Abstract
Background
Vitamin D is essential for a wide range of physiological processes including immune function and calcium homeostasis. Recent investigations have identified candidate genes which are strongly linked to concentrations of 25-hydroxyvitamin D. Since there is insufficient UVB radiation to induce year-round cutaneous synthesis of vitamin D at latitudes distant from the equator it is likely that these genes were subject to forces of natural selection. We used the fixation index (FST) to measure differences in allele frequencies in 993 individuals from ten populations to identify the presence of evolutionary selection in genes in the vitamin D pathway. We then explored the length of haplotypes in chromosomes to confirm recent positive selection.
Results
We find evidence of positive selection for DHCR7, which governs availability of 7-dehydrocholesterol for conversion to vitamin D3 by the action of sunlight on the skin. We show that extended haplotypes related to vitamin D status are highly prevalent at Northern latitudes (Europe 0.72, Northeast Asia 0.41). The common DHCR7 haplotype underwent a recent selective sweep in Northeast Asia, with relative extended haplotype homozygosity of 5.03 (99th percentile). In contrast, CYP2R1, which 25-hydroxylates vitamin D, is under balancing selection and we found no evidence of recent selection pressure on GC, which is responsible for vitamin D transport.
Conclusions
Our results suggest that genetic variation in DHCR7 is the major adaptation affecting vitamin D metabolism in recent evolutionary history which helped early humans to avoid severe vitamin D deficiency and enabled them to inhabit areas further from the equator.
Keywords: Evolutionary selection, Vitamin D, DHCR7, Fixation index, Long range haplotype test
Background
Vitamin D is essential in calcium homeostasis and bone health, with deficiency causing rickets and osteomalacia. In recent decades, vitamin D deficiency has also been implicated in the pathogenesis of other conditions, including cancers, autoimmune diseases, infections and cardiovascular diseases [1].
The main source of vitamin D is the ultraviolet irradiation of 7-dehydrocholesterol in the skin, which produces vitamin D3 (cholecalciferol). 7-dehydrocholesterol is thus a precursor both for vitamin D3 and for cholesterol – formation of the latter being catalysed by DHRC7 (Figure 1). Vitamin D can also be obtained from the diet, primarily from oily fish, although dietary sources are usually inadequate for normal requirements. Vitamin D from the skin and dietary sources is converted to 25-hydroxyvitamin D (25[OH]D), which is the major circulating vitamin D metabolite and generally used as a measure of vitamin D status [1]. This reaction is catalysed by microsomal (CYP2R1, CYP3A4) and mitochondrial (CYP27A) 25-hydroxylases. 25(OH)D is bound to vitamin D binding protein (GC) in the circulation. Genetic variants of GC differ with respect to their affinity for 25(OH)D. This, in turn, influences the availability of 25(OH)D to cells which convert it to 1,25-dihydroxyvitamin D (1,25[OH]2D) which is the biologically active metabolite [2].
Figure 1.
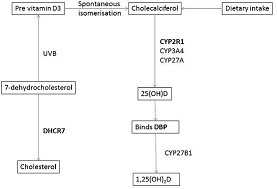
Metabolic pathways involving genes linked to 25-hydroxyvitamin D levels.DHCR7 encodes 7-dehydrocholesterol reductase, which converts 7-dehydrocholesterol to cholesterol, thereby reducing availability for vitamin D synthesis in the skin. CYP2R1 encodes 25-hydroxylase, which converts Vitamin D to 25(OH)D. GC encodes Vitamin D Binding Protein (DBP) which is a glycosylated alpha-globulin that transports vitamin D metabolites from the gut and skin to target end-organs. Proteins linked to 25-hydroxyvitamin D levels are indicated in bold.
Exposure to UVB in sunlight is the prime determinant of serum 25(OH)D concentration, but this effect is modified by biological factors such as skin pigmentation [3] and age-related reductions in cutaneous synthesis [4].
Twin and family studies suggest that heritability is responsible for a significant degree of variability in 25(OH)D levels [5,6], and recent genome-wide association studies have identified specific genes associated with 25(OH)D concentrations [7-10]. Single nucleotide polymorphisms (SNPs) in or near three genetic loci are strongly associated with 25(OH)D levels: GC[7,8,10], on Chromosome 4, which encodes Vitamin D binding protein; DHCR7[7,8], on Chromosome 11, which encodes 7-dehydrocholesterol reductase; and CYP2R1[7,8], on Chromosome 11, which encodes vitamin D 25-hydroxylase.
Given the pleiotropic functions of vitamin D in maintaining human health, and the established evidence for genetic influence on circulating 25(OH)D levels, we looked for evidence that vitamin D status has played a part in the adaptive evolution of humans. To this end, we searched for signals of natural selection in these three genes strongly associated with 25(OH)D levels.
Strategies for finding signatures of selection include: a) identification of a high frequency of function-altering mutations; b) a reduction in genetic diversity; c) high-frequency derived alleles; d) allele frequency differences between populations; e) unusually long haplotypes [11,12]. We focused on the latter two strategies, as these detect selective events in more recent times, and in specific populations. Thus we aimed to track the period in evolutionary history when early humans spread from Africa to northern latitudes where sunlight is more limited and cutaneous synthesis of vitamin D is very much reduced or absent in winter months [1].
Firstly, we investigated whether any of the markers associated with circulating 25(OH)D levels had outlying fixation index (FST) values conditional on their expected heterozygosity (He). FST is a commonly used statistic for measuring population differentiation; such differences in allele frequency among human populations will have accumulated after the major migrations out of Africa 50,000 to 75,000 years ago [12]. We ranked the pairwise-FST values of individual SNPs associated with vitamin D status against those of all SNPs on the remainder of the chromosome upon which the gene was located, thereby using these remaining alleles to generate an empirical distribution of chromosome-wide FST values. This allowed us to assess the evidence for positive selection at these loci, whilst avoiding confounding by population demographic history [13].
