infected with WT M. tuberculosis for 4 hr and immunostained with anti-LC3 antibodies. Results are the means ± SEM of three independent experiments.
NDP52, in addition to binding LC3, interacts with NAP1 and SINTBAD to recruit Tank-binding kinase (TBK1) (Ryzhakov and Randow, 2007). TBK1 is necessary for transcriptional induction of type I interferon (IFN) during WT M. tuberculosis infection (Stanley et al., 2003) but is also required for selective targeting of salmonella to autophagy (Wild et al., 2011a). To determine whether TBK1 is also important in autophagy targeting of M. tuberculosis, we stained infected BMDMs with antibodies specific for the activated, phosphorylated form of TBK1. As shown in Figures 2H and 2I, activated TBK1 colocalized to WT M. tuberculosis but not to ESX-1 mutants, and dual-labeling experiments showed that the majority of the TBK1+ bacteria colocalized with LC3 (Figures 2J and 2K). Importantly, BMDMs from Tbk1−/− mice had a 60% reduction in their ability to target LC3 to M. tuberculosis during infection (Figure 2L), demonstrating that the kinase plays an important role in both transcriptional and autophagic innate immune responses to virulent M. tuberculosis.
LC3-Positive M. tuberculosis Cells Are Ubiquitinated
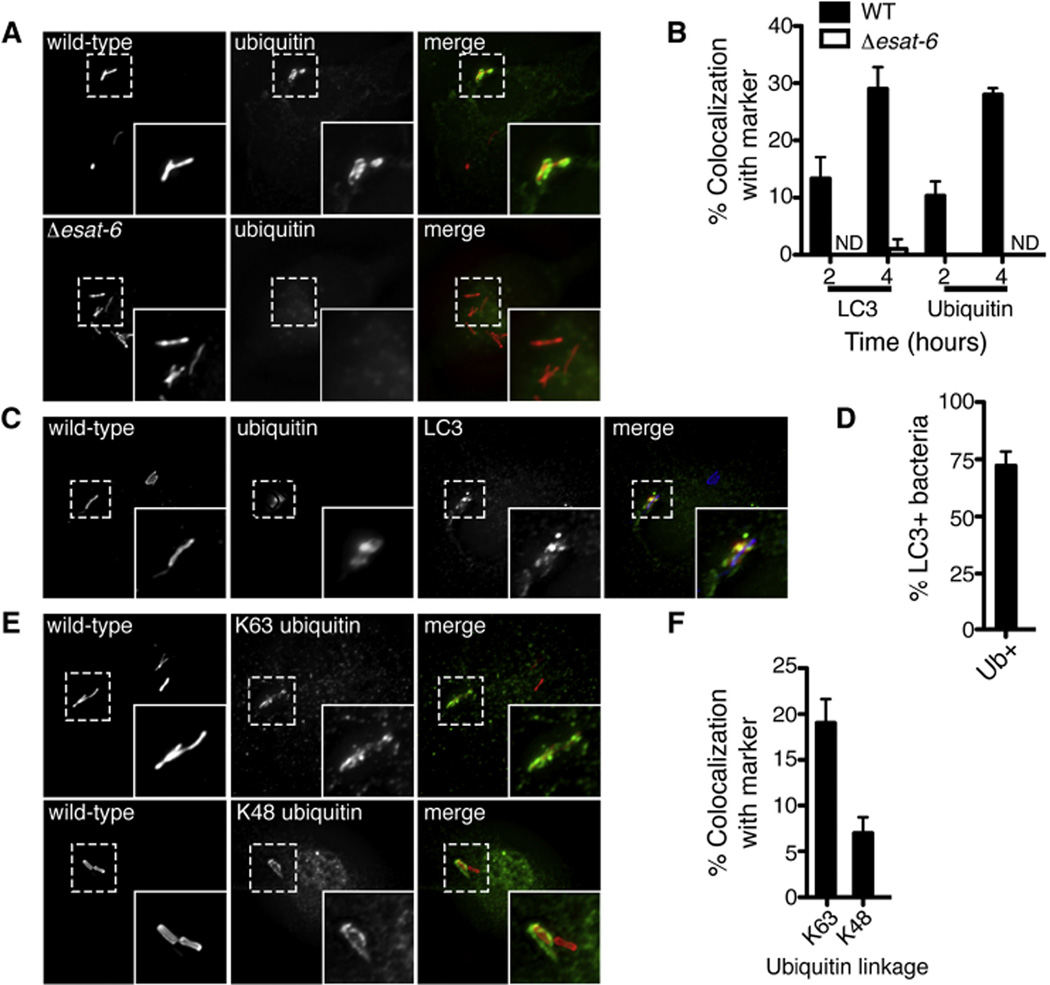
Because both p62 and NDP52 are recruited to ubiquitinated substrates, we tested whether M. tuberculosis colocalized with host ubiquitin by immunostaining with anti-ubiquitin antibodies that recognize conjugated mono- and polyubiquitin. As shown in Figures 3A and 3B, ~30% of WT M. tuberculosis colocalized with ubiquitin at 4 hr post-infection, whereas ESX-1 mutant bacteria did not colocalize with ubiquitin. Coimmunostaining for both ubiquitin and LC3 revealed that ~70% of all WT M. tuberculosis that were coated with ubiquitin also recruited LC3 (Figures 3C and 3D). Using antibodies that recognize specific linkages of polyubiquitin chains, we found that ~20% of the total M. tuberculosis population was associated with K63-linked ubiquitin, whereas ~7% was K48 linked (Figures 3E and 3F). Importantly, ubiquitin recruitment was unaffected in Atg5− macrophages, indicating that ubiquitin acquisition precedes the recruitment of LC3 and that the observed LC3 colocalization is specific to the ubiquitin-mediated autophagy pathway (Figures S2A and S2B).
Figure 3. Mycobacterial ESX-1 System Is Required for Ubquitin Colocalization.
(A) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT or Δesat-6 M. tuberculosis (red) and immunostained with antiubiquitin antibodies (green).
