INTRODUCTION
There are numerous studies suggesting that eosinophils are associated with asthma exacerbations.1 In a study that compared asthma therapeutic adjustments based on the status of airway eosinophilia versus guideline-based management, reduced rates of asthma exacerbations were observed in the patients whose therapy was directed at the reduction of airway eosinophilia.1 In several clinical trials using anti-IL-5 therapy for the treatment of severe eosinophil-prominent asthma, the reduction of eosinophil levels in the airway and blood was associated with reduced rates of asthma exacerbations.2-5 In experimentally-induced rhinovirus (HRV) infection, baseline sputum eosinophilia was associated with increased severity of respiratory illness.6 These findings suggest that eosinophils directly contribute to the development and severity of exacerbations of asthma.
Viral infections, specifically HRV, are the most common cause of asthma exacerbations in both children and adults.7, 8 A recent study demonstrated that allergic sensitization is a risk factor for HRV-induced wheezing in children, which has been linked to the development of asthma.9 Therefore, understanding the mechanism of association between allergic inflammation and innate immunity in the setting of HRV infection may identify targets for the prevention of asthma exacerbations. Type I and III interferons play a key role in the innate immune response to viral infection. Defects in the expression of these cytokines have been hypothesized to contribute to increased susceptibility of asthma subjects to viral infections.10, 11
It is now recognized that eosinophils are not simply effector cells that release toxic granule proteins, but they also act as regulatory cells capable of influencing and often enhancing local inflammation.12 In considering the close clinical relationship between eosinophilia and virus-induced exacerbations, we hypothesized that eosinophils inhibit anti-viral responses of airway cells. To test this in an in vitro model, we examined the effects of human eosinophils on interferon expression by BEAS-2B epithelial cells in response to the toll-like receptor (TLR)-3 ligand poly-IC and in response to HRV infection.
METHODS
Cell Purification and Culture
Eosinophils were isolated from peripheral blood obtained from human subjects who provided written consent to participate in a University of Wisconsin Institutional Review Board approved protocol. Subjects with severe or uncontrolled allergic disease or those using systemic steroids were excluded. The peripheral blood was subjected to density centrifugation and immunomagnetic bead negative selection as previously described, which includes exposure to newborn calf serum.13 Eosinophils were >98% pure; contaminating cells were typically neutrophils and/or lymphocytes. The BEAS-2B immortalized cell line was cultured in serum free Bronchial Epithelial Cell Growth Medium (BEGM, Lonza, Basel, Switzerland) at 37°C with 5%CO2 and humidified air. The BEGM was prepared according to manufacturer instructions, which includes hydrocortisone at final concentration of 0.5μg/mL.
For co-culture experiments, BEAS-2B cells were plated and grown on 6 well tissue culture plates for 48 hours to ~50-70% confluence. The medium was then replaced and both 1 ×106 /mL eosinophils and 25μg/mL poly-IC were added. To separate eosinophils from BEAS-2B cells, Transwell inserts (Corning, Lowell, MA) were utilized. For the HRV experiments, HRV1A or HRV16 stocks were prepared by purification of virus grown in HeLa cell suspension cultures with sucrose gradients as previously described.14 The HRV1A or HRV16 was added at a multiplicity of infection (MOI) of 5 or 20 plaque forming units per cell (PFU/cell). For eosinophil cultures with HRV1A alone, 1×106 PFU of HRV1A was incubated with 5×105 eosinophils with or without 0.1mM fMLP (induces degranulation) at room temperature for 3 hours. For the recombinant TGF-β1 experiments, 0.1ng/mL recombinant TGF-β1 (R&D Systems, Minneapolis, MN) was added. The cultures were incubated for 24 hours at 37°C for poly-IC experiments or 34°C for virus experiments. Supernatants were colle cted and centrifuged to remove debris. BEAS-2B cell lysate was collected for RNA purification after rinsing plates with PBS to remove eosinophils, with greater than approximately 95% removed.
RNA Measurements
Total RNA was isolated according to manufacturer’s instructions using an RNeasy mini kit (Qiagen, Valencia, CA), including DNase I digestion to remove contaminating DNA. cDNA was synthesized from 0.75 or 1.5 μg purified RNA according to the manufacturer’s instructions using Superscript III (Invitrogen, Carlsbad, CA).
