Chromophore-assisted laser inactivation (CALI) was developed to instantly and specifically inactivate cofilin in cells. Simultaneous CALI and live imaging revealed that the principal role of cofilin in lamellipodia at steady state is to break down F-actin, control filament turnover, and regulate the rate of retrograde flow in lamellipodia.
Abstract
Cofilin is a key regulator of the actin cytoskeleton. It can sever actin filaments, accelerate filament disassembly, act as a nucleation factor, recruit or antagonize other actin regulators, and control the pool of polymerization-competent actin monomers. In cells these actions have complex functional outputs. The timing and localization of cofilin activity are carefully regulated, and thus global, long-term perturbations may not be sufficient to probe its precise function. To better understand cofilin's spatiotemporal action in cells, we implemented chromophore-assisted laser inactivation (CALI) to instantly and specifically inactivate it. In addition to globally inhibiting actin turnover, CALI of cofilin generated several profound effects on the lamellipodia, including an increase of F-actin, a rearward expansion of the actin network, and a reduction in retrograde flow speed. These results support the hypothesis that the principal role of cofilin in lamellipodia at steady state is to break down F-actin, control filament turnover, and regulate the rate of retrograde flow.
INTRODUCTION
The actin cytoskeleton is regulated by a complex network of regulatory molecules that act on distinct aspects of filament assembly and disassembly, including filament nucleation, severing, cross-linking, and end capping, as well as monomer sequestering and localization (Dent and Gertler, 2003; Dent et al., 2011). Many studies on the regulation of actin in migrating cells depend on long-term modifications of protein level or activity through knockdown and overexpression. Because most actin regulators are involved in the fundamental structure and function of the cytoskeleton, an essential component of cell viability, long-term manipulations may be marred by compensatory mechanisms, making it difficult to accurately interpret their results. Moreover, these actin-binding proteins are tightly regulated in space and time, and thus understanding their specific function in cellular events requires spatiotemporal information. The need to determine the functional consequences of localized, instantaneous changes in protein activity has spurred the development of light-mediated techniques to activate or knock down a protein's function.
Chromophore-assisted laser inactivation (CALI) is a loss-of-function technique in which proteins are inactivated with light by irradiating an attached photosensitizer chromophore. The chromophore generates reactive photoproducts that are highly destructive but short lived, thus only affecting the protein to which the chromophore is directly adjacent. CALI allows one to perform instantaneous protein loss-of-function analysis with subcellular precision (Jacobson et al., 2008). One issue with CALI has been in the labeling of the target protein; adding agents that label motifs other than the target can lead to nonspecific collateral damage. A major advance in this area occurred when it was shown that fluorescent proteins can be used as CALI photosensitizers (Rajfur et al., 2002), as the CALI target could be covalently labeled with a genetically encoded chromophore. The pitfall in using fluorescent proteins for CALI has been that they are relatively weak photosensitizers in comparison to other CALI chromophores and thus require a large dose of light that must be delivered by high-powered lasers not found in most imaging systems (Rajfur et al., 2002; Vitriol et al., 2007). This drawback was abated with the generation of KillerRed, a fluorescent protein photosensitizer that can be efficiently used for CALI with more than an order-of-magnitude less light than other fluorescent proteins (Bulina et al., 2006). Here we use KillerRed to inactivate the actin regulatory protein cofilin1.
The actin-depolymerizing factor (ADF)/cofilin family of proteins is a highly conserved group of actin regulatory molecules composed of ADF, cofilin1, and cofilin2 (Bernstein and Bamburg, 2010). Cofilin was once characterized by its ability to increase the rate of actin filament depolymerization by stimulating dissociation of actin from the pointed end (Carlier et al., 1997). Its principal role in actin regulation, however, appears to be through filament severing (Maciver et al., 1991; Andrianantoandro and Pollard, 2006). Severing is believed to occur primarily at the bare and cofilin-decorated filament boundary (Suarez et al., 2011) and at regions of mechanical discontinuity caused by cofilin-dependent changes in actin structure and rigidity (Elam et al., 2013). In addition to severing, cofilin itself can act as a nucleation factor at high concentrations (micromolar range; Chen et al., 2004; Andrianantoandro and Pollard, 2006), induce phosphate release from F-actin (Blanchoin and Pollard, 1999; Suarez et al., 2011), change filament rigidity (McCullough et al., 2011), and remodel filament structure (Pfaendtner et al., 2010; Galkin et al., 2011; Fan et al., 2013), although some of these effects are intertwined with its severing activity.
Whereas cofilin-induced filament severing leads to actin disassembly in a variety of actin structures (Lappalainen and Drubin, 1997; Okreglak and Drubin, 2007; Breitsprecher et al., 2011; Henty et al., 2011), in the lamellipodia it can have opposite functional outputs. Severing was initially believed to break down actin networks and turn over filaments (Svitkina and Borisy, 1999). It also was later shown, however, that cofilin-mediated filament severing could increase filament polymerization by creating new barbed ends (Ichetovkin et al., 2002), which is the preferential site of actin monomer addition. These new barbed ends, together with other factors that stimulate nucleation, polymerization, and stabilization of actin filaments, could lead to actin network growth and leading-edge protrusions (DesMarais et al., 2004; Ghosh et al., 2004). Cofilin filament severing may also promote actin assembly at the leading edge by increasing the pool of polymerization-competent ATP-actin monomers (Kiuchi et al., 2007), which involves a number of other factors to release cofilin and promote nucleotide exchange. This bimodal effect of cofilin on actin is tightly controlled by limiting cofilin activity to specific times and places in the cell through a complex, multilayered array of signaling cascades (Oser and Condeelis, 2009). As a result, a better elucidation of cofilin's function in regulating actin dynamics requires tools that can manipulate its activity with spatiotemporal precision.
A chemically engineered light-sensitive analogue of cofilin was made to address the importance of spatiotemporal cofilin activation. In a series of seminal experiments, it was shown that local activation of cofilin at the cell edge polymerized actin and generated protrusions (Ghosh et al., 2004). This provided strong support for the hypothesis that cofilin promoted actin polymerization rather than filament disassembly. Unfortunately, photoactivatable cofilin was not widely used after the initial study, most likely because it required microinjection of a chemically modified protein into cells and ultraviolet light for uncaging (Ghosh et al., 2002). In this article, we describe a means of inactivating cofilin with a standard laser scanning confocal microscope using KillerRed-CALI. We define parameters that make CALI of KillerRed cofilin successful and show that CALI is specific and does not cause collateral photodamage. We use CALI to demonstrate that instantaneous inactivation of cofilin in the cell inhibits actin filament turnover and has profound effects on the structure and dynamics of the lamellipodial actin network. Our data with CALI support the hypothesis that cofilin's principal role in the lamellipodia is to break down actin filaments and control the rate of retrograde flow.