(B) Quantitative analysis of ubiquitin and LC3 colocalization with WT or Δesat-6 M. tuberculosis at indicated times post-infection. Results are the means ± SEM of three independent experiments.
(C) Fluorescence images of BMDMs infected for 4 hr with mCherry-expressing WT M. tuberculosis (blue) and immunostained with anti-LC3 (green) and anti-ubiquitin antibodies (red).
(D) Quantitative analysis of M. tuberculosis colocalization with ubiquitin and LC3 at 4 hr postinfection. Results are the means ± SEM of three independent experiments.
(E) Fluorescence images of BMDMs infected with mCherry-expressing WT M. tuberculosis and immunostained with anti-K63 and anti-K48 ubiquitin antibodies (green).
(F) Quantitative analysis of K63 and K48 ubiquitin colocalization in BMDMs infected with WT M. tuberculosis at 4 hr post-infection. Results are the means ± SEM of three independent experiments.
See also Figure S2.
Cytosolic DNA Activates Selective Autophagy
Next, we sought to determine the microbial signal(s) necessary for host recognition and targeting of ubiquitin-mediated selective autophagy to M. tuberculosis. We found previously that M. tuberculosis extracellular DNA (eDNA) is exposed to the host cytosol during macrophage infection, activating TBK1 to elicit the production of type I IFNs (Manzanillo et al., 2012). Although how eDNA is liberated and exposed during M. tuberculosis infection is unknown, a strong body of literature supports the notion that eDNA plays important roles in normal bacterial physiology, most notably biofilm formation (Whitchurch et al., 2002). Given the necessity of TBK1 in both autophagic targeting of M. tuberculosis and activation of type I IFNs in response to M. tuberculosis DNA (Stanley et al., 2007), we sought to test whether DNA sensing is necessary and sufficient to mediate autophagy recruitment during infection of macrophages.
We first explored whether transfection of purified doublestranded DNA (dsDNA) into the cytosol could trigger ubiquitinmediated selective autophagy. Indeed, transfection of plasmid DNA into LC3-GFP BMDMs resulted in production of autophagic puncta to an extent similar to that seen with rapamycin treatment (Figure 4A), and induced processing of LC3 in an ATG5-depen-dent manner (Figure 4B). This is in agreement with recent studies showing that dsDNA viruses (HSV-1, HCMV) can induce robust LC3 lipidation (McFarlane et al., 2011; Rasmussen et al., 2011) and suggests that cytosolic dsDNA, in addition to activating tran-scriptional responses, is a potent inducer of autophagy.
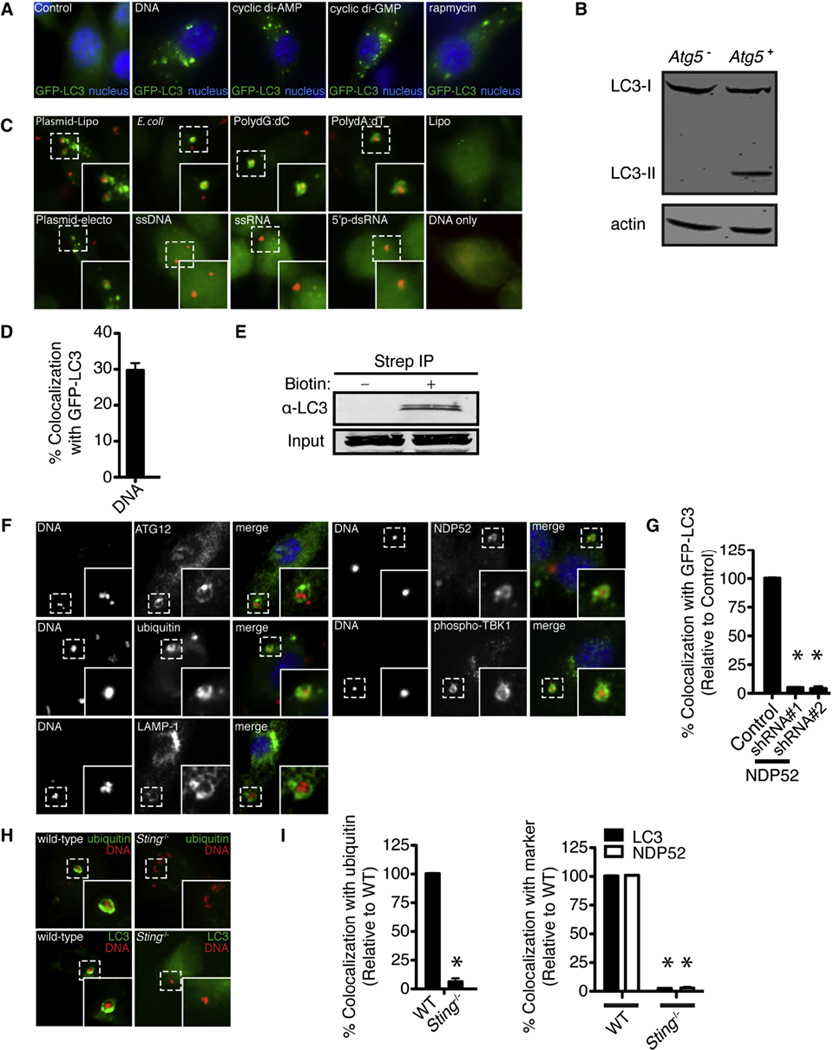
Figure 4. Cytosolic DNA Is Targeted by Ubiquitin-Mediated Selective Autophagy and Requires NDP52 and STING.
(A) Fluorescence images of GFP-LC3 RAW 264.7 cells (green) at 4 hr post-transfection with lipofectamine alone (control), plasmid DNA, cyclic di-AMP, or cyclic di-GMP or treated with rapamycin.
(B) Western blot analysis of LC3 in Atg5+ and Atg5− BMDMs transfected with DNA for 3 hr.
(C) Fluorescence images of GFP-LC3 RAW 264.7 cells at 4 hr post-transfection with Cy3-labeled nucleic acid. Plasmid DNA was introduced by either lipofection (plasmid-lipo) or electroporation (plasmid-electro), and all the other nucleic acid species were introduced by lipofection. Cells were also treated with lipofectamine reagent alone (lipo) or DNA without transfection reagent (DNA only).