Next, we looked for the presence of extended haplotypes in these loci, comparing haplotype length with all core haplotypes of similar frequency on each chromosome. We evaluated the length of homozygosity of these haplotypes across extended chromosomal distances. Finally, we compared the extent of homozygosity of these haplotypes with others of similar frequency, in order to verify that the haplotypes had risen rapidly to high frequency before recombination events had time to disrupt linkage disequilibrium.
Results
Genetic markers near DHRC7 show signals of positive selection and marked differences in allele frequencies between populations
We used the HapMap3 (International Haplotype Map Project, Phase 3, release 2) dataset to examine 15 of the 18 SNPs significantly associated with circulating 25(OH)D levels in recent meta-analyses conducted in populations of European descent [7]. Three of the SNPs, rs17467825, rs4945008 and rs10741657 were not in the HapMap3 dataset. The mean 25(OH)D concentrations by genotype for each SNP in the 1958 British Birth Cohort are shown in Additional file 1: Table S1.
Three SNPs near DHCR7- rs12785878, rs7944926 and rs3794060 - were identified as candidates for positive selection, and three near CYP2R1 - rs2060793, rs1993116 and rs7116978 - for balancing selection using a neutrality test (Table 1). We analysed the allele frequencies of these SNPs in ten Hapmap3 populations: Africans in Southwest USA (ASW); Utah residents with ancestry from Northern and Western Europe (CEU); Chinese in Denver, Colorado (CHB); Gujerati in Houston, Texas (GIH); Japanese in Tokyo and Han Chinese in Beijing (JPT + CHB); Luhya in Kenya (LWK); Mexicans in Los Angeles (MEX); Maasai in Kenya (MKK); Toscans in Italy (TSI); Yoruba in Nigeria (YRI) (Tables 2 and 3).
Table 1.
Sample He and FST values at 5 GC, 5 DHCR7 and 5 CYP2R1 SNPs calculated in LOSITAN
| Loci | He | FST | P* |
|---|---|---|---|
|
DHCR7 SNPs | |||
| rs3794060 |
0.463422 |
0.265612 |
0.997845 |
| rs7944926 |
0.467318 |
0.25473 |
0.996788 |
| rs12785878 |
0.467134 |
0.254663 |
0.99678 |
| rs12800438 |
0.491453 |
0.162411 |
0.922132 |
| rs4944957 |
0.502397 |
0.127569 |
0.794576 |
|
CYP2R1 SNPs | |||
| rs7116978 |
0.457296 |
0.007108 |
0.004184 |
| rs2060793 |
0.468166 |
0.000742 |
0.001499 |
| rs1993116 |
0.44101 |
0.021711 |
0.024486 |
| rs12794714 |
0.434566 |
0.109713 |
0.674206 |
| rs10500804 |
0.435266 |
0.110771 |
0.681743 |
|
GC SNPs | |||
| rs2282679 |
0.306487 |
0.075355 |
0.451885 |
| rs3755967 |
0.310383 |
0.068546 |
0.395229 |
| rs1155563 |
0.331944 |
0.107236 |
0.671098 |
| rs2298850 |
0.280963 |
0.094884 |
0.590703 |
| rs7041 | 0.445035 | 0.142463 | 0.858668 |
*P (Simulated FST < sample FST). The outlier loci above the 0.975 and below the 0.025 quantiles are shown in bold, and indicate positive and balancing selection, respectively.
Table 2.
Allele frequencies of DHCR7 SNPs identified as having undergone positive selection in ten Hapmap populations
| SNP | Allele |
Allele frequencies |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASW | CEU | CHD | GIH | JPT + CHB | LWK | MEX | MKK | TSI | YRI | ||
| rs3794060 |
C |
0.711 |
0.274 |
0.587 |
0.832 |
0.589 |
0.941 |
0.612 |
0.923 |
0.225 |
0.884 |
| T |
0.289 |
0.726 |
0.413 |
0.168 |
0.411 |
0.059 |
0.388 |
0.077 |
0.775 |
0.116 |
|
| rs7944926 |
A |
0.684 |
0.277 |
0.587 |
0.832 |
0.584 |
0.941 |
0.612 |
0.92 |
0.223 |
0.844 |
| G |
0.316 |
0.723 |
0.413 |
0.168 |
0.416 |
0.059 |
0.388 |
0.08 |
0.777 |
0.156 |
|
| rs12785878 | G |
0.684 |
0.274 |
0.587 |
0.832 |
0.586 |
0.941 |
0.612 |
0.92 |
0.225 |
0.844 |
| T | 0.316 | 0.726 | 0.413 | 0.168 | 0.414 | 0.059 | 0.388 | 0.08 | 0.775 | 0.156 | |
Table 3.