RESULTS
Labeling cofilin with KillerRed does not alter its localization to actin
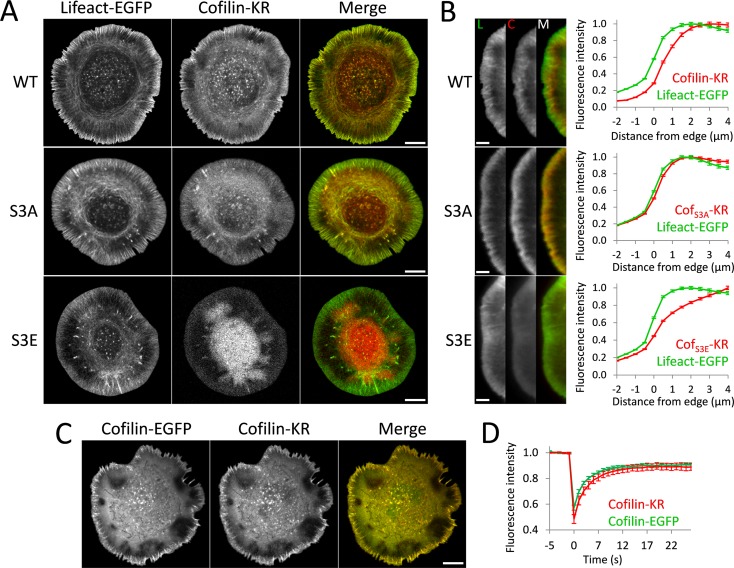
We made DNA constructs in which human cofilin1 (termed cofilin for the rest of the article), cofilinS3A (active and cannot be phosphorylated), or cofilinS3E (inactive, cannot bind actin; Moriyama et al., 1996) is labeled with the fluorescent protein photosensitizer KillerRed (KR; Bulina et al., 2006). An issue in any study using KR is whether it will affect the physiology of the protein to which it is attached. KR is an obligate dimer (Bulina et al., 2006; Pletnev et al., 2009); historically, multimer fluorescent proteins have been poor labeling reagents because they often cause mislocalization or aggregation when expressed. We thus examined the cellular distribution of cofilin-KR with respect to F-actin, which was labeled with Lifeact–enhanced green fluorescent protein (EGFP; Riedl et al., 2008). We observed that both wild-type (WT) and S3A mutant of cofilin-KR did localize appropriately to actin within the cell, including to the lamellipodia (Figure 1). S3E exhibited a diffuse pattern, which is consistent with its inability to bind actin cytoskeleton. As previously shown for endogenous cofilin by immunostaining (Svitkina and Borisy, 1999), WT cofilin-KR was excluded from the first 1–2 μm of the lamellipodia, whereas S3A occupied the entire lamellipodial region (Figure 1B). We further verified that KR did not alter cofilin localization by coexpressing it with the well-characterized cofilin-EGFP (Huang et al., 2008). The two constructs showed identical localization within the cell (Figure 1C). We also assessed the intracellular mobility of cofilin-KR and cofilin-EGFP with fluorescence recovery after photobleaching (FRAP). Both showed identical turnover rates and immobile fractions (Figure 1D and Supplemental Figure S1). Coupled with the results discussed later, we conclude that cofilin can be effectively labeled with KR without altering its localization or activity and behaves similarly to other widely used fluorescent protein–tagged cofilin in cells.
FIGURE 1:
Labeling cofilin with KillerRed does not alter its subcellular localization or turnover. (A) Representative images of cofilin-KR from various constructs used in this study. Cells were cotransfected with cofilin-KR and Lifeact-EGFP constructs to label F-actin. Wild-type cofilin (WT) and active cofilinS3A (S3A) labeled with KR display characteristic localization to actin filaments, including the lamellipodia, whereas inactive cofilinS3E (S3E) displays a reduced association with actin structures. Scale bars, 10 μm. (B) Colocalization of cofilin-KR with F-actin at the leading edge. Left, close-up images of the lamellipodia for Lifeact-EGFP (L), cofilin-KR (C), and a merged image (M). Scale bars, 5 μm. Right, graphs showing edge analysis of Lifeact-EGFP and cofilin-KR fluorescence. Values are normalized to the maximum-intensity value of the profile. Wild-type cofilin (WT) is excluded from the first 1–2 μm of the lamellipodia, whereas active cofilin (S3A) is present at the immediate cell edge. Inactive cofilin (S3E) is diffusely distributed throughout the cell. The number of cells used for this analysis is as follows: WT (14), S3A (14), S3E (11). Error bars represent 95% confidence intervals. (C) Cofilin-KR and cofilin-EGFP were coexpressed, showing a nearly identical localization profile. Scale bars, 10 μm. (D) Average recovery curves for FRAP analysis of cofilin-KR and cofilin-EGFP (n = 20 for each). Error bars represent 95% confidence intervals.
CALI of cofilin reduces actin mobility
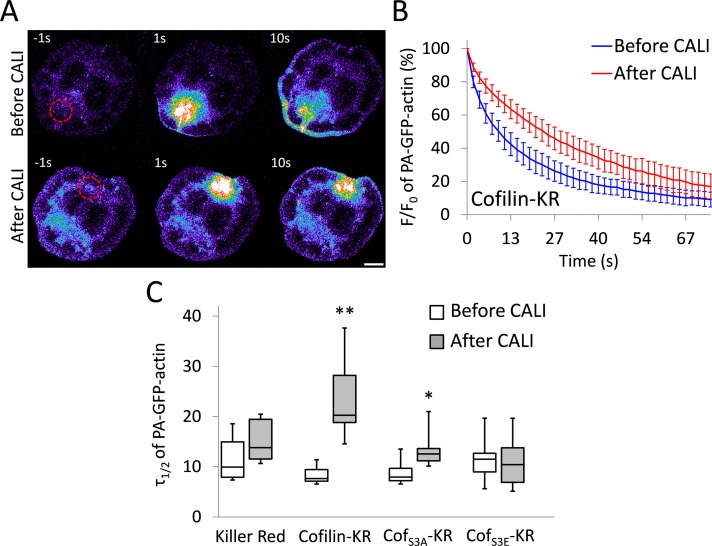
To show that CALI of cofilin-KR can reduce its enzymatic activity, we performed an in vivo assay that measures cofilin-dependent actin filament severing. Previously, it was shown that cofilin regulates the mobile pool of actin in the cell (Kiuchi et al., 2007). Using a photoactivatable fluorescent protein–tagged actin construct, Kiuchi et al. (2007) demonstrated that when cofilin activity was reduced, so was the diffusion of actin away from a photoactivated region. This mobilization of actin was dependent on the filament-severing ability of cofilin. We sought to reproduce these data with CALI. To do this, we performed experiments in which cofilin-KR constructs were coexpressed with photoactivatable GFP–β-actin (hereafter referred to as PA-GFP-actin). A 10-μm cytoplasmic region was photoactivated before and after irradiation of KR (Figure 2A and Supplemental Movie S1). We observed a significant reduction in the mobility of PA-GFP-actin after CALI with cofilin-KR, as evidenced by assessing the average time for the PA-GFP-actin fluorescence to decay to 50% of the initial value (τ1/2). After irradiation of cofilin-KR, τ1/2 nearly tripled, from 7.9 ± 0.9 to 21.7 ± 7.0 s (Figure 2, B and C). We also saw a marked decrease in actin mobility after cofilinS3A-KR was irradiated, although CALI of cofilin-KR had a more pronounced effect (Figure 2C). Irradiation of KR by itself or cofilinS3E-KR caused no change in actin mobility (Figure 2C), demonstrating that KR must be attached to cofilin and that cofilin must be bound to actin for the reduced actin mobility to occur. As an additional control, we used monomeric red fluorescent protein (mRFP) to CALI cofilin. mRFP is not as efficient at generating reactive oxygen species (ROS) as KR (Waldeck et al., 2012), and thus the effect of CALI on actin mobility should be reduced, given the same dose of light. In support of this, CALI of cofilin-mRFP resulted in only a small decrease in actin mobility (τ1/2 changed from 6.0 ± 0.9 to 8.2 ± 1.3 s after CALI). There was also no effect after CALI of cofilinS3A-mRFP or cofilinS3E-mRFP (Supplemental Figure S2). From these data, we concluded that CALI of cofilin-KR is specific, effective, and works by reducing cofilin's ability to sever actin filaments. This conclusion is consistent with work showing that cofilin has reduced actin-binding and -severing activity when exposed to ROS (Klemke et al., 2008; Klamt et al., 2009).
