The role of the 3′-untranslated region of the mouse Cryptochrome 1 (mCry1) gene is studied at the posttranscriptional level. The results suggest that miR-185 plays a role in the fine-tuned regulation of mCRY1 protein expression by controlling the rhythmicity of mCry1 mRNA translation.
Abstract
Mammalian circadian rhythm is observed not only at the suprachiasmatic nucleus, a master pacemaker, but also throughout the peripheral tissues. Investigation of the regulation of clock gene expression has mainly focused on transcriptional and posttranslational modifications, and little is known about the posttranscriptional regulation of these genes. In the present study, we investigate the role of microRNAs (miRNAs) in the posttranscriptional regulation of the 3′-untranslated region (UTR) of the mouse Cryptochrome 1 (mCry1) gene. Knockdown of Drosha, Dicer, or Argonaute2 increased mCry1-3′UTR reporter activity. The presence of the miRNA recognition element of mCry1 that is important for miR-185 binding decreased mCRY1 protein, but not mRNA, level. Cytoplasmic miR-185 levels were nearly antiphase to mCRY1 protein levels. Furthermore, miR-185 knockdown elevated the amplitude of mCRY1 protein oscillation. Our results suggest that miR-185 plays a role in the fine-tuned regulation of mCRY1 protein expression by controlling the rhythmicity of mCry1 mRNA translation.
INTRODUCTION
The mammalian molecular circadian clock system is composed of feedback loops and transcriptional, translational, and posttranslational regulatory steps (Lowrey and Takahashi, 2004). The core molecular clock is established by a positive limb, composed of heterodimers of the transcription factors CLOCK and BMAL, which drives the rhythmic expression of the negative limb proteins Period (PER1–3) and Cryptochrome (CRY1/2; Dibner et al., 2010). Accumulation of PER and CRY proteins results in their translocation to the nucleus, where they inhibit BMAL/CLOCK transcriptional activity, thereby repressing their own expression and initiating a new cycle. The Cryptochrome1 (Cry1) gene, a true circadian oscillator, maintains its rhythmicity in the absence of environmental cues. Mouse Cry1 (mCry1) transcripts persist when mice are kept in a “free-running” state, such as constant darkness (Liu et al., 1999). This has also been observed for other clock genes. Recently posttranscriptional regulation has been recognized as being important for the fine-tuning of the circadian system (Kim et al., 2007; Kojima et al., 2007; Woo et al., 2009; Lee et al., 2012b).
MicroRNAs (miRNAs) act as regulators in fine-tuning gene expression. They comprise an abundant group of endogenous, small, noncoding RNAs and predominantly act at the mRNA level to regulate the expression of protein-coding genes through translational repression and/or degradation of their target mRNAs (Bartel, 2004; Guo et al., 2010). The short (20–22 nucleotides [nt]) miRNAs bind to Argonaute (AGO) proteins and guide the AGO-associated RNA-induced silencing complex (RISC) to mRNA targets (Hutvagner and Simard, 2008). AGO2 then mediates the miRNA-dependent interaction of the RISC complex with the target mRNA (Meister et al., 2004). AGO2, the catalytic center of RISC, induces the endonucleolytic cleavage of perfect or nearly perfect base pairings of miRNA and target mRNA or 11 or 12 contiguous Watson–Crick pairs to the center of the miRNA (Meister et al., 2004; Shin et al., 2010). In contrast, imperfect miRNA–mRNA interactions generally cause translational repression or mRNA destabilization (Bushati and Cohen, 2007; Filipowicz et al., 2008). Bioinformatic analyses suggest that each miRNA is capable of controlling hundreds of target genes in humans, and it was reported that >60% of protein-coding genes are under selective pressure to maintain pairing to miRNAs (Friedman et al., 2009). This indicates that miRNAs have a great potential to regulate the majority of protein-coding genes. Thus the identification of miRNA targets is necessary to understand their mechanisms of action in various physiological responses. However, relatively few miRNA targets have been experimentally validated, even though many miRNAs have been identified. Here we report that miR-185 regulates CRY1 protein expression by controlling the rhythmicity of Cry1 translation.