Allele frequencies of CYP2R1 SNPs identified as having undergone balancing selection in ten Hapmap populations
| SNP | Allele |
Allele frequencies |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASW | CEU | CHD | GIH | JPT + CHB | LWK | MEX | MKK | TSI | YRI | ||
| rs7116978 |
T |
0.395 |
0.363 |
0.352 |
0.411 |
0.349 |
0.359 |
0.397 |
0.23 |
0.297 |
0.333 |
| C |
0.605 |
0.637 |
0.648 |
0.589 |
0.651 |
0.641 |
0.603 |
0.77 |
0.703 |
0.667 |
|
| rs2060793 |
A |
0.412 |
0.394 |
0.352 |
0.431 |
0.347 |
0.367 |
0.414 |
0.305 |
0.343 |
0.342 |
| G |
0.588 |
0.606 |
0.648 |
0.569 |
0.653 |
0.633 |
0.586 |
0.695 |
0.657 |
0.658 |
|
| rs1993116 | A |
0.289 |
0.398 |
0.347 |
0.426 |
0.348 |
0.2 |
0.397 |
0.212 |
0.338 |
0.245 |
| G | 0.711 | 0.602 | 0.653 | 0.574 | 0.652 | 0.8 | 0.603 | 0.788 | 0.662 | 0.755 | |
The distribution of allele frequencies was similar within each population for the three polymorphisms near DHCR7 reflecting the strong linkage disequilibrium in this region of the human genome. Strikingly, the major and minor alleles were reversed for all three SNPs in the Europeans (CEU and TSI) when compared to other populations. Large differences in allele frequencies between populations may signal a locus that has undergone positive selection in one geographical area.
An empirical distribution of FST values for all the SNPs on chromosome 11 in the HapMap3 dataset was constructed. The values were ranked from highest to lowest. Pairwise FST values for the DHCR7 SNPs rs12785878, rs7944926 and rs3794060 between the European and African, and the European and Gujarati populations were above the 95th percentile relative to 70,973 SNPs on Chromosome 11. FST values were also above the 95th percentile for ASW vs LWK and ASW vs MKK at rs7944729 (Tables 4 and 5). Such high FST values can be generated when the direction or strength of selection differs among populations.
Table 4.
Pairwise FST values for rs1279187 and rs7944926
| GIH | LWK | MKK | YRI | |
|---|---|---|---|---|
|
ASW |
not significant |
0.20 (95.8th) |
0.20 (97.3rd) |
not significant |
|
CEU |
0.47 (98.1st) |
0.61 (99.5th) |
0.62 (99.9th) |
0.48 (97.2nd) |
| TSI | 0.52 (99.9th) | 0.66 (99.7th) | 0.67 (99.9th) | 0.53 (98.9th) |
Table 5.
Pairwise FST values for rs3794060
| GIH | LWK | MKK | YRI | |
|---|---|---|---|---|
|
ASW |
not significant |
0.17 (95.6th) |
0.18 (97.1st) |
not significant |
|
CEU |
0.47 (98.1st) |
0.61 (99.5th) |
0.63 (99.9th) |
0.53 (98.6th) |
| TSI | 0.52 (99.9th) | 0.66 (99.7th) | 0.68 (99.9th) | 0.58 (99.2nd) |
Haplotype analysis suggests that DHCR7 markers linked to high vitamin D status have undergone positive selection in Northeast Asia
Another signal of a recent selective sweep is a long chromosomal region with strong linkage disequilibrium around the selected site. This large haplotype block was observed in the European, Northeast Asian, Mexican and Gujarati populations (Figure 2). The block, which varied between 63 kb and 102 kb, depending on the specific population, included all five SNPs associated with vitamin D status near DHCR7 (rs7944926, rs12785878, rs12800438, rs4944957 and rs3794060). This was in contrast to the African populations, which had more haplotype blocks that were shorter, and no block contained all five SNPs (Figure 2). This observation indicates that the underlying haplotype structures in this region and the evolutionary forces driving these structures are different between the African and other populations, with the rate of recombination (compared to genetic drift) being higher in Africans.
Figure 2.
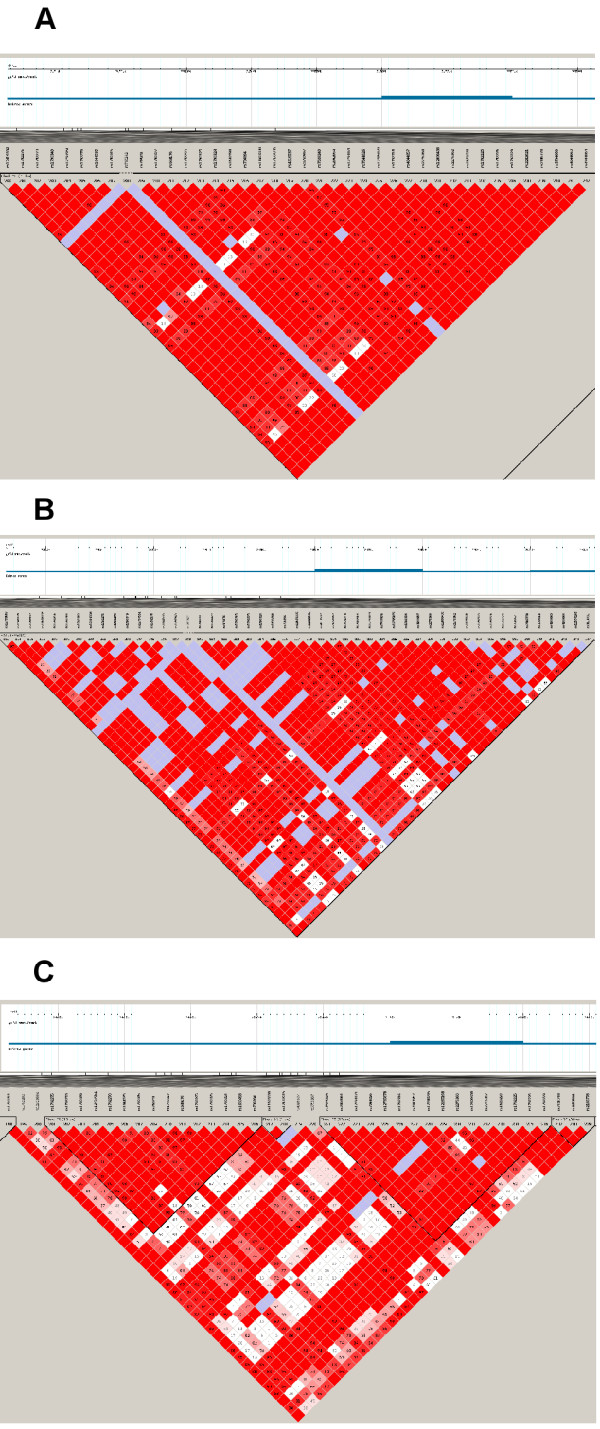
Linkage disequilibrium of SNPs in and around DHCR7 and NADSYN1 for A) JPT + CHB, B) CEU and C) YRI. The numbers within each square indicate the D’ values. Squares are colour-coded as follows: white: D’ < 1, LOD < 2; blue: D’ = 1, LOD < 2; pink: D’ < 1, LOD ≥ 2; and bright red: D’ = 1, LOD ≥ 2.