FIGURE 2:
CALI of cofilin reduces actin filament turnover and monomer mobility. (A) Representative images of cells expressing PA-GFP-actin in which a 10-μm circular region (shown as a red-dashed circle) of the cytoplasm was photoactivated before and after CALI of cofilin-KR. The image is pseudocolored to emphasize changes in fluorescence intensity, of which hot and cool colors indicate high and low intensities, respectively. Scale bar, 10 μm. (B) Average PA-GFP-actin fluorescence decay curves displayed as percentage of initial fluorescence value (F/F0) before and after CALI of cofilin-KR. There was a significant decrease in PA-GFP-actin mobility after CALI of cofilin-KR. Error bars represent 95% confidence intervals. (C) Box-and-whisker plot showing τ1/2 for all constructs before and after CALI. Plots denote 95th (top whisker), 75th (top edge of box), 25th (bottom edge of box), and 10th (bottom whisker) percentiles and the median (bold line in box). The number of cells used for this analysis is as follows: KR (7), cofilin-KR (9), cofS3A-KR (10), and cofS3E (10). **p < 0.0001, *p < 0.01 (Student's t test).
CALI of cofilinS3A increases F-actin in the lamellipodia and decreases the rate of retrograde flow
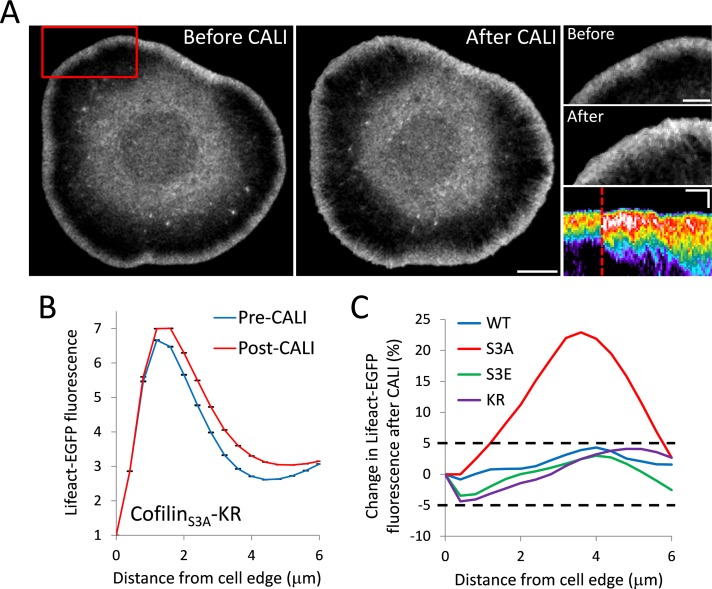
We next sought to use CALI to investigate cofilin's role in regulating the structure and dynamics of actin in lamellipodia. We conducted whole-cell CALI by expressing cofilin-KR, cofilinS3A-KR, and cofilinS3E-KR in Cath.a differentiated (CAD) cells together with Lifeact-EGFP. Peripheral F-actin was quantified with an automated image analysis routine in which the edge of the cell was identified and then fluorescence intensity was measured inward (see Materials and Methods and Supplemental Figure 3). Cells expressing cofilinS3A-KR showed a dramatic increase in F-actin after CALI, with the lamellipodial actin network growing backward toward the cell center (Figure 3 and Supplemental Movie 2). This result closely resembled the phenotype found in Drosophila S2 cells after cofilin depletion with RNA interference (RNAi; Iwasa and Mullins, 2007). The most significant increase in lamellipodial F-actin occurred 3–4 μm behind the leading edge (Figure 3, B and C), supportive of the hypothesis that cofilin's major role is in turning over actin filaments at the back of the lamellipodia. Similar results were observed when EGFP-actin was used to highlight the lamellipodia (Supplemental Figure 4), indicating that the observed changes in lamellipodial F-actin after CALI were not a result of increased Lifeact binding to the F-actin vacated by inactivated cofilinS3A-KR. Cells expressing cofilin-KR, the inactive cofilinS3E-KR, or KR by itself, however, showed no obvious change in lamellipodial F-actin quantity or distribution (Figure 3C and Supplemental Figure S5). We also used a semiautomated analysis routine using kymographs to validate our findings (Supplemental Figure S6). Kymograph analysis allowed us to include more cells in the analysis because we were able to isolate protruding regions of cells that did not have lamellipodia around their entire periphery. Kymograph analysis also revealed a large and significant increase in F-actin after CALI of cofilinS3A-KR and no obvious change after CALI of cofilin-KR or KR by itself. It did show a small but significant decrease in F-actin after CALI cofilinS3E-KR (Supplemental Figure S7). The complete lack of change in F-actin in the lamellipodia after CALI of WT-cofilin was unexpected because it yielded the most significant changes to actin monomer mobility (Figure 2).
FIGURE 3:
CALI of cofilinS3A increases F-actin in the lamellipodia. (A) Representative images of Lifeact-EGFP of a CAD cell before and after CALI of cofilinS3A-KR. Scale bar, 10 μm. Right, close-up images of the region indicated by the red box. Scale bars, 5 μm. Bottom right, representative kymograph of Lifeact-EGFP from a cofilinS3A-KR experiment. The kymograph is pseudocolored to emphasize changes in fluorescence intensity. Time of CALI is indicated with a dotted red line. Scale bars, 3 μm (vertical) and 3 min (horizontal). (B) Average leading edge profile of Lifeact-EGFP before and after CALI of cofilinS3A-KR (n = 6 cells). After CALI, there is a significant increase in Lifeact fluorescence. Error bars represent 95% confidence intervals. (C) Average percentage change in Lifeact-EGFP fluorescence intensity as a function of distance from the cell edge after CALI for the indicated constructs. Only cofilinS3A-KR (S3A) shows a change in intensity >5% at any distance, with the largest increase being ∼20% at 4 μm from the leading edge.