RESULTS
CRY1 expression is regulated by the miRNA machinery
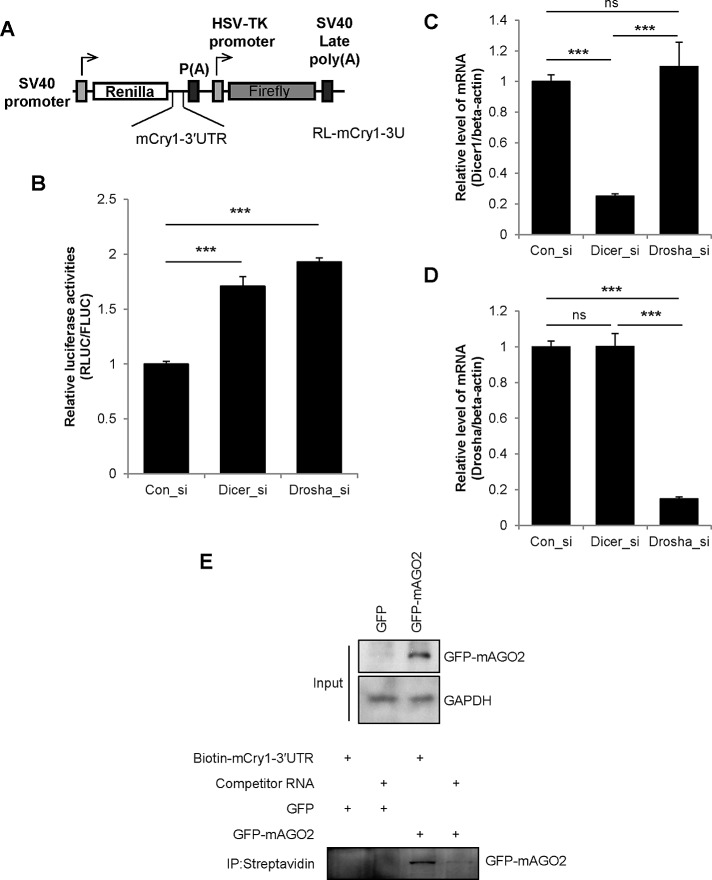
Although miRNA binding is not restricted to the 3′ untranslated region (UTR) of mRNA (Chi et al., 2009; Hafner et al., 2010), 3′UTR is the most preferential target of miRNAs. Therefore a Renilla luciferase reporter that was fused to mCry1-3′UTR was used to investigate the role of miRNAs in the regulation of Cry1 expression (Figure 1A). A small interfering RNA (siRNA) knockdown approach was used to down-regulate the expression of AGO2, Dicer, and Drosha, key proteins in miRNA genesis and function. Downregulation of Dicer or Drosha mRNA stabilized mCry1-3′UTR reporter activity (Figure 1B). siRNA-mediated knockdown of Dicer and Drosha mRNA was confirmed by real-time PCR (Figure 1, C and D). Furthermore, siRNA-mediated reduction of AGO2 expression increased the luciferase activity of the mCry1-3′UTR reporter (Supplemental Figure S1, A and B). These results suggested that the miRNA machinery regulates mCry1. However, it is possible that miRNAs may mediate this effect indirectly. miRNAs regulate diverse biological functions; therefore alterations in other miRNA functions may have indirectly contributed to the stabilization of mCry1-3′UTR reporter activity. To obtain more direct evidence of the regulatory role of miRNAs on mCry1 expression, we determined AGO2 protein binding with mCry1-3′UTR by RNA affinity precipitation. Overexpressed green fluorescent protein (GFP)–AGO2 was coprecipitated with biotin-labeled mCry1-3′UTR mRNA (Figure 1E). In contrast, 3′UTR-bound AGO2 was dramatically decreased by nonlabeled mCry1-3′UTR. Overexpressed GFP did not coprecipitate with the biotin-labeled mCry1-3′UTR. This finding suggested that AGO2 might specifically interact with the 3′UTR of mCry1. Collectively these results indicated that miRNAs might play a role in the regulation of mCry1 expression.
FIGURE 1:
Cry1 expression is regulated by the miRNA machinery. (A) Schematic diagram of the reporter plasmid containing the full-length 3′UTR of mCry1. Full-length mCry1-3′UTR was fused to the Renilla luciferase reporter gene. Firefly luciferase was used as a transfection control. (B) Microporation was used to cotransfect NIH 3T3 cells with the Renilla luciferase mCry1-3′UTR reporter and siRNAs specific for Dicer1 (Dicer_si) or Drosha (Drosh_si). Nonspecific siRNA (Con_si) was used as a control. After 24 h, cells were harvested and dual-luciferase assays were performed using firefly luciferase as a transfection control. The activity of Con_si was set to 1 (n = 4; ***p < 0.0001). The relative mRNA levels of (C) Dicer1 and (D) Drosha were quantified by real-time PCR and normalized to mActb (n = 4; ***p < 0.0001). (E) The in vitro–transcribed mCry1-3′UTR construct was labeled with biotin-UTP and incubated with cytoplasmic extracts of GFP-mAGO2–overexpressing NIH 3T3 cells. Streptavidin affinity-purified samples were separated by SDS–PAGE and subjected to immunoblotting with anti-GFP or anti-GAPDH antibodies.
The miR-185–binding region of mCry1-3′UTR acts as a cis-element in translation repression
To investigate the potential involvement of miRNAs in the posttranscriptional regulation of mCry1-3′UTR, we applied several miRNA target prediction algorithms in a screening process (MIRanda, MIRBase, and TargetScan). Of interest, the three prediction algorithms identified miR-185 as a candidate miRNA with the potential to bind to the 3′UTR of mCry1 (Figure 2A). Indeed, mCry1-3′UTR and miR-185 were predicted to form nearly perfect duplexes (Figure 2B). Many miRNAs regulate gene expression by base pairing with miRNA recognition elements found in their mRNA targets to induce mRNA degradation (Hutvagner and Zamore, 2002) or translational repression (Zeng et al., 2002). To verify the interaction between miR-185 and mCry1-3′UTR, we performed luciferase reporter assays using a reporter construct that contained the SV40 promoter, which triggers Renilla luciferase expression, and the TK promoter, which triggers firefly luciferase expression (internal control). One or two copies of the mCry1-3′UTR fragment that can be targeted by miR-185 (1×185B or 2×185B) were inserted between the luciferase stop codon and the synthetic poly(A) tail (Figure 2C). miR-185 repressed the luciferase activity of the RL-2×185B reporter by 50–60% compared with the empty vector control (RL; Figure 2D). Two miRNA-binding sites in close proximity have a synergistic effect (Bartel, 2009); however, this does not reflect the endogenous situation. Therefore the experiment was repeated with a reporter containing only one miR-185–binding site (RL-1×185B). Luciferase activity of the RL-1×185B reporter was decreased ∼30% in comparison with the RL reporter. Reporter mRNA levels were not different between the RL-2×185B and RL reporters (data not shown). These data provide evidence that miR-185 may target the 3′UTR of mCry1. miR-28 and miR-708 were also identified as candidate miRNAs with the potential to interact with mCry1-3′UTR (Figure 2A); however, mCry1-3′UTR fragments that can be targeted by miR-28 or miR-708 did not affect mCry1-3′UTR luciferase activity (data not shown).