A signature of positive selection is indicated by unusually long haplotypes and high population frequency [14]. Long range haplotypes persist for relatively short periods of time, since, after 30,000 years random recombination events tend to break down the haplotype, leaving fragments that are too short to detect [12]. We therefore next employed a long range haplotype test – Extended Haplotype Homozygosity (EHH), to assess whether these alleles had risen in prevalence so rapidly that there had been insufficient time for recombination to break down linkage disequilibrium with alleles at neighbouring loci [14].
A core region was defined, comprising 17 adjacent SNPs in DHCR7 and a neighbouring gene NADSYN1 (rs7944926, rs12785878, rs1792316, rs4944957, rs12280295, rs12285168, rs2276360, rs12800438, rs2276362, rs1629220, rs1792225, rs1792226, rs1792229, rs7131218, rs2282621, rs2186778 and rs3794060). This core region included the five SNPs associated with circulating 25(OH)D levels, in addition to non-synonymous coding variants in NADSYN1. In none of the African populations – ASW, LWK, MKK or YRI – could a haplotype block comprising these seventeen SNPs be formed, reinforcing the suggestion of relatively high effects of recombination in these populations. For the other six populations, the 17 SNPs defined four core haplotypes denoted DHCR7-CH1 to 4 for core haplotypes 1 to 4 (Table 6).
Table 6.
Core haplotype frequencies in six populations
| Core haplotype | Core alleles |
Core haplotype frequencies in six populations |
|||||
|---|---|---|---|---|---|---|---|
| CEU | CHD | GIH | JPT + CHB | MEX | TSI | ||
|
DHCR7-CH1 |
GTCGTGCAGCCCACTCT |
0.717 |
0.406 |
0.165 |
0.406 |
0.4 |
0.75 |
|
DHCR7-CH2 |
AGCATGGGATCTGCCCC |
0.212 |
0.388 |
0.472 |
0.406 |
0.29 |
0.205 |
|
DHCR7-CH3 |
AGAATGGGACTCACCTC |
0 |
0.141 |
0.352 |
0.109 |
0.3 |
0.028 |
| DHCR7-CH4 | AGCATGGGACCCACTCC | 0 | 0.047 | 0 | 0.074 | 0 | 0 |
Core alleles refer to those of the seventeen SNPs in order from left to right: rs7944926, rs12785878, rs1792316, rs4944957, rs12280295, rs12285168, rs2276360, rs12800438, rs2276362, rs1629220, rs1792225, rs1792226, rs1792229, rs7131218, rs2282621, rs2186778, rs3794060.
For each core haplotype the relative EHH (REHH) was calculated. REHH is the factor by which EHH decays on the core haplotype compared with the decay of EHH on all the other core haplotypes combined. Haplotypes were placed in 20 groups based on their frequency, at 5% intervals. The significance of the REHH values was assessed by comparing their percentiles within these 5% frequency groups for all possible core haplotypes on Chromosome 11. The REHH value at 0.25 cM for DHCR7-CH1 was 4.90 (99th percentile of core haplotypes within 5% of its frequency) for CHD, and 5.03 (99th percentile of core haplotypes within 5% of its frequency) for JPT + CHB (Figure 3).
Figure 3.
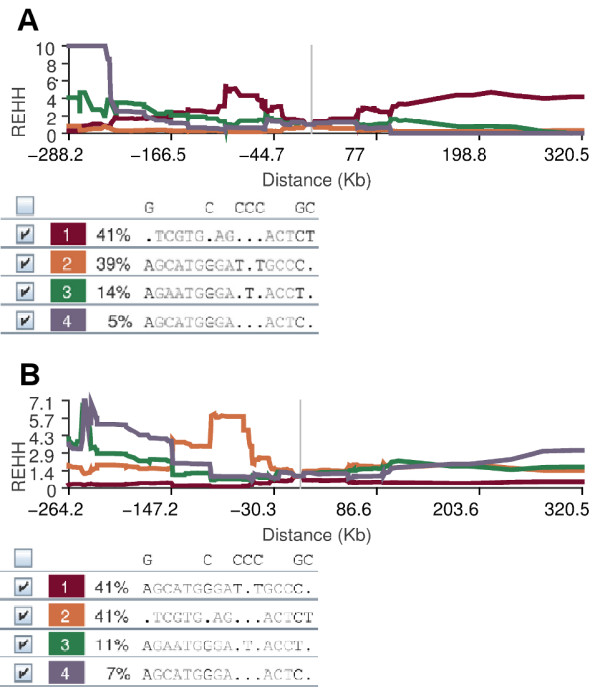
Long range haplotype analysis of DHCR7/NADSYN1 core haplotypes. REHH versus genomic distance plots of DHCR7-CH1 to CH4 for A) CHD and B) JPT + CHB are shown.