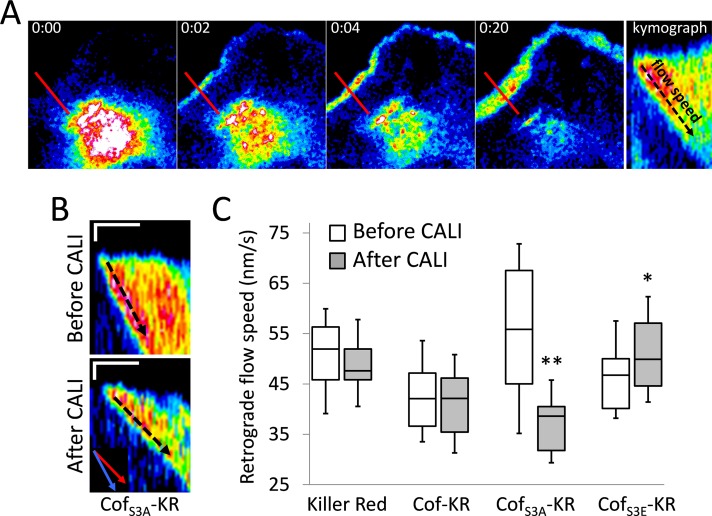
The increase in F-actin in lamellipodia after CALI of cofilinS3A-KR appeared to be accompanied by a decrease in the rate of actin retrograde flow (Supplemental Movie S2). We thus measured the retrograde flow of activated PA-GFP-actin in lamellipodia before and after irradiation of cofilin-KR (Figure 4A). Consistent with other results showing that cofilin activation increases retrograde flow (Delorme et al., 2007; Zhang et al., 2012), we saw a marked increase in flow rate when cells were overexpressing constitutively active cofilinS3A, which was significantly reduced by ∼44% (56 ± 4.0 to 37 ± 3.5 nm/s) after CALI (Figure 4, B and C). In cells expressing either cofilin-KR or KR alone, no change was observed after CALI; there was a slight increase in retrograde flow speed after CALI of cofilinS3E-KR (Figure 4C).
FIGURE 4:
CALI of cofilinS3A decreases retrograde flow of F-actin in the lamellipodia. (A) Schematic showing how retrograde flow rates are obtained. The first four images depict a cell in which PA-GFP-actin has been photoactivated in the cytoplasm. PA-GFP-actin then travels to leading edge, becomes incorporated into the lamellipodia, and gets pulled backward by retrograde flow. A kymograph is made from a region of the lamellipodia (red line), and retrograde flow speed is obtained by measuring the diagonal path of actin (black-dotted arrow), which represents distance over time. (B) Representative kymographs of PA-GFP-actin before and after CALI of cofilinS3A-KR. There is a significant reduction in retrograde flow speed after CALI. The inset in the bottom kymograph shows an overlay of the average retrograde flow speeds from the before (blue) and after (red) CALI kymographs. Scale bars, 1 μm (vertical) and 1 min (horizontal). (C) Box-and-whisker plot showing retrograde flow speeds for all constructs before and after CALI. Plots denote 95th (top whisker), 75th (top edge of box), 25th (bottom edge of box), and 10th (bottom whisker) percentiles and the median (bold line in box). The number of cells and before/after kymographs used for this analysis is as follows: KR (7, 32/30), cof-KR (8, 37/31), cofS3A-KR (9, 39/29), and cofS3E-KR (8, 33/34). **p < 0.0001, *p < 0.05 (Student's t test).
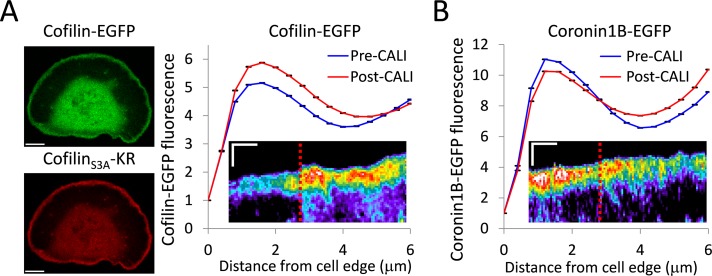
CofilinS3A outcompetes wild-type cofilin for lamellipodia localization
The lack of any phenotype in the lamellipodia after CALI of wild-type cofilin was perplexing. It is of note that these experiments were performed by overexpressing KR constructs. Endogenous cofilin is not labeled with a photosensitizer chromophore, and thus it cannot be inactivated with light. It is possible that cofilinS3A-KR preferentially binds to actin in the lamellipodia, excluding endogenous cofilin, and creates a local concentration of inactivatable protein that is sufficient to produce a loss-of-function effect after CALI. To address this, we performed CALI of cofilinS3A while expressing wild-type cofilin-EGFP. If cofilinS3A were preferentially binding to the actin in the lamellipodia, we would expect to see an increase in wild-type cofilin after CALI of cofilinS3A. After irradiation of cofilin-KR, there was indeed a significant increase of cofilin-EGFP localization to the lamellipodia (Figure 5A). This result also provides additional evidence that cofilin-binding sites, as well as the immediate binding partners of cofilin that could be affected by collateral damage, are not being destroyed by irradiation of cofilinS3A-KR, since wild-type cofilin is still recruited to the lamellipodia after CALI.
FIGURE 5:
CofilinS3A outcompetes wild-type cofilin for lamellipodia localization. (A) Left, coexpression of cofilin-EGFP and cofilinS3A-KR. Scale bars, 10 μm. Right, graph depicting the average leading edge profile of cofilin-EGFP before and after CALI of cofilinS3A-KR (n = 7 cells). After CALI, there is a significant increase in cofilin-EGFP fluorescence. Error bars represent 95% confidence intervals. The graph inset shows a representative kymograph of cofilin-EGFP from a cofilinS3A-KR CALI experiment. (B) Cells were cotransfected with coronin1B-EGFP and cofilinS3A-KR. The graph depicts average leading-edge profile of coronin1B-EGFP before and after CALI of cofilinS3A-KR (n = 6 cells). After CALI, there is a significant loss of coronin1B-EGFP fluorescence from the leading edge. Error bars represent 95% confidence intervals. The inset shows a representative kymograph of coronin1B-EGFP from a cofilinS3A-KR CALI experiment. Kymographs are pseudocolored to emphasize changes in fluorescence intensity. Time of CALI is indicated with a dotted red line. Kymograph scale bars, 3 μm (vertical) and 3 min (horizontal).
To be sure that the increase in wild-type cofilin localization was not a general, nonspecific effect from increasing F-actin after CALI, we performed CALI of cofilinS3A-KR while expressing another leading-edge actin-binding protein, coronin1B. CALI of cofilinS3A-KR caused a small depletion of Coronin1B from the lamellipodia (Figure 5B). Thus actin regulators are not simply being recruited to the lamellipodia en masse due to the increase in F-actin levels after CALI of cofilinS3A. This indicates that the increase in cofilin-EGFP to lamellipodia is in fact due to its occupation of binding sites left vacant by inactivated cofilinS3A.