FIGURE 2:
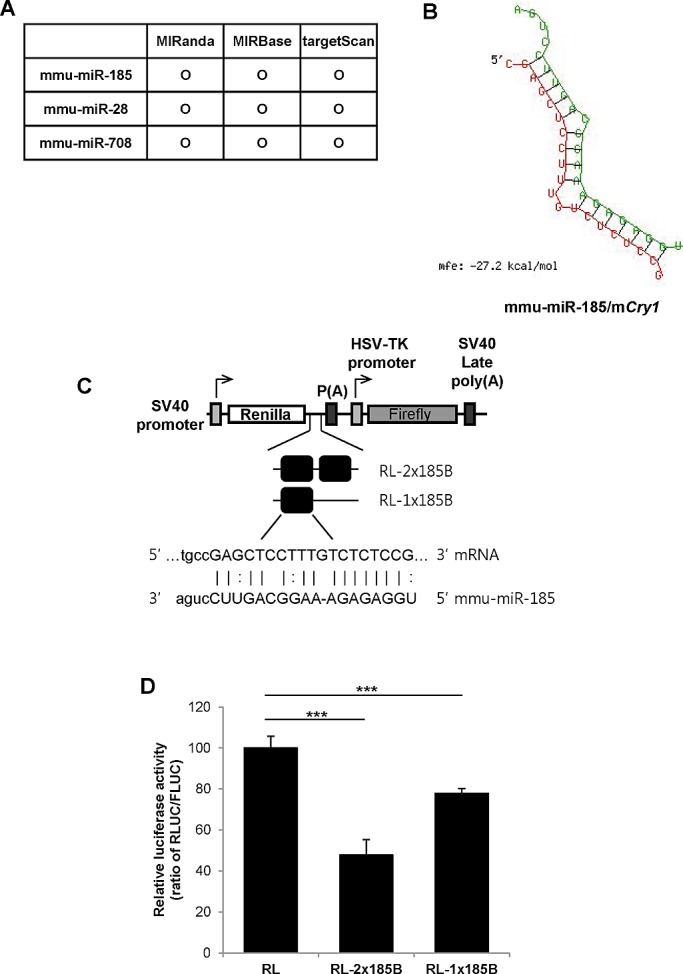
The miR-185–binding region of mCry1-3′UTR acts as a cis element in translation repression. (A) miRNA target prediction algorithms (MIRanda, MIRBase, and TargetScan) were applied to screen for miRNAs with the potential to bind the 3′UTR of mCry1. (B) Predictions of mCry1-3′UTR (lower strand) and miR-185 (upper strand) hybrids were performed using RNAhybrid software (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid; Rehmsmeier et al., 2004). (C) Schematic representation of the reporter constructs. One or two copies of the miR-185 recognition element of mCry1-3′UTR (miR-185 MRE) were fused to the Renilla luciferase reporter gene. Firefly luciferase was used as a transfection control. (D) Luciferase activity was determined in NIH 3T3 cells transfected with RL (control), RL-2×185B, or RL-1×185B plasmids. The relative luciferase activity (ratio of RLUC/FLUC) was set to 100. Results shown are the mean ± SEM (n = 4; ***p < 0.0001).