Genetic variants in vitamin D binding protein (GC) and 25 hydroxylase (CYP2R1) show no evidence of recent evolutionary selection
FST analysis for SNPs near CYP2R1 showed FST values below the 2.5% quantile compared to all other SNPs on the chromosome, an effect which could be produced by balancing selection. However, long-range haplotype analysis found no evidence for recent selective sweeps in any HapMap3 population for the core haplotypes containing the SNPs associated with vitamin D status. None of the SNPs in GC associated with circulating 25(OH)D levels showed evidence of selection in either FST or long range haplotype analysis.
Discussion
We searched for evidence of positive evolutionary selection in genetic variants strongly linked to higher circulating 25(OH)D levels in recent, large-scale genome-wide association studies [7-10]. We now show for the first time unique signatures of recent positive selection at the DHCR7 genetic locus. The same haplotype carrying these polymorphisms has risen to very high frequencies in both Europe (0.72) and in Northeast Asia (0.41). DHCR7 converts 7-dehydrocholesterol, which is a precursor for Vitamin D, into cholesterol. Thus reduced activity variants of DHRC7 would increase the availability of 7-dehydrocholesterol for cutaneous vitamin D synthesis. This implies that DHCR7 mutations associated with higher vitamin D status conferred a survival advantage which allowed early humans to avoid severe deficiency when migrating to northern latitudes.
Rare loss-of-function mutations in DHCR7 have been previously described and are associated with Smith-Lemli-Opitz syndrome (SLOS), in which affected homozygous individuals have greatly elevated serum 7-dehydrocholesterol levels and correspondingly low serum cholesterol [15]. It has previously been suggested that these rare variants might confer a survival advantage to heterozygotes by increasing 7-dehydrocholesterol availability and hence vitamin D synthesis [15].
One suggested mechanism by which this might occur is reduction in cephalopelvic disproportion due to rickets which in the past was a common cause of obstructed labour [16]. Low vitamin D status is also associated with low sperm count in men and with disorders of ovulation in women [17]. In addition to these effects on reproductive fitness, mortality from infectious diseases, particularly those affecting children and young adults could be a powerful selective force. Examples of such diseases linked to low vitamin D status have been clearly demonstrated in archaeological specimens from Homo erectus engaged in northerly migration [18].
Here we show that highly prevalent polymorphisms with a much smaller effect on 25(OH)D levels than those causing SLOS may nevertheless have exerted a strong selective force in human evolution by similar mechanisms. Whilst lower cholesterol levels arising from reduced DHCR7 activity may be advantageous in reducing the risk of heart disease and stroke, these effects are seen in later life and hence less likely to be a selective force in evolution. Thus vitamin D status rather than cholesterol level is more likely to be a driving force for natural selection.
Despite the higher prevalence of alleles associated with increased 25(OH)D levels in Europeans compared to other populations including Northeast Asians, a detailed examination of DHCR7 haplotype homozygosity found no evidence of a recent selective sweep in Europeans. However, REHH for the haplotype associated with higher 25(OH) D levels was significant when compared to other SNPs of similar frequency on chromosome 11 in the Northeast Asian populations. The significant pairwise FST values between European and non-European populations suggest that there was an evolutionary drive in European populations for an allele which increased circulating 25(OH)D levels more than 70,000 years ago, but that the mutation did not achieve fixation. Although pairwise FST values between Northeast Asian and other populations were not significant, the finding of significant REHH values in Northeast Asian populations suggest that a selective event subsequently occurred approximately 30–50,000 years ago.
This suggests that mutations associated with higher vitamin D status were positively selected at higher latitudes in both continents over different periods due to a variety of environmental pressures, as they proffered an evolutionary advantage. These genetic variants then rose quickly to high frequencies in humans living in areas distant from the equator.
The differences in timing of the selective events between the European and Northeast Asian populations may in part reflect the diets of the respective populations in the late Pleistocene and early Holocene. The inhabitants of Europe during this period are thought to have had a diet that was rich in mammal and fish meat, which have a high vitamin D content [19-21]. In East Asia, in contrast, plant foods may have become increasingly important in the human diet with intensive exploitation of certain types of flora [22,23]. The comparatively vitamin D-rich diet of the European inhabitants at this time may be the reason the DHCR7 mutation did not reach fixation, while the low vitamin D content in the diet of the East Asians may have added to the selection pressure on DHCR7 over this period.
7-dehydrocholesterol is found in high concentrations in the epidermis [24]. Chen, et al. showed that the conversion of epidermal 7-dehydrocholesterol in hypopigmented skin is much more efficient than in highly pigmented skin. This suggests that sufficient previtamin D3 can be synthesized in Caucasians but not in people with heavily pigmented skin after a brief exposure to summer noon sunlight even at high latitudes [25]. Dependency upon sunlight for vitamin D synthesis may also explain why 7-dehydrocholesterol levels are three to eight times higher in non-feathered skin areas of birds such as the legs and feet when compared to body skin which is covered with feathers [26].
These studies show that humans and animals have adapted mechanisms to exploit optimal methods of synthesiszing vitamin D in response to their environment. The increased frequencies of DHCR7 alleles associated with higher vitamin D status in the hypopigmented populations of Europe and Northeast Asia may represent yet another adaptation which conferred a survival advantage allowing early humans to avoid severe deficiency when migrating to northern latitudes.
Ethnic differences in vitamin D status have long been recognised [27-30], although the evolutionary factors responsible for the biological differences have not been elucidated. SLC24A5, a gene associated with skin pigmentation, shows clear evidence of adaptive selection in those of European ancestry but interestingly is not associated with vitamin D status in recent genome wide association studies [31,32]. This highly unexpected finding may suggest that factors other than vitamin D status are driving natural selection on the basis of skin colour. However vitamin D genome-wide analyses to date have been conducted mainly in European populations and thus the full range of SLC24A5 genotypes may not have been fully explored.