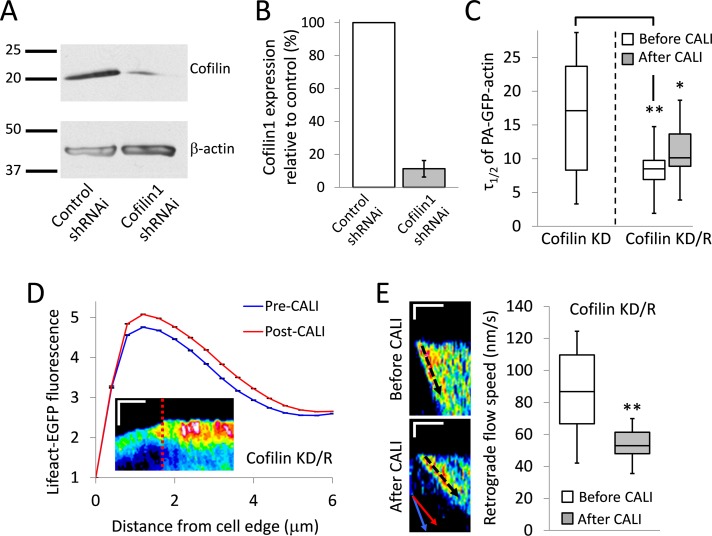
Knockdown of endogenous cofilin allows for effects in lamellipodia after CALI of wild-type cofilin
Although the S3A/wild-type cofilin competition assay explained why CALI of cofilinS3A-KR is effective in overexpression experiments, it did not address the issue of whether the loss-of-function phenotypes seen in lamellipodia after CALI cofilinS3A-KR are the result of creating an artificial environment with abnormally high levels of active cofilin. To address this, we performed cofilin-KR CALI experiments in cells in which endogenous cofilin had been stably knocked down with short hairpin RNAi (shRNAi; Figure 6, A and B). We were able to achieve ∼89% cofilin1 knockdown (Figure 6B). Cofilin knockdown (cofilin KD) cells displayed significantly reduced actin mobility (Figure 6C), as expected from our CALI experiments (Figure 2) and previous work (Kiuchi et al., 2007). This phenotype was rescued with expression of cofilin-KR (cofilin knockdown/rescue [KD/R] in Figure 6C). As with the case of cofilin-KR CALI in cells in which it is overexpressed, CALI of cofilin-KR caused a substantial reduction in actin mobility/filament turnover (Figure 6C).
FIGURE 6:
Knockdown of endogenous cofilin1 allows for effects in the lamellipodia after CALI of wild-type cofilin. (A, B) Cofilin1 knockdown with shRNAi verified by Western blot. Sample gel (A) and bar graph showing quantification (B). The Western blot was repeated three times. Error bars indicate SD. (C) Box-and-whisker plot showing τ1/2 for PA-GFP-actin mobility assay in cells in which endogenous cofilin had been knocked down with shRNAi (cofilin-KD, n = 13) or cells in which endogenous cofilin had been knocked down and then rescued with cofilin-KR (cofilin KD/R, n = 14). Actin mobility in cofilin KD/R cells was assessed before and after CALI. **p < 0.01, *p < 0.05 (Student's t test). (D) Average leading-edge profile of Lifeact-EGFP before and after CALI of cofilin-KR in cofilin KD/R cells (n = 7 cells). After CALI, there is a significant increase in Lifeact-EGFP fluorescence. Error bars represent 95% confidence intervals. The graph inset shows a representative kymograph of Lifeact-EGFP from a cofilin KD/R CALI experiment. The kymograph is pseudocolored to emphasize changes in fluorescence intensity. Time of CALI is indicated with a dotted red line. Scale bars, 3 μm (vertical) and 5 min (horizontal). (E) Left, representative kymographs of PA-GFP-actin before and after CALI of cofilin-KR in cofilin-KD cells (cofilin KD/R). The inset in the bottom kymograph shows an overlay of the average retrograde flow speeds from the before (blue) and after (red) CALI kymographs. There is a significant reduction in retrograde flow speed after CALI. Scale bar, 1 μm (vertical) and 3 min (horizontal). Right, a box-and-whisker plot showing retrograde flow speeds in cofilin KD/R cells before and after CALI (n = 11 cells, 52/50 kymographs from before/after CALI). Double asterisk indicates p < 0.0001 (Student's t test). All box-and-whisker plots denote 95th (top whisker), 75th (top edge of box), 25th (bottom edge of box), and 10th (bottom whisker) percentiles and the median (bold line in box).
Contrary to overexpression experiments, CALI of cofilin-KR in KD/R cells resulted in a significant increase in F-actin in the lamellipodia (Figure 6D), as well as a significant reduction in the rate of retrograde flow (Figure 6E). These results mirrored those found in experiments in which cofilinS3A-KR was overexpressed, further indicating that CALI cofilinS3A-KR was effective in lamellipodia because it outcompetes endogenous cofilin for binding there (Figure 5A) and not because it creates an artificial pre-CALI environment. The requirement for knockdown/rescue for cofilin-KR provides critical evidence that CALI of cofilin in lamellipodia is specific.
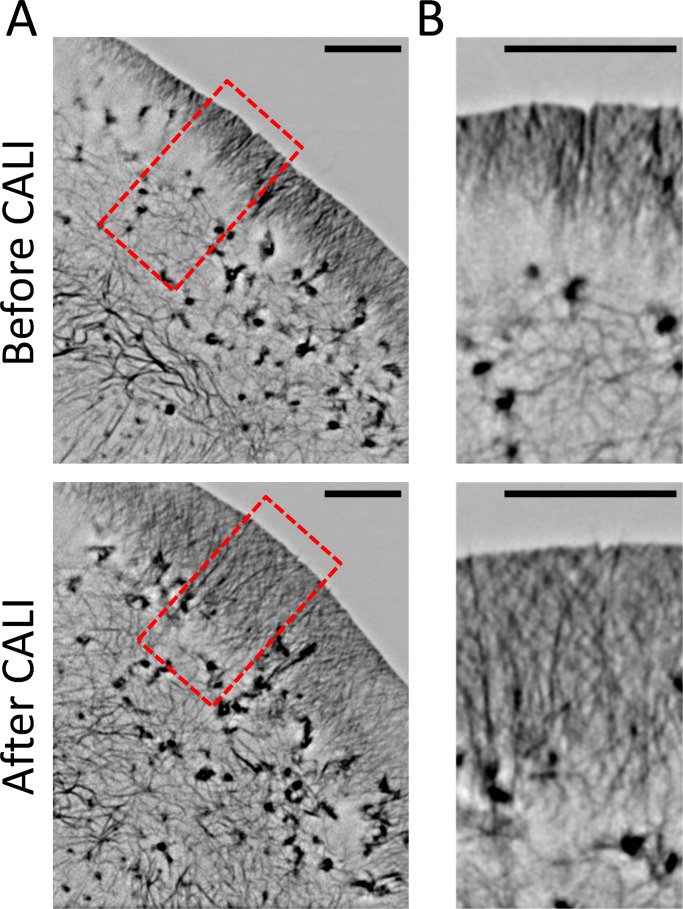
Live-cell superresolution imaging demonstrates an increase in lamellipodial F-actin after CALI of cofilin
To provide greater detail about how CALI of cofilin alters the structure of lamellipodia, we performed CALI of cofilin-KR in cofilin-KD cells together with live-cell three-dimensional structured illumination microscopy (3D-SIM). 3D-SIM allows one to double the resolution of light microscopy to ∼100 nm, is compatible with commonly used fluorophores, and allows for fast-imaging speeds (Schermelleh et al., 2010). For these experiments, we also used Lifeact-EGFP to label F-actin. 3D-SIM imaging confirmed the increase in the size of the lamellipodia (Figure 7 and Supplemental Movie S3) and revealed the presence of long filaments interspersed throughout it. Again, these data support the hypothesis that cofilin works in lamellipodia to sever actin filaments to turn them over, restrict the size of lamellipodia, and control the rate of retrograde flow.