miR-185 overexpression represses translation
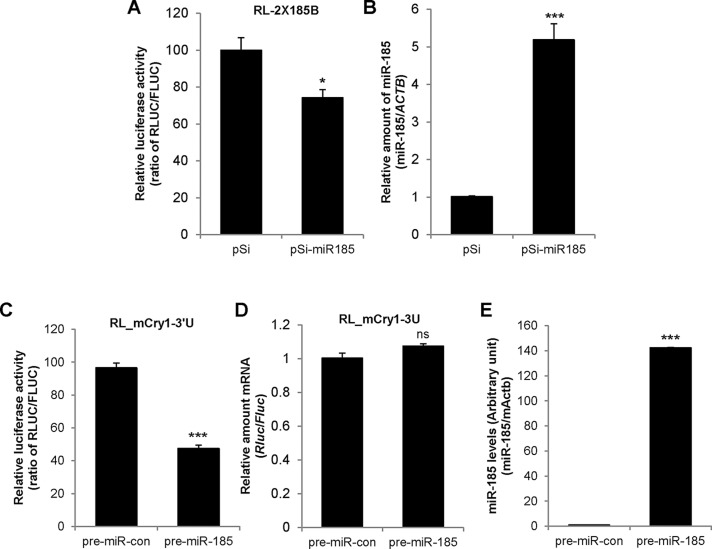
Next we investigated the effect of miR-185 on the functional regulation of mCry1-3′UTR by inducing miR-185 overexpression with the mmu-miR-185 expression vector (pSi-miR185). miR-185 overexpression resulted in 30% inhibition of RL-2×185B reporter activity compared with the pSi empty vector control (Figure 3A). miR-185 levels were increased approximately fivefold by the mmu-miR-185 expression vector compared with the empty vector control (Figure 3B). In contrast, the reporter that did not harbor the 3′UTR of the miR-185–binding region was not affected by miR-185 overexpression (Supplemental Figure S2A). The 3′UTR sequence context beyond the binding site is a crucial determinant of miRNA targeting efficacy (Nielsen et al., 2007; Didiano and Hobert, 2008; Sun et al., 2010). Because the mCry1-3′UTR reporter contained the miR-185 binding–site region and not the full-length 3′UTR, it may have induced artificial effects. Therefore we confirmed the effect of miR-185 on mCry1 regulation using a reporter that contained full-length mCry1-3′UTR. Furthermore, a pre-miRNA transfection system was used in conjunction with the vector system to increase the efficiency of miR-185 overexpression. The reporter activity of the full-length mCry1-3′UTR was decreased 50–60% by pre–miR-185 overexpression (Figure 3C). The overexpression of pre–miR-185 did not affect the mRNA levels of the reporter (Figure 3D) or the luciferase activity of the vector lacking mCry1-3′UTR (Supplemental Figure S2B). The overexpression of miR-185 was confirmed using real-time PCR. The miR-185 levels were dramatically increased by pre–miR-185 (Figure 3E). Therefore miR-185 overexpression decreased the activity of a luciferase reporter that harbors either the miR-185–binding region or full-length mCry1-3′UTR. Taken together, our data suggest that miR-185 suppresses mCry1 expression via 3′UTR-mediated regulation.
FIGURE 3:
miR-185 overexpression represses translation. (A) NIH 3T3 cells were cotransfected with RL-2×185B plasmids and control pSi or miR-185-expressing plasmids (pSi-miR185). After 24 h, dual-luciferase assays were performed. The RLUC/FLUC ratio of pSi was set to 100 (n = 4; *p < 0.05, p = 0.0189). (B) Real-time PCR was used to quantify miR-185 mRNA levels in NIH 3T3 cells cotransfected with RL-2×185B plasmids and pSi or pSi-miR185 plasmids. miR-185 mRNA levels were normalized to mActb mRNA levels. Relative miR-185 levels of pSi were set to 1 (***p < 0.0001; unpaired t test). (C) NIH 3T3 cells were cotransfected with RL-mCry1-3U (full-length mCry1-3′UTR) and pre–miR-con or pre–miR-185 plasmids. After 24 h, luciferase assays were performed (n = 4; ***p < 0.0001). (D) Real-time PCR was used to determine the Rluc mRNA levels in NIH 3T3 cells cotransfected with RL-mCry1-3U (full-length mCry1-3′UTR) and pre–miR-con or pre–miR-185. Rluc mRNA levels were normalized to Fluc mRNA levels. The relative luciferase activity (RLUC/FLUC) of pre–miR-con was set to 1 (n = 6; p = 0.0587). (E) The overexpression of miR-185 was confirmed by real-time PCR (n = 4; ***p < 0.0001). Data shown in C–E represent the mean ± SEM.
Mutation of the miR-185–binding site increases translation
A short perfect match complemented by imperfect matches in close vicinity to the target mRNA sequence is the basic prerequisite for miRNA targeting in metazoans and is considered the most important feature for target recognition by miRNAs in mammals (Nielsen et al., 2007; Bartel, 2009). The minimal core 5′ sequence that is essential for target-site recognition is called the seed sequence. The seed sequence is considered to be a 6- to 8-nt-long substring within the first 8 nt at the 5′ end of the miRNA (Lewis et al., 2003; Ellwanger et al., 2011). To examine the specificity of miR-185 binding to mCry1-3′UTR, we made four point mutations in the miR-185–binding region of the full-length 3′UTR and inserted it into the RL vector (RL-185mut; Figure 4A). Although the mRNA levels of RL-185mut and wild-type full-length mCry1-3′UTR (RL-mCry1-3′U) were not dramatically different, the mutant reporter showed an approximately fourfold increase in luciferase activity (Figure 4, B and C). Furthermore, the overexpression of pre–miR-185 decreased the reporter activity of RL-mCry1-3′U but not that of RL-185mut (Figure 4, D and E). These findings suggest that miR-185 specifically binds to the 3′UTR of mCry1 to regulate mCry1 expression.