Taqman primer and probe sets for target genes (IFN-α1, IFN-β1 and IFN-λ1) and housekeeping/reference genes (β-actin and cyclophilin A) were obtained from a commercial source (Applied Biosystems, Foster City, CA). Samples were run in duplicate with each reaction containing Universal Master Mix (Applied Biosystems), the primer and probe set and cDNA for a total reaction volume of 25μl. Fluorescence was measured for each cycle using the ABI PRISM 7500 Sequence Detection System or the Step One Plus Real-Time PCR System (both by Applied Biosystems).
Virus Measurements
HRV1A was incubated with eosinophils in RPMI for 3 hours at room temperature. Samples were microfuged and supernatants collected for measurement of infectivity and viral RNA levels. Viral infectivity was measured by a plaque-forming assay as previously described.15 Briefly, HeLa cell monolayers in 60mm plates were infected with 10-fold dilutions of culture supernatants at room temperature for 1 hour, agar and nutrient overlays were added, then incubated at 35°C for 3 days for plaque develo pment. Cells were fixed and stained to quantify plaques which are expressed as PFU/mL. Viral RNA levels were measured with quantitative polymerase chain reaction with a pan-HRV primer set using an ABI Prism 7500 (Applied Biosystems) as previously described.16, 17 The HRV1A virus quantification was based on a standard curve of cDNA corresponding to known amount of HRV1A virions.
Protein Measurements
IFN-λ1 (IL-29) and TGF-β1 protein in culture supernatants was measured according to manufacturer’s instructions using commercial ELISA kits (R&D Systems). Notably, activation of latent TGF-β1 in supernatants was carried out using 1N HCL followed by neutralization with 1.2 N NaOH/0.5 M HEPES prior to assay performance. Samples were run in duplicate and the sensitivity ranged from 1.7-15.4 pg/mL.
Data Analysis
Statistical analyses were performed using SigmaPlot software (Systat Software, Chicago, IL). RNA data were analyzed following log transformation. Comparisons were made using a paired t-test or Wilcoxon sign ranked test for parametric and nonparametric data, respectively. Statistical significance was defined as p < .05.
RESULTS
Eosinophil Donor Characteristics
As shown in Table 1, the eosinophils used for all the experiments were purified from 27 donors, 18 to 49 (mean of 32) years old (14 female). All had allergic rhinitis, 17 had a diagnosis of asthma, and none with atopic dermatitis. The donors generally had mild disease, mostly managed with antihistamine. Only one donor was using an inhaled steroid controller medication. The mean eosinophil count was 203 cells/μL. Of note, different subsets of these donors were utilized for each experiment and demographics of the respective subsets were similar.
Table 1.
Eosinophil Donor Characteristics
| Characteristic | Values |
|---|---|
| Total Donors | 27 |
| Age | 32 (range 18 to 49) |
| Sex | |
| Male | 13 (48%) |
| Female | 14 (52%) |
| Allergy Diagnoses | |
| Allergic Rhinitis | 27 (100%) |
| Asthma | 17 (63%) |
| Atopic Dermatitis | 0 (0%) |
| Allergy Medications* | |
| Antihistamine | 17 (77%) |
| Nasal corticosteroid | 2 (9%) |
| Leukotriene modifier | 1 (4%) |
| Inhaled corticosteroid | 1 (4%) |
| Peripheral Eosinophil Count (cells/μL) | 203 (range 53 to 546) |
Medication data not available for 5 of the donors.
Effect of Eosinophils on Poly-IC Stimulation of BEAS-2B Cells
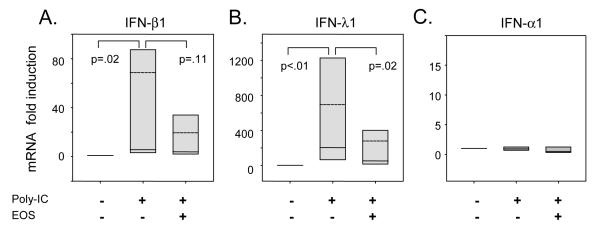
BEAS-2B cells were stimulated with poly-IC, and mRNA expression was induced for IFN-β1 (69±44 fold, p=.02) and IFN-λ1 (690±370 fold, p<.01) (Figure 1A-B, respectively). However, IFN-α1 expression was essentially unchanged (1.5 fold) with poly-IC stimulation (Figure 1C). Co-culture with eosinophils, in the absence of transwells with potential for cell-cell contact, suppressed poly-IC stimulated mRNA expression of IFN-λ1 (2.5 fold decrease, p=.02), and a similar trend was observed for IFN-β1 (3.6 fold decrease, p=.11). Co-culture of eosinophils did not affect the poly-IC-induced mRNA expression of IFN-α1.