The three genes that we studied contribute a relatively modest proportion of the variation in circulating 25(OH)D levels and of these only DHCR7 shows marked differences in allele frequencies across populations. Whilst these small differences in vitamin D status appear to be sufficient to drive processes of natural selection, it seems likely that other important genetic mechanisms are still to be discovered, some of which may involve interactions between genes. Environmental factors such as sun exposure, latitude and diet will clearly play an important part and may also interact with genetic influences on vitamin D status [33].
With the exception of rs7041 in GC, the SNPs identified in the genome-wide association studies as being associated with vitamin D status are found in non-coding regions of the genome. These regions may exercise regulatory functions on relevant genes or alternatively the polymorphisms identified may be in linkage disequilibrium with neighbouring functional polymorphisms [34,35]. Further studies are required to determine how these polymorphisms effected the association with vitamin D status.
Because of the strong linkage disequilibrium in this genomic region it is possible that the positive selection that we detected relates not to the DHCR7 gene, but to NADSYN1, which is situated close to DHCR7 in the genome and was included in the core haplotype. NADSYN1 encodes NAD synthetase, an enzyme that catalyzes the final step in the biosynthesis of nicotinamide adenine dinucleotide (NAD) from nicotinic acid adenine dinucleotide (NaAD). NAD is an important cofactor in redox reactions and is also involved in post-translational modification of proteins; however there is no known biological connection between this gene and vitamin D metabolism.
It is notable that whilst the major alleles for SNPs in GC and DHCR7 are associated with higher 25(OH)D levels in Europeans, the opposite was true for four of the SNPs near CYP2R1 (Additional file 1: Table S1). This makes the suggestion of balancing selection acting near the CYP2R1 locus plausible, although the biological reasons for this are unclear.
Neither GC nor CYP2R1 showed evidence of positive selection in our analysis. One explanation for this may be that selection in the GC or CYP2R1 genes took place earlier than that in the DHCR7 gene, and hence was not detected by the methods used in this study. Alternatively, it may be the level of vitamin D3 rather than 25(OH)D that influences survival and brings about evolutionary selection. If this were the case, then the availability of 7-dehydrocholesterol to promote synthesis of vitamin D in the skin would be more important than 25 hydroxylase activity or the contribution of D binding protein (Figure 1). Whilst this conclusion would be contrary to most current opinion there is some evidence that 25-hydroxylation may occur in a range of target tissues [36,37].
Conclusions
This observation of positive evolutionary selection highlights the physiological importance of vitamin D in humans and sheds new light on the crucial role of DHCR7 in the synthetic pathway. The finding that the same molecular variant is selected in both northern Europe and Northeast Asia is highly unusual and strengthens the argument that individuals with low DHCR7 activity had a survival advantage at northern latitudes despite variations in other environmental factors in different continents. The unique signal of evolutionary selection, amongst all the genetic variants linked to higher 25(OH)D levels, points to the primacy of DHCR7 amongst the vitamin D synthetic enzymes and will help to direct future research towards a hitherto neglected area of vitamin D metabolism.
Methods
Genotype data
Phased genotype data were obtained from release 2 of the HapMap 3 dataset [38]. Only paired chromosomes were studied. These were from 993 individuals: 53 of African ancestry in Southwest USA (ASW); 113 from Utah with Northern and Western European ancestry from the Centre d’Etude du Polymorphisme Humain (CEPH) collection (CEU); 85 from the Chinese population in Metropolitan Denver, Colorado (CHB); 88 from the Gujerati population in Houston, Texas (GIH); 170 from the Japanese population in Tokyo and the Han Chinese population in Beijing (JPT + CHB); 90 from the Luhya population in Webuye, Kenya (LWK); 50 of Mexican ancestry in Los Angeles (MEX); 143 from the Maasai population in Kinyawa, Kenya (MKK); 88 from the Tuscan population in Italy (TSI); and 113 from the Yoruba population in Ibadan, Nigeria (YRI).
FST vs. heterozygosity
An FST -outlier detection approach [39] using the LOSITAN [40] software package was implemented to identify SNPs which may have undergone positive or balancing selection. The expected distribution of Wright’s inbreeding coefficient, FST, against the expected heterozygosity, He, was constructed under an island model of migration with neutral markers. This was used to identify outlier loci with excessively high or low FST compared to neutral expectations, which would be candidates for selection. 10,000 simulation replicates were used (infinite allele model) for a sample size of 993 individuals.
Pairwise FST
Pairwise FST values to assess population differentiation were calculated as described by Weir and Cockerham with Nei’s correction for sample size [41-43] between the ten populations. Values were ranked from highest to lowest, with those above the 95th percentile regarded as significant.
Haplotype block definition
Haplotype blocks were constructed using the LD-based empirical block definition proposed by Gabriel, et al. [44]. Haploview 3.0 [45] was used to estimate and view the detailed blocks and their underlying haplotype structure.
EHH and REHH
The EHH statistic uses the decay of linkage disequilibrium to estimate the age of the haplotypes. It is defined as the probability that any two randomly chosen chromosomes carrying a particular core haplotype have the same extended haplotype from the core region to a distance x. The relative EHH (REHH), which compares the EHH of a core haplotype to that of other core haplotypes at the same locus, corrects for local variation in recombination rates [14]. EHH and REHH values were calculated for core regions which contained the SNPs of interest.