(D) Quantitative analysis of Cy3-labeled plasmid DNA with GFP-LC3 RAW 264.7 cells at 4 hr post-transfection. Results are the means ± SEM of three independent experiments.
(E) Western blot analysis of LC3 after streptavidin immunoprecipitation of cell lysates 4 hr post-transfection of dsDNA or biotintylated dsDNA.
(F) Fluorescence images of BMDMs transfected with Cy3-labeled plasmid DNA (red) and immunostained with anti-ATG12, anti-ubiquitin, anti-LAMP-1, anti-NDP52, or anti-phospho-TBK1 antibodies (green).
(G) Quantitative analysis of GFP-LC3 colocalization with Cy3-labeled plasmid DNA at 4 hr post-transfection after shRNA knockdown of NDP52 in GFP-LC3 RAW 264.7 cells. Results are the means ± SEM of three independent experiments. Data are expressed as a percentage relative to control knockdown cells (n = 3 per group, *p < 0.005).
(H) Fluorescence images of WT and Sting−/− BMDMs at 4 hr post-transfection with Cy3-labeled plasmid DNA (red) and immunostained with anti-ubiquitin or anti-LC3 antibodies.
(I) Quantitative analysis of ubiquitin, NDP52, and LC3 colocalization with Cy3-labeled plasmid DNA at 4 hr post-transfection in WT and Sting−/−BMDMs. Results are the means ± SEM of three independent experiments. Data are expressed as a percentage relative to control knockdown cells (n = 3 per group, *p < 0.005).
See also Figure S4.
Previous work showed that transfected dsDNA is not distributed heterogeneously within the cell but instead exists in discrete cytoplasmic foci (Hagstrom et al., 1997). We took advantage of this phenomenon to determine whether dsDNA induced a general, nonspecific autophagic response in cells or whether the nucleic acid itself was targeted for selective autophagy. As seen in Figures 4C and 4D, Cy3-dsDNA appeared within LC3-positive vesicles during lipofection or electroporation of GFP-LC3 RAW 264.7cells but not in cells incubated with DNA in the absence of transfection reagents. Immunopreciptation experiments with biotinylated DNA resulted in the recovery of an LC3-DNA protein complex (Figure 4E), suggesting that cytosolic DNA recruits LC3-positive autophagic vesicles after transfection. Furthermore, DNA also appeared within ATG12- and LAMP-1 positive vesicles (Figure 4F), suggesting that cytosolic DNA is targeted to autophagosomes that subsequently fuse with the lysosome. Transfection of either dsDNA species such as poly(dG:dC), poly(dA:dT), E. coli genomic DNA, or plasmid DNA resulted in robust induction of autophagy as well as targeting to autophagosomes, whereas single-stranded DNA (ssDNA) was not targeted (Figures 4C and Figure S5A). These requirements for autophagy induction by DNA are identical to the immunostimulatory properties of cytosolic DNA (Stetson and Medzhitov, 2006). Furthermore, dsRNA and ssRNA did not result in autophagic autophagosome formation or targeting (Figures 4C and Figure S4A), despite their ability to activate a similar immune response in host cells (Takeuchi and Akira, 2010), showing that the induction of autophagy is specific to dsDNA.
Cytosolic DNA Is Directly Targeted for Autophagy via Ubiquitin-Mediated Selective Autophagy and STING
Because TBK1 kinase activity is stimulated by cytosolic DNA and the protein physically interacts with NDP52, we hypothesized that cytosolic DNA also activates ubiquitin-mediated selective autophagy. As shown in Figure 4F, ubiquitin, TBK1, and NDP52 all colocalized to DNA puncta by 4 hr after transfection. GFP-LC3-positive dsDNA puncta also colocalized with ubiquitin, and both K48 and K63 linkages were observed (Figures S4C–S4F). Furthermore, knockdown of NDP52 resulted in reduced targeting of DNA into LC3 autophagosomes (Figure 4G), further suggesting that DNA is targeted to LC3-positive vesicles via ubiquitin-mediated selective autophagy.
STING is an essential adaptor protein that functions upstream of TBK1 in the interferon stimulatory DNA pathway (Barber, 2011). STING colocalizes with LC3 during DNA stimulation (Saitoh et al., 2009) and is required for LC3 lipidiation in response to herpes virus DNA (Rasmussen et al., 2011). Thus, we next sought to determine whether STING directly mediates ubiquitin-selective autophagy in response to cytosolic DNA. In contrast to WT BMDMs, Sting−/− macrophages were unable to recruit ubiquitin, LC3, and NDP52 to transfected dsDNA (Figures 4H and 4I). Furthermore, direct activation of STING with either cyclic di-AMP or cyclic di-GMP (Burdette et al., 2011) activated GFP-LC3 puncta formation to a similar extent as cytosolic DNA stimulation and rapamycin treatment (Figure 4A), suggesting that direct activation of STING is sufficient to induce autophagsome formation. Taken together, we conclude that activation of STING serves a critical role during the initial steps of ubiquitin-mediated autophagy of DNA.
Cytosolic DNA Sensing Mediates Targeting of M. tuberculosis to Autophagosomes
Because STING is activated within macrophages by M. tuberculosis (Manzanillo et al., 2012), we hypothesized that mycobacterial DNA is the ligand that triggers ubiquitin-mediated selective autophagy of M. tuberculosis during infection. As shown in Figures 5A and 5B, Sting−/− BMDMs displayed reduced ubiquitin-M. tuberculosis colocalization and were severely defective in targeting LC3 to bacteria. In addition, M. tuberculosis colocalization with NDP52 and phospho-TBK1 was undetectable during infection of Sting−/− BMDMs (Figure 5C). Ubiquitin and LC3 localization to M. tuberculosis was normal in macrophages deficient for inflammasomes (Asc1−/−and Nalp3−/−; Shi et al., 2012), TLR signaling (Myd88−/− Trif−/−), or type I IFN signaling (Irf3−/−Irf7−/−, Ifnar1−/−), indicating that targeting is independent of other pathways activated by cytosolic nucleic acid (Figure S5). Taken together, our data suggest that STING is necessary for directing the formation of the ubiquitin signals that route M. tuberculosis to the ubiquitin-mediated selective autophagy pathway.