FIGURE 4:
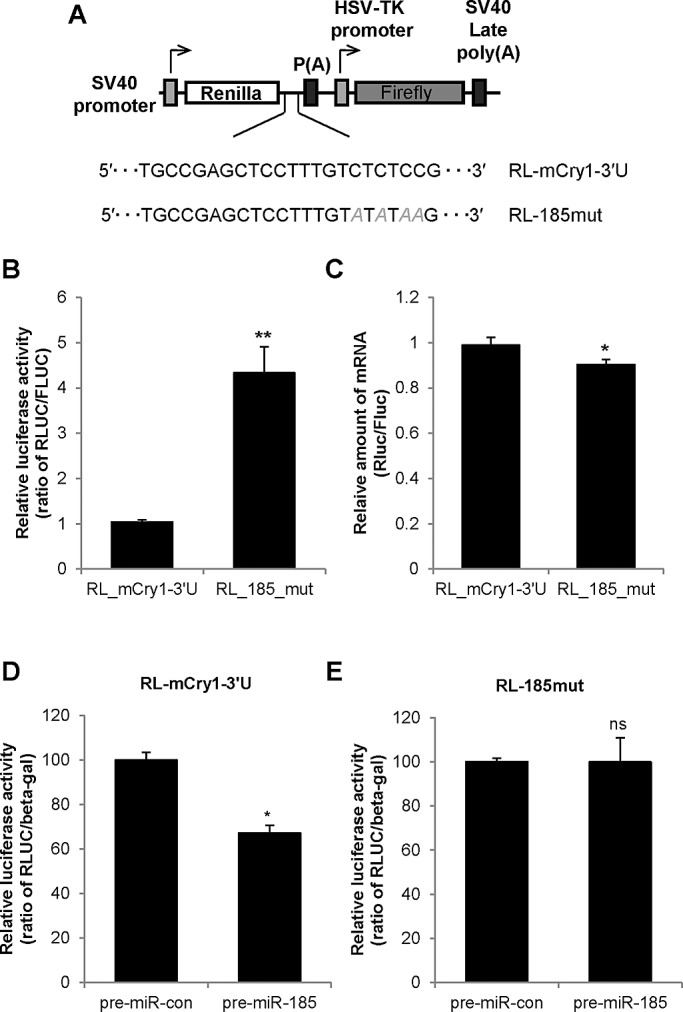
Modulation of the miR-185–binding site increases translation. (A) The naive (wild-type) full-length mCry1-3′UTR sequence containing the miR-185 target site was amplified and ligated into a pRL reporter vector (pRL-mCry1-3U). A plasmid containing the full-length mCry1-3′UTR with a selected point mutation in the miR-185 target sequence (pRL-185-mut) was also constructed. (B) Luciferase reporter assays were performed using NIH 3T3 cells transfected with pRL-mCry1-3U or pRL-185-mut. The relative luciferase activity of RL-mCry1-3U was set to 1 (n = 4; p = 0.0047). (C) Total RNA extracted from NIH 3T3 cells transfected with pRL-mCry1-3U or pRL-185-mut were subjected to real-time PCR to determine the relative Rluc mRNA levels (ratio of Rluc/Fluc; n = 4; p = 0.0465). (D) Luciferase assays were performed in NIH 3T3 cells cotransfected with pRL-mCry1-3U and pre–miR-con or pre-miR-185 (n = 3; p = 0.0165). (E) Luciferase assays were performed in NIH 3T3 cells cotransfected with pRL-185-mut and pre–miR-con or pre–miR-185 (n = 3; p = 0.9928). Data shown represent the mean ± SEM.
Cytoplasmic miR-185 oscillation regulates CRY1 expression
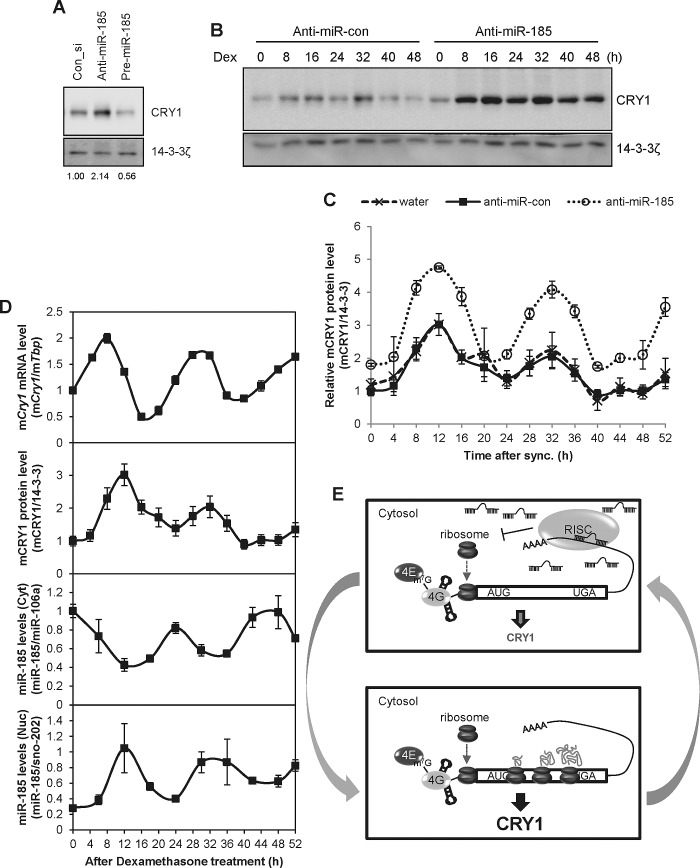
We demonstrated that miR-185 regulates mCry1 expression by using a reporter linked to the 3′UTR of mCry1. Therefore, to determine whether miR-185 also regulated endogenous Cry1 expression, we transfected NIH 3T3 cells with anti–miR-185, a chemically modified, single-stranded nucleic sequence designed to specifically bind to and inhibit endogenous miR-185 or pre–miR-185. Inhibition of miR-185 with anti–miR-185 increased CRY1 protein levels, whereas overexpression of pre–miR-185 decreased CRY1 protein levels (Figure 5A). Furthermore, anti–miR-con, but not anti–miR-185, did not significantly change the expression of mCRY1 compared with water treatment (Supplemental Figure S3A). To assess the physiological role of miR-185–mediated mCry1 translational regulation, we determined the effect of anti–mir-185 on the circadian expression of CRY1 in NIH 3T3 cells. To do this, we used dexamethasone to synchronize the rhythmicity of the cells (Lee et al., 2012b). Again, inhibition of miR-185 by anti–miR-185 resulted in higher mCRY1 protein levels (Figure 5, B and C). In contrast, anti–miR-185 did not affect the phase of CRY1 expression, suggesting that miR-185 is critical for mCry1 RNA translation (Supplemental Figure S3B). Because miR-185 was capable of regulating endogenous CRY1 protein levels, we hypothesized that miR-185 levels may oscillate with circadian time. Therefore we determined the expression profile of miR-185 in synchronized NIH 3T3 cells. Synchronized cells showed rhythmic mCry1 mRNA expression (Supplemental Figure S4A); however, miR-185 did not exhibit an oscillation pattern (Supplemental Figure S4B). Because miRNA-mediated posttranscriptional regulation, especially translational repression, mainly occurs in the cytoplasm, we examined the cytoplasmic levels of miR-185. Synchronized NIH 3T3 cells were found to have a rhythmic mCry1 mRNA expression profile with phase-delayed mCRY1 protein oscillation (Figure 5D, first and second from top). Of interest, in synchronized cells, the amount of cytoplasmic miR-185 fluctuated according to the specific circadian time period (Figure 5D, third from top). Moreover, the pattern of cytoplasmic miR-185 levels was nearly antiphase to that of mCRY1 protein levels and nuclear miR-185 levels (Figure 5D, top and bottom). These data provide evidence that cytosolic miR-185 has circadian rhythmicity antiphase to the mCRY1 protein profile and that its expression suppresses endogenous mCRY1 expression.