Figure 1.
Effect of eosinophil co-culture on IFN and cytokine expression of poly-IC stimulated BEAS-2B cells. BEAS-2B cells were cultured for 24 hours with or without poly-IC 25μg/mL and eosinophils (EOS) as indicated. RNA was extracted from the BEAS-2B cells attached to the culture plate. Quantitative PCR results are presented as fold inductions of unstimulated BEAS-2B cells for the following genes: A, IFN-β1 (n=7); B, IFN-λ1 (n=8); and C, IFN-α1 (n=8). Fold-inductions of mRNA are shown with box plots with the median (solid line) and mean (dashed line) values. Error bars represent the 5-95th percentile. P values for statistical comparisons are shown.
To determine whether the eosinophil mediated suppression of poly-IC stimulated gene expression from the BEAS-2B cells was due to soluble mediators or dependent on cell contact, the co-cultures were performed in the presence of a semi-permeable membrane (Transwell) to physically separate the cell populations. Despite the lack of cell-to-cell contact, eosinophil mediated suppression of IFN-β1 and IFN-λ1 was evident (Online supplement, Figure E1). This suggests that eosinophils suppress BEAS-2B interferon expression via a soluble mediator.
Effect of Eosinophils on HRV Infection of BEAS-2B Cells
Next, we tested for effects of eosinophils on HRV-induced interferon expression using a minor receptor (LDLR) group, HRV1A, and major receptor (ICAM-1) group, HRV16, virus. There was greater induction of IFN-β1 (338 fold vs 247 fold) and IFN-λ1 (2010 fold vs 890 fold) with the use of HRV1A compared to HRV16 (Online supplement, Figure E2). Furthermore, the eosinophil-mediated suppression of their expression was observed with two different concentrations of virus (multiplicities of infection (MOI) = 5 and 20 PFU/cell); however, it was more pronounced at the higher concentration. Therefore, we chose to perform further experiments with HRV1A at an MOI of 20 PFU/cell, which is consistent with our previous studies.18, 19
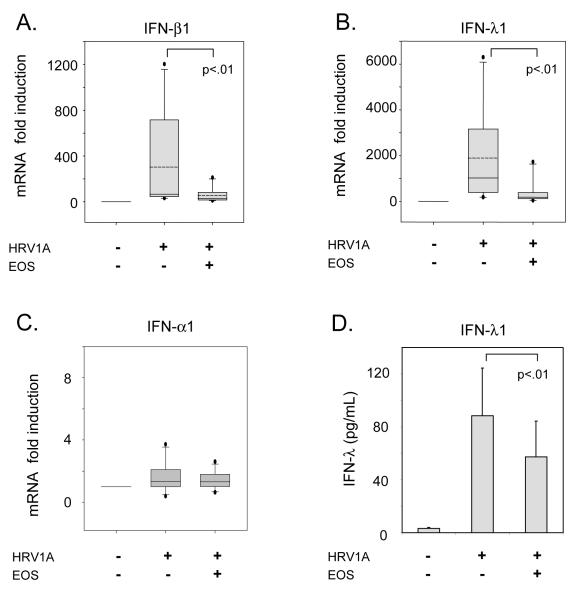
Similarly, as shown in Figure 2A-C, HRV1A infection of BEAS-2B cells also induced both IFN-β1 (304±133 fold) and IFN-λ1 (1890±630 fold). Co-culture with eosinophils suppressed HRV-1A-induced IFN-β1 mRNA (53±20 fold, p<.01) and IFN-λ1 mRNA (380±160 fold, p<.01) compared to responses of epithelial cell monocultures. IFN-α1 was also examined; however, its expression was not greatly induced (3 fold) and there was no significant effect upon co-culture with eosinophils. To determine whether the changes in gene expression corresponded to changes in protein production, IFN-λ1 and IFN-β secretion was examined. Infection of BEAS-2B with HRV1A induced IFN-λ1 protein secretion (89±36 pg/mL) that was suppressed by eosinophil co-culture (1.6 fold decease, p<.01, Figure 2D). IFN-β protein levels were below the limits of detection by ELISA.