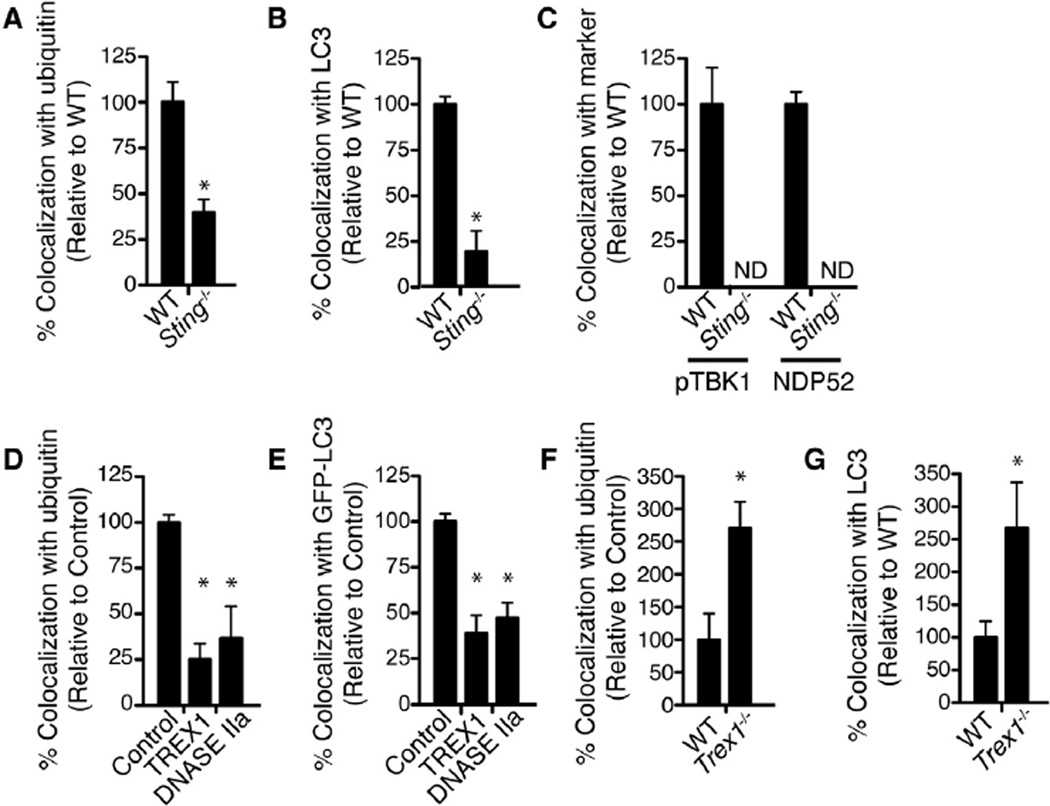
Figure 5. STING and Cytosolic DNases Modulate Autophagic Targeting ofM. tuberculosis.
(A) Quantitative analysis of ubiquitin colocalization with WT M. tuberculosis at 4 hr post-infection in WT or Sting−/− BMDMs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of ubiquitin-positive cells relative to WT BMDMs (n = 3 per group, *p < 0.003).
(B) Quantitative analysis of LC3 colocalization with WT M. tuberculosis at 4 hr post-infection in WT or Sting−/− BMDMs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of LC3-positive cells relative to WT BMDMs (n = 3 per group, *p < 0.003).
(C) Quantitative analysis of phospho-TBK1 and -NDP52 colocalization with WT M. tuberculosis at 4 hr post-infection in WT or Sting−/− BMDMs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of marker-positive cells relative to WT BMDMs.
(D) Quantitative analysis of ubiquitin colocalization with WT M. tuberculosis at 4 hr post-infection in RAW cells stably overexpressing TREX1 or DNASE IIa. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of ubiquitin-positive cells relative to control (vector only) cells (n = 3 per group, *p < 0.019).
(E) Quantitative analysis of GFP-LC3 colocalization with WT M. tuberculosis at 4 hr post-infection in GFP-LC3 RAW 264.7 cells stably overexpressing TREX1 and DNASE IIa. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of GFP-LC3-positive cells relative to control (vector only) cells (n = 3 per group, *p < 0.003).
(F) Quantitative analysis of ubiquitin colocalization with WT M. tuberculosis at 4 hr post-infection in WT or Trex1−/−. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of ubiquitin-positive cells relative to WT BMDMs (n = 3 per group, *p < 0.005).
(G) Quantitative analysis of LC3 colocalization with WT M. tuberculosis at 4 hr post-infection in WT or Trex1−/− BMDMs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of GFP-LC3-positive cells relative to WT BMDMs (n = 3 per group, *p < 0.02).
See also Figure S5.
Host DNases Affect Autophagic Targeting of M. tuberculosis
To further demonstrate the role of DNA in targeting M. tuberculosis to autophagosomes, we determined whether host DNases could modulate autophagic targeting of bacilli during infection. DNases such as TREX1 and DNASE IIa have been shown to negatively regulate the innate immune response to cytosolic DNA (Okabe et al., 2009; Stetson et al., 2008), and TREX1 negatively regulates the induction of type I IFN during M. tuberculosis infection (Manzanillo et al., 2012). Overexpression of TREX1 or DNASE IIa within macrophages greatly reduced colocalization of M. tuberculosis with ubiquitin (Figure 5D) and LC3 (Figure 5E) during infection. Furthermore, M. tuberculosis infection of Trex1−/− BMDMs resulted in an increase in targeting of M. tuberculosis to these markers (Figures 5F and 5G). These data reveal an unprecedented role of host DNases in modulating selective autophagy and provide corroborative evidence that M. tuberculosis DNA serves as a molecular signal for selective autophagy.