FIGURE 5:
Cytoplasmic miR-185 oscillation regulates CRY1 expression. (A) CRY1 and 14-3-3ζ expression in NIH 3T3 cells transfected with control siRNA (Con_si), anti-miR-185, or pre–miR-185 were determined by immunoblotting. (B) Anti–miR-con and anti–miR-185–transfected NIH 3T3 cells were synchronized with dexamethasone treatment for 2 h. Then cells were harvested at the indicated time points and subjected to immunoblotting for CRY1 and 14-3-3ζ expression. (C) The relative mCRY1 levels in water-treated (x marks/dashed line), anti–miR-con–transfected (closed squares/solid line), and anti–miR-185–transfected (open circles/dotted line) NIH 3T3 cells. CRY1 levels were normalized to 14-3-3ζ and plotted. mCRY1 protein levels were not significantly different at any time point between water-treated and anti–miR-con–transfected cells (p > 0.05). mCRY1 protein levels were significantly different between the anti–miR-con–transfected and anti–miR-185–transfected groups except at the 0-, 4-, 20-, 24-, and 40-h dexamethasone treatment time points. (D) The rhythmic mCry1 mRNA profiles of anti–miR-con– and anti–miR-185–transfected NIH 3T3 cells (top). Real-time PCR results were calculated using the ratio of mCry1/mTbp. The protein profiles of water-treated, anti–miR-con–transfected, and anti–miR-185–transfected NIH 3T3 cells were quantified and plotted by time (second from top). Cytoplasmic and nuclear miR-185 levels in synchronized NIH 3T3 cells transfected with anti–miR-con and anti–miR-185 (bottom). Cells were treated with dexamethasone for the indicated time points, and cytoplasmic and nuclear extracts were obtained under RNase-free conditions. miR-185, miR-106a, and sno-202 levels were measured by real-time PCR. The cytoplasmic miR-185 levels were normalized to miR-106a, whereas nuclear miR-185 levels were normalized to sno-202. Data shown represent the mean ± SEM. (E) The proposed model for cytosolic miR-185–mediated rhythmic mCry1 translational regulation. Increased cytosolic miR-185 binds to the 3′UTR of mCry1 and inhibits mCry1 translation. When cytosolic miR-185 levels are reduced, miR-185–mediated mCry1 translational inhibition does not occur, and mCRY1 protein is increased.
DISCUSSION
CRY1 is a core gene involved in the molecular clock system. Although the periodicity of transcription is crucial for maintaining rhythmic expression of clock genes, posttranscriptional and posttranslational regulation are also important for the fine-tuned regulation of the rhythmic cycle. The posttranscriptional regulation of mRNA stability and translational efficiency are often mediated by cis elements in mRNA that interact with trans-acting factors, such as RNA-binding proteins or microRNAs. The importance of UTR in the regulation of clock genes has been acknowledged (Garbarino-Pico and Green, 2007; Kojima et al., 2011).
Here we showed that miR-185 interacts with the mCry1 3′UTR to regulate mCry1 expression in a posttranscriptional manner. Of interest, cytoplasmic miR-185 levels were nearly ant-phase to mCRY1 protein levels, and miR-185 knockdown elevated the amplitude of mCRY1 protein oscillation. Our results suggest that miR-185 plays a role in fine-tuned regulation of mCRY1 protein expression by controlling the rhythmicity of mCry1 mRNA translation rate.