Figure 2.
Effect of eosinophil co-culture on IFN expression of HRV1A infected BEAS-2B cells. BEAS-2B cells were infected with HRV1A at 20 PFU/cell for 24 hours with or without eosinophils (EOS) as indicated. RNA was extracted from the BEAS-2B cells attached to the culture plate. Quantitative PCR results are presented for A, IFN-β1, B, IFN-λ1 and C, IFN-α1 as fold inductions of unstimulated BEAS-2B cells (n=11). Fold-inductions of mRNA are shown with box plots with the median (solid line) and mean (dashed line) values. Error bars represent the 5-95th percentile. Percentage expression is shown as the means with error bars representing the standard error of the mean. D, BEAS-2B cells were infected with HRV1A at 20 PFU/cell for 24 hours with or without eosinophils (EOS) as indicated. Supernatants were collected and ELISA results for IFN-λ protein are shown as the means with error bars representing the standard error of the mean. P values for statistical comparisons are shown.
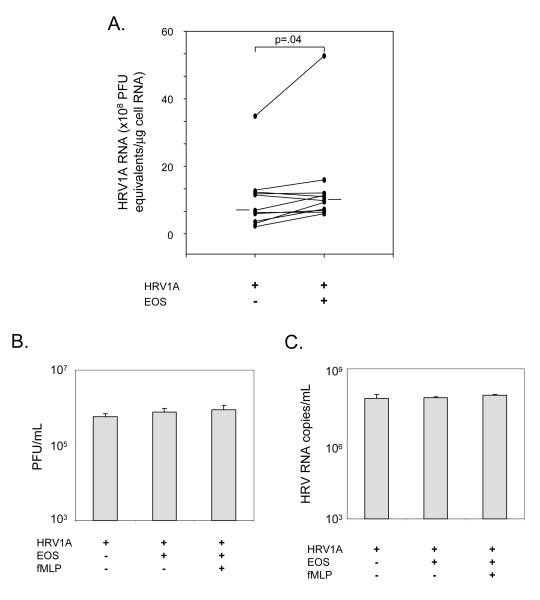
Since one role of interferons is to block virus replication, we examined whether the co-culture of eosinophils and corresponding suppression of IFN-β1 and IFN-λ1 affected HRV1A virus levels in infected BEAS-2B cells. There was a significant 34% increase in the quantity of HRV1A virus RNA in infected BEAS-2B cells upon co-culture with eosinophils (p=.04, Figure 3A).
Figure 3.
Effect of eosinophil co-culture on HRV1A. A, BEAS-2B cells were infected with HRV1A at 20 PFU/cell for 24 hours with or without eosinophils (EOS) as indicated. RNA was extracted from the BEAS-2B cells and RT-PCR performed for viral RNA levels with a standard curve to determine HRV1A RNA levels (PFU equivalents) per μg of cellular RNA. Median HRV1A RNA levels are shown as horizontal bars. P value for statistical comparison is shown. Supernatants were collected for analyses of B, plaque forming units (PFU), and C, copies of HRV RNA from cultures of HRV1A (MOI 1), eosinophils (EOS, 1×106 cells/mL), and fMLP (0.1 mM) as indicated incubated for 3 hours.
Eosinophils Do Not Directly Affect HRV Infectivity
To determine whether eosinophils have anti-HRV activity that could diminish interferon responses, we cultured HRV1A with resting eosinophils, or eosinophils that were treated with fMLP to induce degranulation. The virus suspension was then re-tested for HRV1A infectivity (Figure 3B) and HRV1A RNA levels (Figure 3C). Eosinophils had no significant effects on HRV infectivity. Furthermore, we confirmed that EDN was released and there was increased RNase activity in the culture supernatants with eosinophils (Figure E3).
Role of TGF-β1 in Eosinophil-mediated Suppression of Interferon Expression
To determine whether TGF-β could suppress IFN-β1 and IFN-λ1 mRNA expression in our system, recombinant TGF-β1 was added to BEAS-2B cells infected with HRV1A for 24 hours (Figure 4A). TGF-β1 had no effect on IFN-β1mRNA, but significantly suppressed IFN-λ1 mRNA (65% decrease).
Figure 4.