The significance of REHH was tested using empirical data from the HapMap3 dataset of the entire chromosome on which the core region was situated. REHH values were calculated for core haplotypes in all haplotype blocks for the relelvant chromosome. The haplotypes were then placed into 20 bins based on their frequency. Analysis was carried out using Sweep software (Varilly P, Fry B, and Sabeti P, http://www.broadinstitute.org/mpg/sweep/).
1958 Birth cohort
Serum 25(OH)D concentrations were measured using an automated IDS OCTEIA enzyme-linked immunosorbent assay (ELISA) (Dade-Behring BEP2000 analyzer), standardized according to the mean from Vitamin D External Quality Assessment Scheme (DEQAS) [46]. Genetic data for the cohort were obtained from by Affymetrix 6.0 and Illumina 550 K Infinium methods from two genome wide sub-studies (Wellcome Trust Case–control Consortium 2 and Type 1 Diabetes Genetics Collaboration) [47,48].
Abbreviations
ASW: African ancestry in Southwest USA; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; CHD: Chinese in Metropolitan Denver, Colorado; cM: centiMorgan; EHH: Extended haplotype homozygosity; GIH: Gujarati Indians in Houston, Texas; GWAS: Genome wide association study; JPT + CHB: Japanese in Tokyo, Japan + Han Chinese in Beijing, China (MERGED); LWK: Luhya in Webuye, Kenya; MEX: Mexican ancestry in Los Angeles, California; MKK: Maasai in Kinyawa, Kenya; REHH: Relative extended haplotype homozygosity; SNP: Single nucleotide polymorphism; TSI: Toscans in Italy; YRI: Yoruba in Ibadan, Nigeria (West Africa).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RW, ARM, VK and CJG contributed to study design. VK and EH took part in data analysis. VK, RW, ARM, EH and CJG interpreted the results. VK, RW and ARM drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Geometric mean of 25-hydroxyvitamin D by SNP genotypes in the 1958 British Birth Cohort (n = 5,233).
Contributor Information
Valerie Kuan, Email: valeriekuan@yahoo.co.uk.
Adrian R Martineau, Email: a.martineau@qmul.ac.uk.
Chris J Griffiths, Email: c.j.griffiths@qmul.ac.uk.
Elina Hyppönen, Email: elina.hypponen@unisa.edu.au.
Robert Walton, Email: r.walton@qmul.ac.uk.
References
- Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357:266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP) Hum Genet. 1993;92(2):183–188. doi: 10.1007/BF00219689. [DOI] [PubMed] [Google Scholar]
- Clemens TL, Adams JS, Henderson SL, Holick MF. Increased skin pigment reduces the capacity of skin to synthesise vitamin D3. Lancet. 1982;1:74–76. doi: 10.1016/s0140-6736(82)90214-8. [DOI] [PubMed] [Google Scholar]
- MacLaughlin J, Holick MF. Aging decreases the capacity of human skin to produce vitamin D3. J Clin Invest. 1985;76:1536–1538. doi: 10.1172/JCI112134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D’Agostino RB Sr, Ordovas JM, O'Donnell CJ, Dawson-Hughes B, Vasan RS, Booth SL. Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr. 2009;63:458–464. doi: 10.1038/sj.ejcn.1602959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snellman G, Melhus H, Gedeborg R, Olofsson S, Wolk A, Pedersen NL, Michaëlsson K. Seasonal genetic influence on serum 25-hydroxyvitamin D levels: a twin study. PLoS One. 2009;4:e7747. doi: 10.1371/journal.pone.0007747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O’Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL. et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, Li Q, Weinstein SJ, Purdue M, Virtamo J, Horst R, Wheeler W, Chanock S, Hunter DJ, Hayes RB, Kraft P, Albanes D. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19:2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman CD, Meyers KJ, Ziegler JT, Taylor KD, Palmer ND, Haffner SM, Fingerlin TE, Wagenknecht LE, Rotter JI, Bowden DW, Langefeld CD, Norris JM. Genome-wide association study of vitamin D concentrations in Hispanic Americans: the IRAS family study. J Steroid Biochem Mol Biol. 2010;122:186–192. doi: 10.1016/j.jsbmb.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su J, Lange N, Brehm JM, Damask A, Soto-Quiros M, Avila L, Celedón JC, Canino G, Cloutier MM, Hollis BW, Weiss ST, Litonjua AA. Genome-wide association analysis of circulating vitamin D levels in children with asthma. Hum Genet. 2012;131:1495–1505. doi: 10.1007/s00439-012-1185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive Natural Selection in the Human Lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Black WC 4th, Baer CF, Antolin MF, DuTeau NM. Population genomics: Genome-wide sampling of insect populations. Annu Rev Entomol. 2001;46:441–469. doi: 10.1146/annurev.ento.46.1.441. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Kelley RI, Hennekam RCM. The Smith Lemli-Opitz syndrome. J Med Genet. 2000;37:321–335. doi: 10.1136/jmg.37.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz JM, Gilbert-Barness E, Ackerman J, Lowichik A. Cholesterol and development: the RSH (“Smith-Lemli-Opitz”) syndrome and related conditions. Pediat Path Molec Med. 2002;21:153–181. doi: 10.1080/15227950252852078. [DOI] [PubMed] [Google Scholar]
- Lerchbaum E, Obermayer-Pietsch B. Vitamin D and fertility: a systematic review. Eur J Endocrinol. 2012;166(5):765–778. doi: 10.1530/EJE-11-0984. [DOI] [PubMed] [Google Scholar]