FIGURE 7:
Live-cell superresolution imaging of actin demonstrates an increase in lamellipodial F-actin after CALI of cofilin. (A) Representative images of Lifeact-EGFP before and after CALI of cofilin-KR in cofilin-KD cells taken with 3D-SIM imaging. (B) Close-up images of the region indicated by the red box in A. After CALI, the lamellipodia significantly extended inward and displayed numerous long filaments interspersed throughout. Scale bars, 5 μm.
DISCUSSION
CALI offers a rare opportunity to compare the same cells with and without a key target protein without the complication of molecular compensation, akin to using a specific inhibitor. In this study we implemented a fluorescent protein–based CALI approach that uses the efficient photosensitizer KillerRed, allowing one to selectively and instantly inactivate cofilin1. This work provides arguably the first demonstration of cofilin's real-time regulation of actin turnover in living cells. Consistent with previous studies (Hotulainen et al., 2005; Iwasa and Mullins, 2007), our results confirm that cofilin's primary role in regulating the steady state of lamellipodia is to disassemble F-actin and regulate filament turnover, as CALI of cofilin causes an immediate increase of F-actin there. Several pieces of evidence indicate that the F-actin increase is caused by a decrease of filament disassembly and not from the formation of new filaments. First, the increase in F-actin occurs primarily 3–4 μm behind the leading edge. Second, 3D-SIM imaging reveals the presence of numerous longer filaments interspersed throughout the lamellipodia after CALI (Figure 7). Finally, the increase in wild-type cofilin localization after CALI of cofilinS3A (Figure 5) indicates that existing filaments are elongating, since wild-type cofilin preferentially binds aged, ADP-actin filaments rather than newly polymerized actin (Yeoh et al., 2002).
It was also interesting to see that CALI of cofilin resulted in immediate slowdown of F-actin retrograde flow. This finding is consistent with a recent study demonstrating an essential role for ADF/cofilin in regulating retrograde flow in neurogenesis (Flynn et al., 2012) and other studies showing an increase in retrograde flow downstream of cofilin activation (Delorme et al., 2007; Zhang et al., 2012). Of importance, the decrease in retrograde flow rate suggests that the increase of F-actin at the back of the lamellipodia actin network after cofilin CALI is not simply a result of more F-actin being moved backward but instead is caused by loss of filament disassembly. In vitro it has been shown that filament severing by cofilin stochastically alters filament dynamics to favor turnover (Michelot et al., 2007) and can turn over branched actin networks in bulk (Reymann et al., 2011). Increasing the dynamic nature of the actin network in lamellipodia downstream of cofilin activity via enhanced filament turnover and retrograde flow appears to be important for protrusion formation and cell motility (Hotulainen et al., 2005; Delorme et al., 2007; Zhang et al., 2012), although the inverse has also been shown during growth cone guidance (Wen et al., 2007). The differences here may lie in the amount of cofilin activation and specific context in which it is activated.
We do not feel that our data conflict with models in which cofilin filament severing is used to create barbed ends and polymerize new actin (Oser and Condeelis, 2009). Much work has been done to show that the polymerization-inducing aspect of cofilin activity is limited to small subcellular compartments downstream of local signaling events. Furthermore, to be fully effective, cofilin-induced actin polymerization requires a synergy with nucleation factors such as Arp 2/3 (Ichetovkin et al., 2002). Filament severing by cofilin also stimulates actin assembly downstream of extracellular signals by increasing the monomer pool (Kiuchi et al., 2007). It is also certainly plausible that localized cofilin-induced polymerization can occur simultaneously with cofilin-induced filament breakdown and turnover, depending on the specific environment in which cofilin is activated. One piece of evidence in our data supporting the dual role of cofilin is that simply overexpressing cofilinS3A increases F-actin in lamellipodia (Supplemental Figure S8), which coincides with its role in promoting F-actin polymerization. However, global inactivation of cofilinS3A with CALI causes F-actin to increase even more (Figure 3), which supports its role in disassembling actin filaments. Unfortunately, due to the extremely rapid turnover of cofilin (Figure 1E and Supplemental Figure S1), we were not able to locally inactivate cofilin to perform the reciprocal experiment of local cofilin photoactivation (Ghosh et al., 2004). Even pulse irradiating a 5-μm region of the leading edge led to complete photobleaching of KR throughout the cell before a gradient could be achieved. It is possible that increased laser power could be used to generate a local phenotype, but based on the mobility of cofilin, it would require an irradiation radius of at least 20 μm (Vitriol et al., 2007).
With this work we demonstrate a specific, instantaneous inactivation of cofilin1 using genetically encoded constructs and a standard laser scanning confocal microscope. Because use of KR to inactivate target proteins has been rather limited since its debut, we hope that this article will provide useful ideas and considerations that will make potential KR-CALI experiments a success. We provided extensive data showing that the KR inactivation of cofilin was specific, including irradiation of KR by itself, CALI of an inactive form of cofilin, use of a less powerful photosensitizer to show dose dependence (Supplemental Figure S2), demonstration that cofilin-binding sites were not being destroyed with CALI (Figure 5), and demonstration of the requirement of knockdown/rescue for WT-cofilin. The knockdown/rescue results are particularly important, as in the case of overexpression, WT-cofilin still maintains all of the same binding partners and localization. If CALI of wild-type cofilin did indeed cause nonspecific collateral damage to the tightly packed environment of the lamellipodia, it should have been evident in both the overexpression and knockdown/rescue experiments. In this sense, overexpression of WT-cofilin-KR becomes an excellent control for CALI specificity in the lamellipodia.