Effect of recombinant TGF-β1 and levels of TGF-β1 upon co-culture with eosinophils. A, BEAS-2B cells were infected with HRV1A at 20 PFU/cell for 24 hours with or without 0.1ng/mL of recombinant TGF-β1. Quantitative PCR results for IFN-β1 and IFN-λ1 (n=3) are shown as fold inductions of unstimulated BEAS-2B cells presented as box plots, with solid line as the median and dashed line as the mean. P values for statistical comparisons are shown. B, BEAS-2B cells were untreated or infected with HRV1A at an MOI of 20 PFU/cell for 24 hours with or without eosinophils (EOS) as indicated. ELISA results for the culture supernatants are shown as the mean of TGF-β1 (n=9), with error bars representing the standard error of the mean. P values for statistical comparisons are shown.
We next tested whether eosinophils induced BEAS-2B cells to produce TGF-β, and whether this induction was enhanced with HRV infection. As shown in Figure 4B, BEAS-2B cells secreted similar amounts of TGF-β with or without HRV infection (67±8 pg/mL vs 60±9 pg/mL). Co-culture with eosinophils increased TGF-β1 secretion either with HRV1A infection (60±9 pg/mL to 110±17 pg/mL, p<.01) or without HRV1A infection (67±8 pg/mL to 113±18 pg/mL, p=.01).
DISCUSSION
Our study examined the effects of eosinophils on the expression of interferons from BEAS-2B epithelial cells. We demonstrated that eosinophils can suppress poly-IC and HRV-stimulated expression of IFN-β1 and IFN-λ1 from BEAS-2B cells, without requirement for cell-cell contact. Consequently, eosinophil co-culture increased HRV replication in BEAS-2B cells. The diminished expression of IFN-β1 and IFN-λ1 is therefore not attributable to eosinophils directly inhibiting HRV infectivity. Rather, our studies suggest that eosinophil secretion of TGF-β1, and perhaps other eosinophil-derived mediators, can suppress virus-induced interferon responses in epithelial cells.
It has been proposed that reduced interferon responses may contribute to virus-induced exacerbations of asthma,20 and that asthma may be associated with a deficiency in type I and type III interferons.10, 11 Other studies, however, have not found asthma-related differences in interferons.18 TGF-β can inhibit the expression of IFN-α and IFN-β from HRV infected fibroblasts.21 A recent study also demonstrated TGF-β inhibition of IFN-β and IFN-λ.22 Our data suggest that TGF-β1 production by airway eosinophils in asthma could provide another mechanism for in vivo deficiency in type I and III interferons. Reduced interferon responses could in turn lead to increased viral replication, more extensive lower airway virus infection and inflammation, and increased likelihood of airway obstruction and an exacerbation of symptoms in patients with asthma. Although we were only able to detect the decrease in IFN-λ1 protein, it is possible a similar reduction is occurring with IFN-β, which is typically expressed at lower levels than IFN-λ.22
Eosinophils have anti-viral effects under some circumstances.23-26 For example, the eosinophil granule proteins eosinophil-derived neurotoxin (EDN) and eosinophil cationic protein (ECP) exhibit ribonuclease activity which can diminish the infectivity of respiratory syncytial virus (RSV).26 It has been shown that activated eosinophils express ICAM-1 which can bind HRV16 and subsequently present HRV antigens to T-cells.27 Thus, eosinophils could simply sequester HRV and prevent their interaction with the co-cultured BEAS-2B cells. In addition, eosinophil granule protein ribonuclease activity caused decreased infectivity of respiratory syncytial virus (RSV).26 However, whether EDN and ECP ribonuclease activities can diminish infectivity of HRV had not previously been examined. We were not able to detect any antiviral effects in vitro in either activated or resting eosinophils.
Eosinophils secreted TGF-β1 and/or stimulated epithelial secretion of additional TGF-β1 at about the same level regardless of the presence of HRV. In our experiments, we did not prime or pre-activate the eosinophils. In an allergic airway, the airway eosinophils are likely primed or activated resulting in an increased capacity to release TGF-β. Since the stimulation of eosinophils by cytokines, including IL-3, IL-4, and IL-5, can increase TGF-β release,28 we speculate more dramatic suppression of IFN-λ1 may occur in vivo.