It has been estimated that miRNAs target >5300 human genes (Lewis et al., 2003). Multiple miRNAs may regulate one gene, indicative of cooperative translational control, and similarly, a single miRNA may regulate multiple target genes, reflecting target multiplicity (Enright et al., 2003; Yoon and De Micheli, 2006). Therefore, although we identified one miRNA involved in the regulation of Cry1 expression, other miRNAs may also be involved. Indeed, combinatorial control of a single target by multiple miRNAs is an important feature because the effects of a single miRNA can be marginal. Therefore the identification of other miRNAs that act on mCry1 may help to elucidate the mechanism of rhythmic mCry1 expression.
Gene silencing mediated by miRNA has two modes: mRNA degradation and translational repression (Lim et al., 2005). In our study, miR-185 was found to regulate mCry1 expression by translational repression. However, miRNA-mediated mRNA degradation may additionally modulate the rhythmic mCry1 mRNA profile. It was reported that 3′UTR-mediated mRNA decay plays a role in the circadian mCry1 mRNA regulatory mechanism (Woo et al., 2010), and the oscillation of mCry1 mRNA is well established (Lee et al., 2001). Therefore it is possible that other miRNAs may also regulate mCry1 mRNA stability, which may consolidate fine-tuned posttranscriptional regulation of rhythmic circadian clock genes.
In the present study, there were some differences in the miR-185–binding site of mCry1-3′UTR predicted by TargetScan compared with rats and humans. Based on the output of the prediction program and sequence analysis, it was difficult to determine whether the miR-185–binding site was conserved. The sequences of full-length Cry1-3′UTR in mice, rats, and humans are markedly different. Furthermore, significant differences in the circadian expression of clock genes among mice, rats, and humans have been observed (Yan et al., 2008). The expression of most core clock genes showed a 4- to 5-h phase delay in rats compared with mice and an 8- to 12-h phase delay in humans compared with mice, which probably reflects the gap between nocturnal and diurnal animals (Yan et al., 2008). The different expression patterns in mice and rats suggest that diverse and rewirable clock gene–regulatory networks may exist. We also suggest that the miR-185–binding site of mCry1-3′UTR may increase the complexity and elaboration of clock gene–regulatory networks and therefore contribute to the sophisticated and fine-tuned molecular basis of physiological and behavioral differences among species. The function of miR-185 in rats or humans was not investigated in the present study, however, and thus further studies are needed to determine the function of miR-185 in different species.
Although we initially believed that the miR-185 level may have circadian rhythm because of the rhythmic expression of its target, total miR-185 levels were found to be constant over the time points investigated. In contrast, the amount of cytoplasmic miR-185 showed an antiphase profile to the pattern of mCRY1 protein expression. The posttranscriptional regulation of miRNAs, which can occur at the level of miRNA maturation, stability, or localization, can also influence their expression (Cullen, 2004; Kim, 2005; Hwang et al., 2007). Therefore the mechanism of rhythmic cytosolic miR-185 accumulation needs to be studied further. Of importance, this study revealed a novel mechanism of miRNA-mediated rhythmic mCry1 control that may be important in the regulation of the circadian clock. This regulatory system may induce fine-tuning of the amplitude of mCRY1 protein expression regardless of mRNA oscillation. A similar circadian time-dependent, miRNA-mediated regulation may also be functional in other clock genes. On the basis of our results, we suggest that physiological circadian rhythm is generated through a complex regulation of gene expression that involves a variety of factors acting in rhythmic transcriptional, time-dependent posttranscriptional, and controlled translational and posttranslational regulation. Therefore our study provides insight into the tightly and finely regulated molecular circadian clock system.
MATERIALS AND METHODS
Cell culture and circadian synchronization
NIH 3T3 cells were cultured in DMEM (HyClone, Logan, UT) supplemented with 10% fetal bovine serum (HyClone) and 1% antibiotics (Welgene, Daegu, South Korea) and maintained in a humidified atmosphere containing 5% CO2. The circadian oscillation of NIH 3T3 cells was synchronized by dexamethasone treatment (100 nM) as described previously (Lee et al., 2012b). After 2 h, the dexamethasone-containing medium was replaced with complete medium (Lee et al., 2012b).
Plasmid construction
The mCry1 3′UTR was amplified from the pcNAT-wt610 plasmid as previously reported (Woo et al., 2010). The resultant products were cloned into the XhoI/NotI site of the control pRL vector, which expresses Renilla luciferase (psiCHECK-2 vector; Promega, Madison, WI). The pRL-185mut plasmid was constructed by deletion mutagenesis using DpnI digestion. The plasmid containing a single or dimer 3′UTR fragment (RL-1×185B or RL-2×185B) was constructed by ligation of an annealed oligonucleotide containing one or two 3′UTR fragments into a pRL vector. For in vitro binding assays, the mCry1 3′UTR was amplified, and the resultant PCR products were digested and subcloned into the EcoRI/XbaI site of the pSK′ vector (Kim et al., 2005) to generate pSK′-mCry1-3U. To overexpress miR-185, we generated pSi-miR185. Pre–miR-185 was amplified from mouse genomic DNA, and the resultant products were cloned into the BamHI/HindIII site of the control pSi vector (pSilencer 4.1-CMV neo; Ambion, Austin, TX). Mouse Ago2 coding region was amplified from the cDNA of NIH 3T3 cells, and the resultant products were cloned into the pEGFP-C1 vector (Clontech, Mountain View, CA).