The identification that in some instances, CALI of cofilinS3A was able to mimic loss of cofilin in cofilin-KD cells was also important, as this may be considered for future experiments in which it is not possible to use a knockdown/rescue approach. Some CALI experiments required knockdown/rescue (Vitriol et al., 2007), whereas others did not (Rajfur et al., 2002). We showed that cofilinS3A has a dominant effect on binding actin in the lamellipodia, excluding WT-cofilin (Figure 5). There may be other examples in which CALI of overexpressed cofilinS3A will be successful. It is also of interest that CALI of cofilinS3E had an opposite effect on the actin of the lamellipodia to that of cofilinS3A or WT-cofilin in the KD/R experiments (Figure 5 and Supplemental Figure S7). Although it is often believed to be inert because it is not able to bind actin (Moriyama et al., 1996), there clearly was a cofilinS3E overexpression phenotype on the F-actin content of the lamellipodia (Supplemental Figure S8). In addition, FRAP analysis revealed that cofilinS3E has a significantly lower mobile fraction than wild-type cofilin or cofilinS3A (Supplemental Figure S1). Taking our results together, our interpretation of this is that cofilinS3E is binding an activator of cofilin, which is then released upon CALI to activate the endogenous cofilin pool. This is also an important example of how multiple controls must be done and carefully assessed to confer CALI specificity.
Through specific and instantaneous inactivation of cofilin at the whole-cell level, we were able to show its principal role in regulating filament turnover, the size of the lamellipodia, and the speed of retrograde flow. As KR-CALI is further developed, we hope to reveal more intricacies of cofilin and how it regulates actin in various cellular processes, including the dynamics of growth cone motility (Wen et al., 2007) and the formation and maintenance of the neuromuscular junction (Lee et al., 2009). We believe that we have shown that KR-CALI, in addition to being of great use for determining the spatiotemporal function of cofilin, is a generally accessible technique and an important addition to the cell biologist's toolbox.
MATERIALS AND METHODS
DNA constructs
Cofilin-KR and its mutant forms were made by subcloning KR from pKillerRed-N (Evrogen, Moscow, Russia) into cofilin-GFP constructs that had been previously subcloned into the pcDNA3.1+ expression vector (Invitrogen, Carlsbad, CA). The following additional DNA constructs were used in this study: Lifeact-EGFP (pN1-Lifeact-EGFP; provided by Roland Wedlich-Soldner, Max-Planck Institute of Biochemistry, Martinsried, Germany), PA-GFP-actin (pC1 PA-GFP-β-actin), EGFP-γ-actin (pCS2+-GFP-γ-actin), and EGFP-coronin1B (pLL5.1 EGFP-coronin1B, provided by James Bear (University of North Carolina at Chapel Hill, Chapel Hill, NC). DNA constructs were prepared using Endotoxin-free Maxi Prep kits (Qiagen, Valencia, CA).
Cell culture
CAD cells were cultured in DMEM/F12 (Life Technologies, Carlsbad, CA) supplemented with 8% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). Cells were imaged in DMEM/F12 minus phenol red supplemented with 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Sigma-Aldrich). CAD cells were transfected with FuGENE 6 (Roche, Indianapolis, IN) according to the manufacturer's protocol. For imaging experiments, cells were plated on coverslips coated with 20 μg/ml laminin (Sigma-Aldrich).
For shRNAi knockdown of cofilin, cells were infected with lentiviral particles (Santa Cruz Biotechnology, Santa Cruz, CA) that expressed both an shRNAi hairpin against mouse cofilin1 and a gene for puromycin resistance. Infection was done in the presence of 5 μg/ml Polybrene (Santa Cruz Biotechnology). At 48 h after infection, cells were selected with 10 μg/ml puromycin (Santa Cruz Biotechnology). This concentration was chosen because it killed 100% of uninfected cells within 24 h. After selection, cells were continuously cultured in media containing 10 μg/ml puromycin, except during imaging experiments. Efficacy of knockdown monitored by Western blot. A rabbit polyclonal antibody against cofilin (ab42824; Abcam, Cambridge, MA) was used for Western blotting.
Microscopy
Imaging for colocalization between cofilin-KR constructs and Lifeact-EGFP was performed on an Eclipse Ti inverted microscope (Nikon, Melville, NY) equipped with a QuantEM electron-multiplying charge-coupled device (EMCCD) camera (Photometrics, Tucson, AZ) using a 60×/1.49 numerical aperture (NA) Apochromat (Apo) total internal reflection fluorescence (TIRF) oil immersion objective. Cells were mounted in a custom live-cell chamber and maintained at 37°C with a heated stage adapter (Warner, Hamden, CT).
CALI experiments were performed on a Nikon A1R fully automated laser scanning confocal microscope equipped with an automated z-drive with Perfect Focus, multiple laser lines with acousto-optic tunable filter control, motorized x-y stage, a second resonance scanner for high-speed imaging and photoactivation, an attached cage incubator with CO2 and temperature control, and multiple photomultiplier tube detectors. Cells were mounted in a custom live-cell chamber. All experiments were performed using a 60×/1.49 NA Apo TIRF oil immersion objective. To ensure evenness of expression, all cells chosen for CALI experiments had mean KR fluorescence values within 20% of each other.
CALI was performed by continuously irradiating a region containing the entire cell with the 561-nm laser line for 2 min with the laser power at 100% (9 mW at the microscope objective) and a 78-μs pixel dwell time. Photoactivation of PA-GFP was performed with the 405-nm laser line (laser power 100%, 2.2-μs pixel dwell time) by irradiating a circular region with a 10-μm diameter with a single 100-ms bleaching iteration. Two different regions were chosen for each cell to measure actin monomer mobility both before and after CALI. FRAP of EGFP and KR was performed by irradiating a 5-μm diameter with a single 1-s bleaching iteration with either the 405/488 or the 405/488/561-nm laser lines, respectively (laser power 100%, 2.2-μs pixel dwell time). For all experiments, GFP or EGFP fluorescence was monitored with the 488-nm laser line. Irradiation of cells with the 561-nm laser line for CALI had no photobleaching effect on GFP or EGFP fluorescence.
Superresolution imaging was performed on a Nikon N-SIM microscope equipped with an automated piezo z-drive with Perfect Focus, a motorized x-y stage, and an iXon3 897 EMCCD camera (Andor, Belfast, United Kingdom). Images were acquired in 3D-SIM mode using a 100×/1.49 NA Apo TIRF objective. Cells were mounted in a custom live-cell chamber and maintained at 30°C.
Image analysis
All microscope image sets were obtained using Nikon Elements and then analyzed with ImageJ software (National Institutes of Health, Bethesda, MD). To determine colocalization between cofilin-KR constructs and Lifeact-EGFP, background-subtracted images were exported into ImageJ, where they were transformed into RGB .tif files and analyzed using the Edge Ratio macro as described (Cai et al., 2008; available at www.unc.edu/∼cail/programming.html). Edge intensity values were normalized to the peak fluorescence value for each curve.
To determine actin monomer mobility rates, fluorescence intensities from the photoactivated region and a background region from the opposite side of the cell were exported into Excel software (Microsoft, Redmond, WA). Intracellular background fluorescence intensity values were subtracted from the photoactivated intensity values to account for general changes in cellular fluorescence. Intensity values were normalized to the time point immediately after photoactivation. Double-exponential curve fitting was performed on the fluorescence decay curves with a free online curve-fitting application (http://ZunZun.com/) so that τ1/2 could be obtained.