Ubiquitin-Mediated Autophagy Targets M. tuberculosis to the Lysosome
Because autophagy has been implicated in acute M. tuberculosis restriction by macrophages (Gutierrez et al., 2004), we sought to determine the fate of ubiquitinated bacteria marked for autophagy early after infection. First, we stained M. tuberculosis-infected macrophages with antibodies that recognize the lysosomal marker LAMP-1 to determine whether ubiquitin and LC3-positive bacteria are targeted to lysosomes. During M. tuberculosis infection of Atg5+ BMDMs, ~30% of bacilli were positive for LAMP-1 at 6 hr post-infection (Figures 6A and 6B). In contrast, only 2%–5% of bacilli colocalized with LAMP-1 during M. tuberculosis infection of Atg5− macrophages. Importantly, macrophages deficient for TBK1 or STING also had severe defects in LAMP-1 colocalization to bacteria, indicating that DNA/ubiquitin-mediated targeting leads to delivery of M. tuberculosis to the lysosome (Figures 6A and 6B).
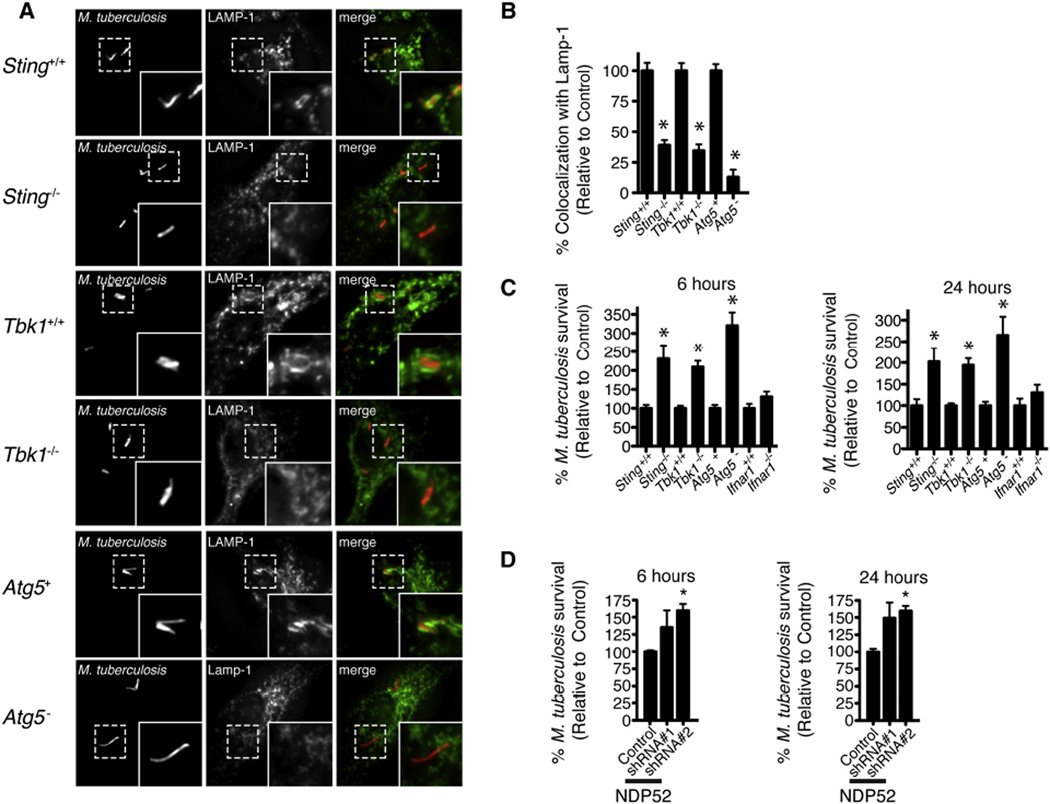
Figure 6. STING-, TBK1-, and ATG5-Dependent Delivery of M. tuberculosis to Lysosomes Limits Bacterial Replication.
(A) Fluorescence images of indicated BMDM genotypes infected for 4 hr with mCherry-expressing WT M. tuberculosis (red) and immunostained with anti-LAMP-1 (green) antibodies.
(B) Quantitative analysis of LAMP-1 colocalization with WT M. tuberculosis at 4 hr post-infection in indicated BMDM genotypes. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of Lamp-1-positive cells relative to control BMDMs (n = 3 per group, *p < 0.012).
(C) BMDMs of the indicated genoytpes were infected with WT M. tuberculosis (multiplicity of infection [moi] 1) for 0, 6, and 24 hr. Bacterial viability was assessed by CFUs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of bacterial survival compared to time zero relative to control macrophages (n = 3 per group, *p < 0.025).
(D) RAW 264.7 cells were infected with WT M. tuberculosis (moi 1) for 0, 6, and 24 hr after shRNA knockdown of NDP52. Bacterial viability was assessed by CFUs. Results are the means ± SEM of three independent experiments. Data are expressed as the percentage of bacterial survival compared to time zero relative to scrambled shRNA (control) (n = 3 per group, *p < 0.046).
To test whether the differences in lysosomal targeting correlated with changes in bacterial survival, we infected BMDMs with WT M. tuberculosis and determined bacterial viability by enumerating colony-forming units (CFUs) at 6 and 24 hr post-infection. Infection of BMDMs deficient for ATG5, STING, or TBK1 resulted in a 2- to 3-fold increase in bacterial survival relative to WT or control BMDMs (Figure 6C), and shRNA knockdown of NDP52 expression had similar effects (Figure 6D) on bacterial numbers. These data demonstrate that targeting of M. tuberculosis to the autophagy pathway is required for the cell-autonomous control of M. tuberculosis replication within macrophages. Moreover, because M. tuberculosis grows extremely slowly and very little replication occurs during the first 24 hr of infection, these data suggest that the population of bacteria targeted by the ubiquitin-mediated autophagy pathway is specifically killed in the lysosome, whereas the nontargeted bacteria remain viable.