Transient transfection
For transient transfection of NIH 3T3 cells with reporter plasmids, miRNA mimics (Ambion), and antagomirs (Ambion), the Neon Transfection System (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions.
Cellular fractionation, RNA quantification, and reporter assay
Cytoplasmic and nuclear fractionation of NIH 3T3 cells was performed as described previously (Hahm et al., 1998; Kim et al., 2007; Lee et al., 2012b). Fractionated or total small miRNAs were prepared using the mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's instructions and subjected to reverse transcription (RT) using specific RT primers (Applied Biosystems, Foster City, CA) with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems).
Total RNA was extracted from NIH 3T3 cells using TRI Reagent (Molecular Research Center, Cincinnati, OH) and reverse transcribed using ImProm-II (Promega) according to the manufacturer's instructions. The mRNA levels of endogenous genes and reporter plasmids were detected by quantitative real-time PCR using a StepOnePlus Real-Time PCR System (Applied Biosystems) with the FastStart Universal SYBR Green Master Mix (Roche, Indianapolis, IN) as described previously (Lee et al., 2012b). The specific primer pairs for mCry1, mTbp, Rluc, Fluc, mAgo2, mDicer, mDrosha, and mβ-actin (mActb) used for real-time PCR are shown in Supplemental Table S1. For miRNA quantification, quantitative real-time PCR was performed using the TaqMan MicroRNA Assay (Applied Biosystems) according to the manufacturer's instructions. A comparative Ct method was used for quantification.
The luciferase assay was performed as previously described (Lee et al., 2012a). The ratios between Firefly and Renilla luciferase activities (RLUC/FLUC) were calculated. The ratio for the empty pRL vector was set to 1.
In vitro RNA synthesis and in vitro binding
For in vitro binding assays, biotin-UTP–labeled RNA was transcribed from the XbaI-linearized pSK-mCry1-3U plasmid using T7 RNA polymerase (Promega). Streptavidin–biotin RNA affinity purification was performed as described previously (Kim et al., 2005). In brief, cytoplasmic extracts prepared from NIH 3T3 cells were incubated with biotinylated-mCry1-3′UTR RNA and subjected to streptavidin resin adsorption. Resin-bound proteins were analyzed by SDS–PAGE.
Immunoblot analysis
Immunoblot analyses were performed using polyclonal anti-CRY1 and monoclonal anti–glyceraldehyde-3-phosphate dehydrogenase (Millipore, Billerica, CA) primary antibodies and horseradish peroxidase–conjugated, species-specific secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Protein bands were visualized using the SUPEX ECL solution kit (Neuronex, Daegu, South Korea) and the LAS-4000 chemiluminescence detection system (Fujifilm, Tokyo, Japan). Acquired images were analyzed using Image Gauge (Fujifilm) according to the manufacturer's instructions.
Statistical analysis
All quantitative data are presented as the mean ± SEM. Comparisons between two groups were analyzed by two-tailed unpaired Student's t tests. For comparisons between more than two groups, a one-way analysis of variance (ANOVA) was used with a post hoc Tukey's test. A p < 0.05 was considered statistically significant. A two-way ANOVA with a post hoc Bonferroni test was used to analyze the effects of anti–miR-185, anti-miR-con, and water treatment on mCRY1 protein levels.
Supplementary Material
Acknowledgments
We thank Choogon Lee for kindly providing the anti-CRY1 antibody. This work was supported by grants from the National Research Foundation of Korea (20110027957, 20110031234, 20120005830, 20110031517), the Brain Korea 21 program, and the World Class University program (R31-10105) funded by the Korean Ministry of Education, Science, and Technology, Ministry of Knowledge Economy, and Korea Institute for Advancement of Technology through Inter-ER Cooperation Projects.
Abbreviations used:
- AGO
Argonaute
- Cry
Cryptochrome
- miRNA
microRNA
- Per
Period
- RISC
RNA-induced silencing complex
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0849) on May 22, 2013.
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Ann Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger DC, Buttner FA, Mewes HW, Stumpflen V. The sufficient minimal set of miRNA seed types. Bioinformatics. 2011;27:1346–1350. doi: 10.1093/bioinformatics/btr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harbor Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm B, Cho OH, Kim JE, Kim YK, Kim JH, Oh YL, Jang SK. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev. Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol. 2005;25:3232–3246. doi: 10.1128/MCB.25.8.3232-3246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev. Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kojima S, et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lee KH, Kim SH, Kim DY, Kim S, Kim KT. Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PloS One. 2012a;7:e37936. doi: 10.1371/journal.pone.0037936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Woo KC, Kim DY, Kim TD, Shin J, Park SM, Jang SK, Kim KT. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol. 2012b;32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu W, Rong Y, Baudry M, Schreiber SS. Status epilepticus induces p53 sequence-specific DNA binding in mature rat brain. Brain Res. Mol Brain Res. 1999;63:248–253. doi: 10.1016/s0169-328x(98)00285-x. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Li H, Rossi JJ. Sequence context outside the target region influences the effectiveness of miR-223 target sites in the RhoB 3′UTR. Nucleic Acids Res. 2010;38:239–252. doi: 10.1093/nar/gkp870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30:197–205. doi: 10.1128/MCB.01154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, De Micheli G. Computational identification of microRNAs and their targets. Birth Defects Res. C Embryo Today. 2006;78:118–128. doi: 10.1002/bdrc.20067. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.