To determine retrograde flow rates, PA-GFP-actin image sets were divided into pre- and post-CALI. Image sets were background subtracted, and then kymographs were drawn at the edge of cells where PA-GFP-actin incorporation into the lamellipodia was clearly visible. Kymographs were drawn in as many regions of the cell as possible. Retrograde flow was measured by drawing a diagonal line that traced the rearward flow of incorporated PA-GFP-actin, representing the distance the actin traveled over time.
To determine the changes in intensity and distribution of actin and actin regulatory proteins after CALI, background-subtracted image sets were exported into Matlab software (MathWorks, Natick, MA) and analyzed using a custom script that automatically detects the cell edge based on a user-input threshold value and then divides the cells into concentric distance bins of user-defined width. For all experiments a distance bin value of 0.4 μm was used. Each CALI movie contained 40 frames before irradiation and 60 frames after irradiation. All movies were manually checked to ensure that edge detection was accurate for the duration of the time lapse. Intensity values from each distance bin from every frame of all movies from each data set were averaged and then exported into Excel for analysis. Edge measurement curves were normalized to the first value of the curve. For edge measurement experiments only cells that exhibited a lamellipodia on at least 80% of their periphery for the entire duration of the movie were used. A second method was used in which kymographs were drawn in lamellipodia regions and then analyzed with a custom Matlab script that automatically detected the edge of the cell with user-defined threshold intensity and then reported intensity values in user-defined distance bins (0.4 μm was used). This method allowed us to include a greater number of cells and exclude regions where there was not a lamellipodium for the entire duration of the movie. Intensity values from each time point of every kymograph were exported into Excel and then normalized as described.
Supplementary Material
Acknowledgments
We thank James E. Bear (University of North Carolina at Chapel Hill) for the kind gift of coronin1B-EGFP and Ken Jacobson (University of North Carolina at Chapel Hill) for critical reading of the manuscript. This project is supported in part by research grants from the National Institutes of Health to J.Q.Z., an F32 fellowship from the National Institutes of Health to E.A.V., and National Institute of Neurological Disorders and Stroke Core Facilities grants to the Viral Vector Core (P30NS055077) and the Integrated Cellular Imaging Microscopy Core (P30NS055077) of Emory Neuroscience.
Abbreviations used:
- CALI
chromophore-assisted laser inactivation
- KD/R
knockdown/rescue
- KR
KillerRed
- PA-GFP
photoactivatable green fluorescent protein
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0156) on May 15, 2013.
REFERENCES
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Koestler SA, Chizhov I, Nemethova M, Mueller J, Goode BL, Small JV, Rottner K, Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J Cell Sci. 2011;124:3305–3318. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Sneider JM, Boyle JA, Minamide LS, Bamburg JR. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43:7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13:646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011;3:a001800. doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam WA, Kang H, De La Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Saunders MG, Haddadian EJ, Freed KF, De La Cruz EM, Voth GA. Molecular origins of cofilin-linked changes in actin filament mechanics. J Mol Biol. 2013;425:1225–1240. doi: 10.1016/j.jmb.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn KC, et al. ADF/cofilin-mediated actin retrograde flow directs neurite formation in the developing brain. Neuron. 2012;76:1091–1107. doi: 10.1016/j.neuron.2012.09.038. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Ichetovkin I, Song X, Condeelis JS, Lawrence DS. A new strategy for caging proteins regulated by kinases. J Am Chem Soc. 2002;124:2440–2441. doi: 10.1021/ja017592l. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Henty JL, Bledsoe SW, Khurana P, Meagher RB, Day B, Blanchoin L, Staiger CJ. Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell. 2011;23:3711–3726. doi: 10.1105/tpc.111.090670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TY, Minamide LS, Bamburg JR, Bokoch GM. Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell. 2008;15:691–703. doi: 10.1016/j.devcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Rajfur Z, Vitriol E, Hahn K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 2008;18:443–450. doi: 10.1016/j.tcb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Ohashi K, Kurita S, Mizuno K. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J Cell Biol. 2007;177:465–476. doi: 10.1083/jcb.200610005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt F, Zdanov S, Levine RL, Pariser A, Zhang Y, Zhang B, Yu LR, Veenstra TD, Shacter E. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol. 2009;11:1241–1246. doi: 10.1038/ncb1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke M, Wabnitz GH, Funke F, Funk B, Kirchgessner H, Samstag Y. Oxidation of cofilin mediates T cell hyporesponsiveness under oxidative stress conditions. Immunity. 2008;29:404–413. doi: 10.1016/j.immuni.2008.06.016. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lee CW, Han J, Bamburg JR, Han L, Lynn R, Zheng JQ. Regulation of acetylcholine receptor clustering by ADF/cofilin-directed vesicular trafficking. Nat Neurosci. 2009;12:848–856. doi: 10.1038/nn.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guerin C, Boujemaa-Paterski R, Staiger CJ, Martiel JL, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Moriyama K, Iida K, Yahara I. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells. 1996;1:73–86. doi: 10.1046/j.1365-2443.1996.05005.x. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Condeelis J. The cofilin activity cycle in lamellipodia and invadopodia. J Cell Biochem. 2009;108:1252–1262. doi: 10.1002/jcb.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaendtner J, De La Cruz EM, Voth GA. Actin filament remodeling by actin depolymerization factor/cofilin. Proc Natl Acad Sci USA. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev S, et al. Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. J Biol Chem. 2009;284:32028–32039. doi: 10.1074/jbc.M109.054973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Dissecting the link between stress fibres and focal adhesions by CALI with EGFP fusion proteins. Nat Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- Reymann AC, Suarez C, Guerin C, Martiel JL, Staiger CJ, Blanchoin L, Boujemaa-Paterski R. Turnover of branched actin filament networks by stochastic fragmentation with ADF/cofilin. Mol Biol Cell. 2011;22:2541–2550. doi: 10.1091/mbc.E11-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, Uetrecht AC, Shen F, Jacobson K, Bear JE. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proc Natl Acad Sci USA. 2007;104:6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldeck W, Heidenreich E, Mueller G, Wiessler M, Toth K, Braun K. ROS-mediated killing efficiency with visible light of bacteria carrying different red fluorochrome proteins. J Photochem Photobiol B Biol. 2012;109:28–33. doi: 10.1016/j.jphotobiol.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Wen Z, Han L, Bamburg JR, Shim S, Ming GL, Zheng JQ. BMP gradients steer nerve growth cones by a balancing act of LIM kinase and Slingshot phosphatase on ADF/cofilin. J Cell Biol. 2007;178:107–119. doi: 10.1083/jcb.200703055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh S, Pope B, Mannherz HG, Weeds A. Determining the differences in actin binding by human ADF and cofilin. J Mol Biol. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- Zhang XF, Hyland C, Van Goor D, Forscher P. Calcineurin-dependent cofilin activation and increased retrograde actin flow drive 5-HT-dependent neurite outgrowth in Aplysia bag cell neurons. Mol Biol Cell. 2012;23:4833–4848. doi: 10.1091/mbc.E12-10-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.