Autophagy Is Essential In Vivo for Control of M. tuberculosis
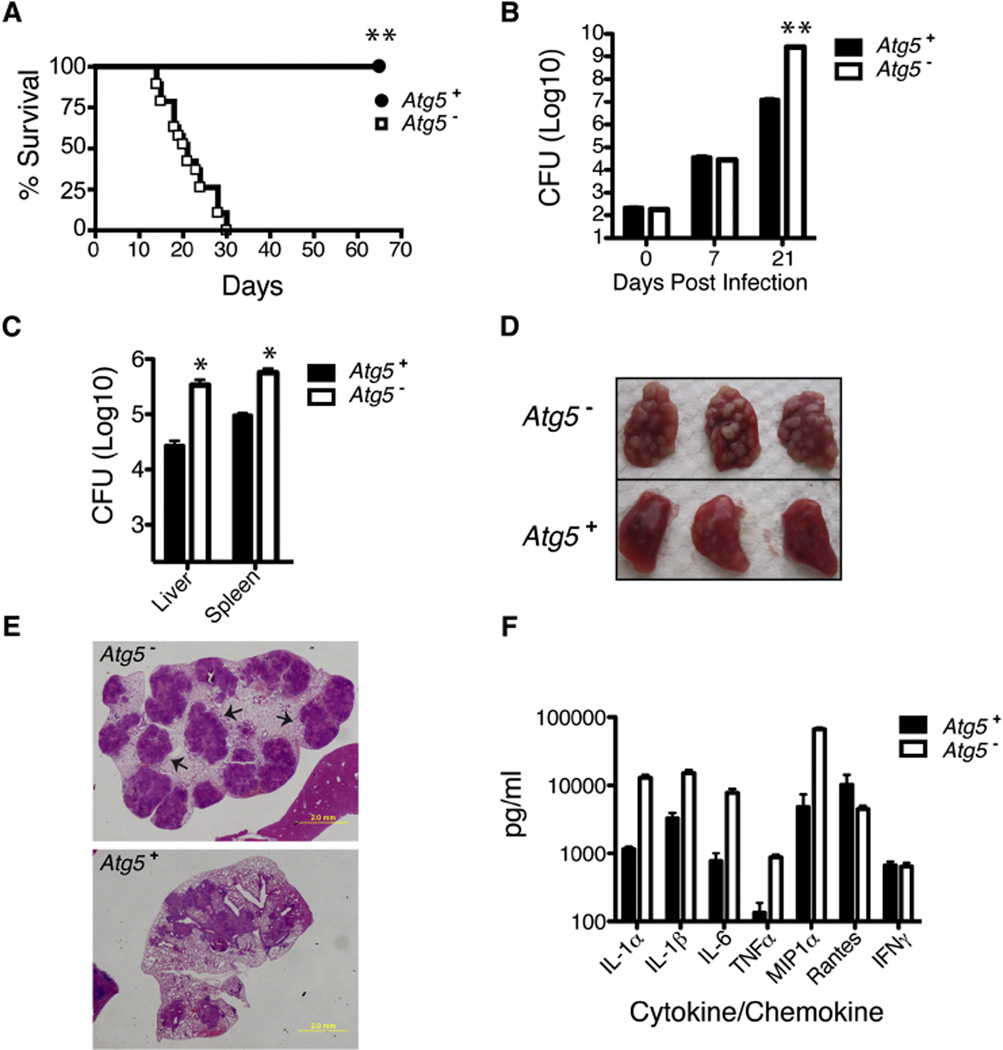
Because previous studies on the role of autophagy during M. tuberculosis infection have been performed with cultured macrophage cells (Deretic et al., 2009), we used a mouse model of tuberculosis infection to determine the contribution of auto-phagy in macrophages to host resistance in vivo. To this end, we performed low-dose aerosol infections of Atg5− mice flox/flox-Lyz-Cre) and monitored both mouse survival and bacterial burdens within infected tissues. Surprisingly, Atg5− mice were extremely sensitive to M. tuberculosis, as all mutant mice succumbed to infection by 4 weeks post-infection, in contrast to Atg5+ mice that displayed no overt signs of distress at this early time point (Figure 7A). In addition, the rate of M. tuberculosis replication was unchecked within Atg5− mice, which resulted in nearly a 1,000-fold increase in bacilli within the lungs and a 10- to 20-fold increase of bacilli within the spleen and liver at 21 days post-infection, relative to control mice (Figures 7B and 7C). The increased bacterial burden in Atg5− mice was accompanied by massive pulmonary abscesses that were obvious both by gross morphology of entire lungs and by staining of tissue sections (Figures 7D and 7E). Furthermore, lungs from Atg5− mice contained 5- to 10-fold higher levels of proinflammatory cytokines TNF-a, IL-1α, IL-1β, and IL-6 within infected lung tissues (Figure 6F). In contrast, production of IFN-y was unchanged between Atg5− and Atg5+ infected mice (Figure 6F), suggesting that the defect in Atg5− mice was not simply due to a failure of T cells to produce this important cytokine. Collectively, these data demonstrate that ATG5-mediated autophagy in monocytes plays a major role in eliciting an effective innate immune response to M. tuberculosis infection in vivo.
Figure 7. ATG5 Is Required In Vivo for Control ofM. tuberculosis.
Atg5fl/fl (Atg5+) and Lyz-Cre-Atg5fl/fl (Atg5−) from a single cohort were infected with 200 CFUs of WT M. tuberculosis via the aerosol route (n = 30 per group).
(A) Atg5+ mice showed significantly improved survival compared to Atg5− mice as calculated by log-rank test (**p < 0.001).
(B and C) Bacterial burdens as measured by CFUs in the lungs 0, 7, and 21 days post-infection (B) and the spleen and liver 21 days post-infection (C) (n = 5 per time point, *p < 0.005, **p < 0.001).
(D) Lungs from Atg5+ and Atg5− mice 21 days post-infection.
(E) H&E staining of lung sections from Atg5+ and Atg5− mice 21 days post-infection. Arrows indicate large pulmonary abscesses not observed in WT mice.
(F) Multiplex ELISA analysis of cytokines from lungs of control and Atg5− mice 21 days postinfection (n = 4 per group).