Most studies of virus-induced inflammatory responses utilize monocultures of epithelial cells, mononuclear cells, or granulocytes which do not reflect cell-cell interactions that occur in vivo. We describe an in vitro co-culture system that can be used as a model to examine cell-cell interactions that occur in patients with allergic asthma during HRV infections. A limitation of this study was the use of the BEAS-2B cell line instead of primary epithelial cells. However, BEAS-2B cells have been useful as a model system, particularly in the study of HRV-induced signaling.29-31 In fact, our findings are in general agreement with a recent study showing that TGF-β mediated suppression of both IFN-λ and IFN-β in primary bronchial epithelial cells.22 Another limitation of in vitro co-culture studies is reliance on reagents that may be important for one cell but have consequences for the other cell type. For example, serum-free media was used for co-cultures which is important to maintain the epithelial phenotype and prevent transition to fibroblasts; however, serum is also typically important for long-term survival and function of eosinophils. Similarly, hydrocortisone was included in the co-culture media for the epithelial cells, but can also affect eosinophil survival and function. We did confirm than the lack of serum and inclusion of hydrocortisone did not affect eosinophil survival; however, it remains possible that there are effects on eosinophil function.
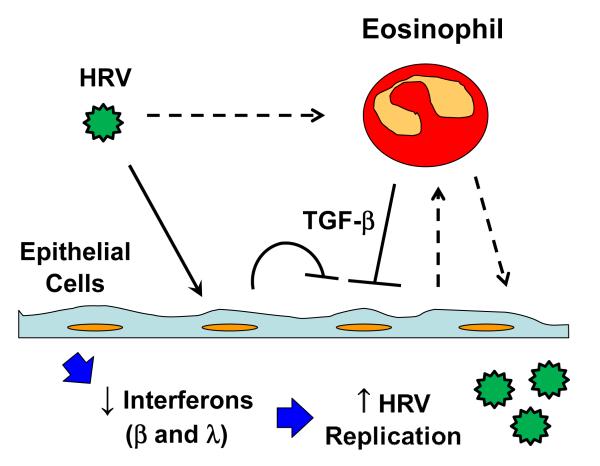
The eosinophil has been postulated to be a regulator of local immunity and/or remodeling/repair, also referred to as the “LIAR” hypothesis.12 With respect to this hypothesis, we and others have previously shown that human eosinophils are capable of enhancing cytokine secretion from activated T-cells.27, 32-34 Our results suggest that eosinophil secretion of cytokines, such as TGF-β, and other eosinophil-dependent mechanisms regulate virus-induced interferon responses, and thereby affect virus replication and clearance of virus. As shown in Figure 5, eosinophils may release TGF-β due to HRV stimulation or signaling from epithelial cells. TGF-β is also released from epithelial cells and this may be further increased by signaling from eosinophils. The net increase in TGF-β and potentially other mediators from eosinophils suppress the expression of IFN-β and IFN-λ. The decreased levels of interferons enable greater virus replication, which in turn can stimulate increased inflammatory responses leading to more severe HRV illnesses in patients with airway eosinophilia. This may, in part, explain the clinical observations of lower asthma exacerbation risk upon reduction of airway eosinophils by inhaled corticosteroids or mepolizumab.
Figure 5.
Schematic of proposed eosinophil regulatory activity. Epithelial cells are shown being infected by HRV in the presence of eosinophils. The eosinophil release of TGF-β along with epithelial cell endogenous TGF-β is shown to suppress the epithelial expression of IFN-β and IFN-λ, which enables increased virus replication that could lead to more severe inflammation and asthma symptoms. The dashed line between HRV and eosinophil represents a possible activation of eosinophils directly by HRV. The dashed lines between the eosinophil and epithelial cells represent possible release of unidentified mediators from epithelial cells to regulate eosinophil function and release of unidentified mediators from eosinophils to further regulate epithelial cell activity.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Beth Schwantes, Michelle Wolff and Evelyn Falibene for their support in the purification of eosinophils. This research was supported by the following NIH grants: HL088594, HL69116 and UL1RR025011.
Funding: This research was supported by the following NIH grants: HL088594, HL69116 and UL1RR025011.