- Kappelman J, Alçiçek MC, Kazanci N, Schultz M, Ozkul M, Sen S. First Homo erectus from Turkey and implications for migrations into temperate Eurasia. Am J Phys Anthropol. 2008;135(1):110–116. doi: 10.1002/ajpa.20739. [DOI] [PubMed] [Google Scholar]
- Richards MP, Jacobi R, Cook J, Pettitt PB, Stringer CB. Isotope evidence for the intensive use of marine foods by Late Upper Paleolithic humans. J Hum Evol. 2005;49(3):390–394. doi: 10.1016/j.jhevol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Richards MO, Hedges REM. Stable isotope evidence for similarities in the types of marine foods used by late Mesolithic humans at sites along the Atlantic coast of Europe. J Archaeol Sci. 1999;26:717–722. doi: 10.1006/jasc.1998.0387. [DOI] [Google Scholar]
- Dobrovolskaya MV. Upper palaeolithic and late stone age human diet. J Physiol Anthropol Appl Human Sci. 2005;24(4):433–438. doi: 10.2114/jpa.24.433. [DOI] [PubMed] [Google Scholar]
- Jablonski NG, Whitfort MJ, Roberts-Smith N, Qinqi X. The influence of life history and diet on the distribution of catarrhine primates during the Pleistocene in eastern Asia. J Hum Evol. 2000;39(2):131–157. doi: 10.1006/jhev.2000.0405. [DOI] [PubMed] [Google Scholar]
- Liu L, Bestel S, Shi J, Song Y, Chen X. Paleolithic human exploitation of plant foods during the last glacial maximum in North China. Proc Natl Acad Sci USA. 2013;110(14):5380–5385. doi: 10.1073/pnas.1217864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holick MF. The cutaneous photosynthesis of previtamin D3: a unique photoendocrine system. J Invest Dermatol. 1981;77(1):51–58. doi: 10.1111/1523-1747.ep12479237. [DOI] [PubMed] [Google Scholar]
- Chen TC, Chimeh F, Lu Z, Mathieu J, Person KS, Zhang A, Kohn N, Martinello S, Berkowitz R, Holick MF. Factors that influence the cutaneous synthesis and dietary sources of vitamin D. Arch Biochem Biophys. 2007;460(2):213–217. doi: 10.1016/j.abb.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch EM, Koch FC. The Provitamin D of the Covering Tissues of Chickens. Poult Sci. 1941;1:33–35. [Google Scholar]
- Pal BR, Marshall T, James C, Shaw NJ. Distribution analysis of vitamin D highlights differences in population subgroups, preliminary observations from a pilot study in UK adults. J Endocrinol. 2003;179:119–129. doi: 10.1677/joe.0.1790119. [DOI] [PubMed] [Google Scholar]
- Stephens WP, Klimiuk PS, Berry JL, Mawer EB. Annual high-dose vitamin D prophylaxis in Asian immigrants. Lancet. 1981;28:1199–1202. doi: 10.1016/s0140-6736(81)91439-2. [DOI] [PubMed] [Google Scholar]
- Harris SS, Dawson-Hughes B. Seasonal changes in plasma 25-hydroxyvitamin D concentrations of young American black and white women. Am J Clin Nutr. 1998;67:1232–1236. doi: 10.1093/ajcn/67.6.1232. [DOI] [PubMed] [Google Scholar]
- Nesby-O’Dell S, Scanlon KS, Cogswell ME, Gillespie C, Hollis BW, Looker AC, Allen C, Doughertly C, Gunter EW, Bowman BA. Hypovitaminosis D prevalence and determinants among African American and white women of reproductive age: third national health and nutrition examination survey, 1988–1994. Am J Clin Nutr. 2002;76:187–192. doi: 10.1093/ajcn/76.1.187. [DOI] [PubMed] [Google Scholar]
- Izagirre N, Garcia I, Junquera C, de la Rua C, Alonso S. A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol. 2006;23:1697–1706. doi: 10.1093/molbev/msl030. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, Schaffner SF, Lander ES, Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y. International HapMap Consortium et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AW. Sunlight, season, skin pigmentation, vitamin D, and 25-hydroxyvitamin D: integral component of vitamin D endocrine system. Am J Clin Nutr. 1998;67:1108–1110. doi: 10.1093/ajcn/67.6.1108. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12(6):634–639. doi: 10.1016/S0959-437X(02)00355-6. [DOI] [PubMed] [Google Scholar]
- Cobb J, Büsst C, Petrou S, Harrap S, Ellis J. Searching for functional genetic variants in non-coding DNA. Clin Exp Pharmacol Physiol. 2008;35(4):372–375. doi: 10.1111/j.1440-1681.2008.04880.x. [DOI] [PubMed] [Google Scholar]
- Sigmundsdottir H, Pan J, Debes GF, Alt C, Habtezion A, Soler D, Butcher EC. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxylase. J Biol Chem. 2003;278(39):38084–38093. doi: 10.1074/jbc.M307028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc Biol Sci. 1996;263:1619–1626. doi: 10.1098/rspb.1996.0237. [DOI] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinforma. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1359–1270. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann Hum Genet. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Hyppönen E, Turner S, Cumberland P, Power C, Gibb I. Serum 25-hydroxyvitamin D measurement in a large population survey with statistical harmonization of assay variation to an international standard. J Clin Endocrinol Metab. 2007;92:4615–4622. doi: 10.1210/jc.2007-1279. [DOI] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, Edkins S, Gray E, Booth DR, Potter SC, Goris A, Band G, Oturai AB, Strange A, Saarela J, Bellenguez C, Fontaine B, Gillman M, Hemmer B, Gwilliam R, Zipp F, Jayakumar A, Martin R, Leslie S, Hawkins S, Giannoulatou E. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geometric mean of 25-hydroxyvitamin D by SNP genotypes in the 1958 British Birth Cohort (n = 5,233).