DISCUSSION
We have identified four critical steps in the pathway by which M. tuberculosis cells are recognized by host macrophages and targeted to autophagosomes. First, the bacterial ESX-1 secretion system initiates the interaction by permeabilizing the phago-somal membrane early after phagocytosis. Second, permeabilization exposes DNA on the surface of the bacteria that is recognized by components of the cytosolic DNA pathway, including STING, to initiate autophagy targeting. Third, a population of the engulfed bacteria become associated with, and surrounded by, host ubiquitin chains. Most of the labeled bacteria are associated with K63-linked chains, but some K48 linkages are also detectable on a lower percentage of bacteria. Our finding that STING is necessary for ubiquitin colocalization of a subpopulation of bacteria suggests that distinct recognition events underlie M. tuberculosis targeting. Fourth, the ubiquitin LC3-binding autophagy adaptors p62 and NDP52 are required to recruit autophagy components to create a phagophore surrounding the bacilli, a process that also requires the TBK1 kinase and ATG5. Once targeted to the ubiquitin-mediated autophagy pathway, bacilli-containing autophagosomes are matured via fusion with lysosomes to create autophagolysosomes. Delivery of this population of bacteria to the lysosome is responsible for limited bacterial killing by macrophages ex vivo, but the entire autophagy pathway in macrophages is a major determinant of host resistance to M. tuberculosis infection in vivo.
Given the modest effect of ATG5 on M. tuberculosis survival in macrophages, it may be surprising that the Atg5− mice are so profoundly susceptible to infection. Indeed, Atg5− mice succumb to infection with kinetics nearly identical to that of mice that completely lack an adaptive immune system or are missing key activators of macrophage activation, such as IFN-y (MacMicking et al., 2003). However, it is becoming clear that autophagy plays a broader role in innate and adaptive immune responses than simply leading to direct killing of microbes in the lysosome. For example, autophagy enhances antigen presentation in dendritic cells (Jagannath et al., 2009) and negatively regulates inflammasome activation (Nakahira et al., 2011; Saitoh et al., 2008; Zhao et al., 2008). Indeed, the pronounced increase in inflammatory cytokines, such as IL-1β during infection of Atg5− mice suggests that inflammasome signaling is augmented in the Atg5− mice during M. tuberculosis infection (Shi et al., 2012). Therefore, whereas delivery of bacteria to the lysosome plays a direct role in acute bacterial restriction, autophagy may play a more pronounced role in overall control via non-cell-autonomous effects on innate and specific immune responses.
In contrast to the major role of ATG5 in controlling M. tuberculosis infection, previous studies demonstrated that macrophages from Atg5− mice are capable of controlling BCG infection (Zhao et al., 2008). Our results suggest that the reason for the apparent dispensability of ATG5 during BCG infection is because the vaccine strain does not naturally induce the ubiquitin-mediated pathway due to a lack of ESX-1 secretion and the concomitant lack of membrane permeabilization. Importantly, reintroduction of ESX-1 secretion to BCG (Pym et al., 2003), or induction of general autophagy by administering rapamycin (Jagannath et al., 2009), increased the potency of vaccination, suggesting that incorporation of autophagy-inducing elements may uniquely benefit vaccination strategies to prevent human tuberculosis. Similarly, mice lacking ATG5 in monocytes had a relatively minor defect in protection from infection with L. monocytogenes (Zhao et al., 2008), especially compared to the dramatic effects with M. tuberculosis. In this case, it is probable that the ability of L. monocytogenes to evade autophagy by nucleating actin renders the bacteria insensitive to the absence of autophagic machinery (Yoshikawa et al., 2009). Moreover, it is likely that the ubiquitin-mediated autophagy pathway acts as an inducible mechanism to traffic intracellular pathogens that permeabilize membranes into lysosomes, whereas nonpathogens that do not perturb membranes traffic to lysosomes via the constitutive endosomal-lysosomal maturation pathway.
It is curious that only one-third of intracellular bacteria are targeted by the ubiquitin pathway and colocalize with LC3. This observation is consistent, however, with classic electron microscopy (EM) studies of M. tuberculosis trafficking in macrophages that showed that although live M. tuberculosis bacilli were able to profoundly inhibit phagosome maturation to lysosomes, approximately one-third of bacilli still trafficked to lysosomes (Armstrong and Hart, 1971). It is likely that the fate of those lysosomal bacteria was mediated by autophagic targeting. Although we do not yet understand the mechanistic basis for the heterogeneous response, one possibility is that M. tuberculosis may employ an autophagy evasion strategy that prevents ubiquitination but is only partially effective. Similarly, though perhaps more appealing, is the possibility that cell-to-cell variations in ESX-1 secretion, and thus variations in membrane permeability from phagosome to phagosome, could account for the heterogeneity in autophagy targeting (Ohol et al., 2010). In either case, heterogeneity of autophagic targeting may be important for successful infection by M. tuberculosis. For example, although bacilli that become targeted to lysosomes may be killed, the resultant increase in proinflammatory responses and elicitation of adaptive immune responses may promote and favor the persistence strategy of the bulk of the bacterial population. Alternatively, it is possible that the heterogeneity is due to host processes; for example, there may be an exclusive interplay between bacterial targeting and membrane repair.
The findings that DNA is a bacterial-derived molecule recognized by host cells to target M. tuberculosis to autophagy, and that STING and TBK1 are required for this process, reveals a surprisingly broad connection between cytosolic DNA detection and innate immune responses to pathogens. Indeed, introduction of exogenous DNA into the cytosol of cells triggers three distinct yet interrelated responses: autophagy, cytokine signaling, and inflammasome activation (Hornung et al., 2009; Roberts et al., 2009). STING is a required factor for both autophagy and cytokine signaling and, either directly (Burdette et al., 2011) or indirectly (via interactions with cytosolic DNA receptors), recognizes DNA and leads to ubiquitination and recruitment of both TBK1 and NDP52. It will be interesting to determine whether other ubiquitin-binding adaptors implicated in autophagy, including NBR1 and optineurin, are also important for this pathway (Kirkin et al., 2009; Wild et al., 2011b). Although the molecular connections between DNA and ubiquitination remain unknown, STING itself is a plausible ubiquitinated target as it colocalizes to bacteria and is ubiquitinated during DNA stimulation (Tsuchida et al., 2010).