Author Roles: SKM was the principal investigator for one of the funding sources, conceived of the study, directed the experiments and prepared the manuscript. PSF performed a majority of the experiments and contributed to preparation of the manuscript. JTK performed some of the experiments and edited the manuscript. WML performed some of the experiments and edited the manuscript. JEG co-directed the HRV experiments and edited the manuscript. NNJ was the principal investigator for two of the funding sources, co-directed the study and edited the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360:1715–21. doi: 10.1016/S0140-6736(02)11679-5. [DOI] [PubMed] [Google Scholar]
- 2.Flood-Page P, Swenson C, Faiferman I, Matthews J, Williams M, Brannick L, et al. A study to evaluate safety and efficacy of mepolizumab in patients with moderate persistent asthma. American Journal of Respiratory and Critical Care Medicine. 2007;176:1062–71. doi: 10.1164/rccm.200701-085OC. [DOI] [PubMed] [Google Scholar]
- 3.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and Exacerbations of Refractory Eosinophilic Asthma. The New England Journal of Medicine. 2009;360:973–84. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair P, Pizzichini MMM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for Prednisone-Dependent Asthma with Sputum Eosinophilia. The New England Journal of Medicine. 2009;360:985–93. doi: 10.1056/NEJMoa0805435. [DOI] [PubMed] [Google Scholar]
- 5.Pavord ID, Korn S, Howarth P, Bleecker ER, Buhl R, Keene ON, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–9. doi: 10.1016/S0140-6736(12)60988-X. [DOI] [PubMed] [Google Scholar]
- 6.DeMore JP, Weisshaar EH, Vrtis RF, Swenson CA, Evans MD, Morin A, et al. Similar colds in subjects with allergic asthma and nonatopic subjects after inoculation with rhinovirus-16. J Allergy Clin Immunol. 2009;124:245–52. doi: 10.1016/j.jaci.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, et al. Community Study of Role of Viral-Infections in Exacerbations of Asthma in 9-11 Year-Old Children. British Medical Journal. 1995;310:1225–9. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson KG, Kent J, Ireland DC. Respiratory Viruses and Exacerbations of Asthma in Adults. British Medical Journal. 1993;307:982–6. doi: 10.1136/bmj.307.6910.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. American Journal of Respiratory and Critical Care Medicine. 2008;178:667–72. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wark PAB, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. The Journal of Experimental Medicine. 2005;201:937–47. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PAB, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nature Medicine. 2006;12:1023–6. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 12.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clinical and Experimental Allergy. 2010;40:563–75. doi: 10.1111/j.1365-2222.2010.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto H, Sedgwick JB, Busse WW. Differential Regulation of Eosinophil Adhesion and Transmigration by Pulmonary Microvascular Endothelial Cells. The Journal of Immunology. 1998;161:971–7. [PubMed] [Google Scholar]
- 14.Lee WM, Wang W. Human rhinovirus type 16: mutant V1210A requires capsid-binding drug for assembly of pentamers to form virions during morphogenesis. J Virol. 2003;77:6235–44. doi: 10.1128/JVI.77.11.6235-6244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WM, Monroe SS, Rueckert RR. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993;67:2110–22. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosser AG, Vrtis R, Burchell L, Lee WM, Dick CR, Weisshaar E, et al. Quantitative and Qualitative Analysis of Rhinovirus Infection in Bronchial Tissues. American Journal of Respiratory and Critical Care Medicine. 2005;171:645–51. doi: 10.1164/rccm.200407-970OC. [DOI] [PubMed] [Google Scholar]
- 17.Denlinger LC, Sorkness RL, Lee WM, Evans M, Wolff M, Mathur S, et al. Lower Airway Rhinovirus Burden and the Seasonal Risk of Asthma Exacerbation. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201103-0585OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochkov YA, Hanson KM, Keles S, Brockman-Schneider RA, Jarjour NN, Gern JE. Rhinovirus-induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal.Immunol. 2010;3:69–80. doi: 10.1038/mi.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Manthei DM, Guadarrama AG, Lenertz LY, Denlinger LC. Rhinovirus-induced IL-1beta release from bronchial epithelial cells is independent of functional P2×7. Am J Respir Cell Mol Biol. 2012;47:363–71. doi: 10.1165/rcmb.2011-0267OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston SL. Innate immunity in the pathogenesis of virus-induced asthma exacerbations. Proc Am Thorac Soc. 2007;4:267–70. doi: 10.1513/pats.200701-030AW. [DOI] [PubMed] [Google Scholar]
- 21.Thomas BJ, Lindsay M, Dagher H, Freezer NJ, Li DS, Ghildyal R, et al. Transforming Growth Factor-beta Enhances Rhinovirus Infection by Diminishing Early Innate Responses. American Journal of Respiratory Cell and Molecular Biology. 2009;41:339–47. doi: 10.1165/rcmb.2008-0316OC. [DOI] [PubMed] [Google Scholar]
- 22.Bedke N, Sammut D, Green B, Kehagia V, Dennison P, Jenkins G, et al. Transforming growth factor-Beta promotes rhinovirus replication in bronchial epithelial cells by suppressing the innate immune response. PLoS One. 2012;7:e44580. doi: 10.1371/journal.pone.0044580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamko DJ, Yost BL, Gleich GJ, Fryer AD, Jacoby DB. Ovalbumin sensitization changes the inflammatory response to subsequent parainfluenza infection: Eosinophils mediate airway hyperresponsiveness, M-2 muscarinic receptor dysfunction, and antiviral effects. Journal of Experimental Medicine. 1999;190:1465–77. doi: 10.1084/jem.190.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg HF, Domachowske JB. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. Journal of Leukocyte Biology. 2001;70:691–8. [PubMed] [Google Scholar]
- 25.Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, et al. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. 2007;110:1578–86. doi: 10.1182/blood-2007-01-071340. [DOI] [PubMed] [Google Scholar]
- 26.Domachowske JB, Dyer KD, Bonville CA, Rosenberg HF. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. Journal of Infectious Diseases. 1998;177:1458–64. doi: 10.1086/515322. [DOI] [PubMed] [Google Scholar]
- 27.Handzel ZT, Busse WW, Sedgwick JB, Vrtis R, Lee WM, Kelly EAB, et al. Eosinophils bind rhinovirus and activate virus-specific T cells. Journal of Immunology. 1998;160:1279–84. [PubMed] [Google Scholar]
- 28.Elovic AE, Ohyama H, Sauty A, Mcbride J, Tsuji T, Nagai M, et al. IL-4-dependent regulation of TGF-alpha and TGF-beta 1 expression in human eosinophils. Journal of Immunology. 1998;160:6121–7. [PubMed] [Google Scholar]
- 29.Zaheer RS, Proud D. Human rhinovirus-induced epithelial production of CXCL10 is dependent upon IFN regulatory factor-1. Am J Respir Cell Mol Biol. 2010;43:413–21. doi: 10.1165/rcmb.2009-0203OC. [DOI] [PubMed] [Google Scholar]
- 30.Skevaki CL, Christodoulou I, Spyridaki IS, Tiniakou I, Georgiou V, Xepapadaki P, et al. Budesonide and formoterol inhibit inflammatory mediator production by bronchial epithelial cells infected with rhinovirus. Clin Exp Allergy. 2009;39:1700–10. doi: 10.1111/j.1365-2222.2009.03307.x. [DOI] [PubMed] [Google Scholar]
- 31.Lau C, Castellanos P, Ranev D, Wang X, Chow CW. HRV signaling in airway epithelial cells is regulated by ITAM-mediated recruitment and activation of Syk. Protein Pept.Lett. 2011;18:518–29. doi: 10.2174/092986611794927910. [DOI] [PubMed] [Google Scholar]
- 32.Liu LY, Mathur SK, Sedgwick JB, Jarjour NN, Busse WW, Kelly EAB. Human airway and peripheral blood eosinophils enhance Th1 and Th2 cytokine secretion. Allergy. 2006;61:589–97. doi: 10.1111/j.1398-9995.2006.01060.x. [DOI] [PubMed] [Google Scholar]
- 33.Weller PF, Rand TH, Barrett T, Elovic A, Wong DTW, Finberg RW. Accessory Cell-Function of Human Eosinophils - HLA-DR-Dependent, MHC-Restricted Antigen-Presentation and IL-1-Alpha Expression. Journal of Immunology. 1993;150:2554–62. [PubMed] [Google Scholar]
- 34.Odemuyiwa SO, Ghahary A, Li Y, Puttagunta L, Lee JE, Musat-Marcu S, et al. Cutting Edge: Human Eosinophils Regulate T Cell Subset Selection through Indoleamine 2,3-Dioxygenase. The Journal of Immunology. 2004;173:5909–13. doi: 10.4049/jimmunol.173.10.5909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.