It is likely that cytoplasmic DNA detection also targets other pathogens that access the cytosol to the autophagy pathway. Although it remains to be seen whether autophagy targeting of other bacterial pathogens depends on DNA sensing and STING, it is becoming clear that this same pathway is also operational to limit infection with dsDNA viruses, which expose their genomes during virion assembly in the cytoplasm (McFarlane et al., 2011; Rasmussen et al., 2011). Furthermore, given the eubacterial origin of the mitochondrion, it is tempting to speculate that a signal for mitophagy may also be exposure of mitochondrial DNA on the surface of damaged organelles.
A common requirement of intracellular pathogens is the ability to penetrate host cell membranes in order to gain access to the cytosol. The mechanisms described here illustrate that it is the combination of pathogen molecule and spatial distribution of pattern recognition receptors that allows host cells to detect membrane puncture and discriminate pathogens from nonpathogens. It is thus a combination of ESX-1 secretion and DNA exposure that constitutes the “pattern of pathogenesis” recognized by host cells to mount innate responses against M. tuberculosis (Vance et al., 2009). In addition to detection of microbial products, cells are also able to target autophagy to intracellular pathogens by directly detecting membrane permeabilization (Thurston et al., 2012). Such overlapping systems may be beneficial for different host cell types and as well to allow cells to detect virulent pathogens that do not expose stimulatory eDNA.
EXPERIMENTAL PROCEDURES
A detailed description of materials and methods can be found in the Extended Experimental Procedures.
Antibodies
The following primary antibodies were used: mouse monoclonal antibodies against poly- and monoconjugated-ubiquitin (Enzo Life Sciences), K63 and K48 ubiquitin (Milipore), LAMP-1 (BD Biosciences), STING (Sigma), and NDP52 (Novus Biologicals) and rabbit polyclonal antibodies against phospho-TBK1 (Cell Signaling), ATG12 (Cell Signaling, and LC3B (Invitrogen). Secondary antibodies used were as follows: Alexa 488-conjugated goat antirabbit and Alexa 488- and Alexa 350-conjugated goat anti-mouse IgG antiserum (Molecular Probes).
Bacterial Strains
The Erdman strain of M. tuberculosis was used as the WT strain and mutant background strain for all M. tuberculosis experiments. BCG Pasteur was a gift from W.R. Jacobs.
Macrophage Infection
For infections with mycobacteria, macrophages were infected as previously described at a multiplicity of infection (moi) of 1 (Manzanillo et al., 2012). For determination of CFUs, macrophage monolayers were lysed in 0.1% Triton X-100 and plated on 7H10 agar plates.
Mouse Infection
Atg5flox/flox-Lyz-Cre and Atg5flox/flox control mice were infected with M. tuberculosis (Erdman) via low-dose aerosol infection (~200 CFUs) as previously described (Ohol et al., 2010).
Statistics
Statistical analysis of data was performed using GraphPad Prism software (Graphpad; San Diego, CA, USA). Two-tailed unpaired Student’s t tests were used for analysis of microscopy images and mycobacterium CFU assays. The Kaplan-Meir method was used to analyze mouse survival. Unless otherwise indicated, all experiments were performed at least three times and presented as the mean ± standard error of the mean (SEM).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Vance, G. Barton, D. Monack, G. Barber, N. Mizushima, H. Virgin, and J. Debnath for mice and reagents. We are grateful to S. Johnson and J. Rubenstein for use of their microscopes. We thank S. Bell for critical reading of the manuscript. This work was supported by NIH grants PO1 AI063302 and RO1 AI081727, the Heiser Foundation Fellowship, and the Genentech Research Fellowship.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.06.040.
REFERENCES
- Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. STING-dependent signaling. Nat. Immunol. 2011;12:929–930. doi: 10.1038/ni.2118. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- Brodin P, Poquet Y, Levillain F, Peguillet I, Larrouy-Maumus G, Gilleron M, Ewann F, Christophe T, Fenistein D, Jang J, et al. High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 2010;6:e1001100. doi: 10.1371/journal.ppat.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteriaassociated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–345. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, De Mazière A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5:e1000430. doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honoré N, Marchal G, Jiskoot W, England P, et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 2007;189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Delgado M, Vergne I, Master S, De Haro S, Ponpuak M, Singh S. Autophagy in immunity against mycobacterium tuberculosis: a model system to dissect immunological roles of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:169–188. doi: 10.1007/978-3-642-00302-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hagstrom JE, Ludtke JJ, Bassik MC, Sebestyén MG, Adam SA, Wolff JA. Nuclear import of DNA in digitonin-permeabilized cells. J. Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr., Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kudchodkar SB, Levine B. Viruses and autophagy. Rev. Med. Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, Rao KV. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell. Microbiol. 2011;13:814–835. doi: 10.1111/j.1462-5822.2011.01581.x. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Cox JS. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect. Immun. 2007;75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 2011;85:4212–4221. doi: 10.1128/JVI.02435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. NatImmunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of autophagy by α-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J. Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 2007;26:3180–3190. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Jamieson A, Cresswell P. GILT is a critical host factor for Listeria monocytogenes infection. Nature. 2008;455:1244–1247. doi: 10.1038/nature07344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect. Immun. 2008;76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, and Brown EJ. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schaible UE, Russell DG. Mycobacterium-containing phagosomes are accessible to early endosomes and reflect a transitional state in normal phagosome biogenesis. EMBO J. 1996;15:6960–6968. [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell. Microbiol. 2006;8:1417–1429. doi: 10.1111/j.1462-5822.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011a;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011b;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KW, Jacobs WR., Jr. Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell. Microbiol. 2011;13:1371–1384. doi: 10.1111/j.1462-5822.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, Lee ZW, Lee SH, Kim JM, Jo EK. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–243. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosomeindependent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.