Abstract
Proteins secreted by the type V secretion system (T5SS), known as autotransporters, are large extracellular virulence proteins localized to the bacterial poles. In this study, we characterized two novel autotransporter proteins of ‘ Candidatus Liberibacter asiaticus’ (Las), and redesignated them as LasAI and LasAII in lieu of the previous names HyvI and HyvII. As a phloem-limited, intracellular bacterial pathogen, Las has a significantly reduced genome and causes huanglongbing (HLB), a devastating disease of citrus worldwide. Bioinformatic analyses revealed that LasAI and LasAII share the structural features of an autotransporter family containing large repeats of a passenger domain and a unique C-terminal translocator domain. When fused to the GFP gene and expressed in E. coli, the LasAI C-terminus and the full length LasAII were localized to the bacterial poles, similar to other members of autotransporter family. Despite the absence of a typical signal peptide, LasAI was found to localize at the cell surface by immuno-dot blot using a monoclonal antibody against the partial LasAI protein. Its surface localization was also confirmed by the removal of the LasAI antigen using a proteinase K treatment of the intact bacterial cells. When co-inoculated with a P19 gene silencing suppressor and transiently expressed in tobacco leaves, the GFP-LasAI translocator targeted to the mitochondria. This is the first report that Las encodes novel autotransporters that target to mitochondria when expressed in the plants. These findings may lead to a better understanding of the pathogenesis of this intracellular bacterium.
Introduction
Autotransporters are large multi-domain virulence factors encoded by genomes of diverse gram-negative bacteria. A typical autotransporter consists of three functional domains: a Sec-dependent N-terminal signal peptide, a secreted passenger domain (α-domain) and a conserved C-terminal translocator domain (β-domain) [1]. The central passenger domain will ultimately be either attached to the cell surface or secreted. This type of self-transporting protein system is referred to as a type V secretion system (T5SS). Known virulence factors secreted by T5SS have been shown to be cytotoxic, contain protease activities, or functions such as adhesions. Based on structural features, autotransporters have recently been classified into three sub-types: classical autotransporters (T5aSS), two-partner secretion system (T5bSS) and trimeric autotransporters (T5cSS) [2]. The signal peptide directs export of the precursor protein across the inner membrane using the Sec machinery and then is cleaved by peptidase. Subsequently, the β-domain inserts into the outer membrane and forms a pore with 12 transmembrane β-strands through which the passenger domain is presumed to be exported [3]. Once the passenger domain is translocated to the cell surface, it is usually cleaved from the translocator domain and released extracellularly. In some cases the passenger domain is not cleaved and remains tightly associated with the cells [4]. The trimeric autotransporters (T5cSS) known as AT-2 are exemplified by the oligomeric coiled-coil adhesions from various pathogenic bacteria, such as YadA of Yersinia [5], Hia of Haemophilus [6], and Hap of Haemophilus [7]. Compared with the conventional translocator domain that typically contains about 300 amino acids, AT-2 contains a short translocator domain of about 70 amino acids that is sufficient for translocation of the passenger domains [8,9]. Deletion of the YadA translocator domain abolishes the ability to insert into the outer membrane [9]. Many AT-2 passenger domains contain large repeat units of about 70 residues. Phylogenetic clustering of these repeat units revealed that they share striking clustering patterns in which some of the repeats are almost identical in sequence [10]. It has been reported that autotransporters from a variety of rod-shaped pathogenic bacteria, including IcsA and SepA of Shigella flexneri, AIDA-I of Escherichia coli, and BrkA of Bordetella pertussis, are localized to the bacterial poles [11]. Recently, it was demonstrated that the YadA translocator localized solely to the mitochondrial outer membrane when expressed in yeast and that four β-stands are sufficient for mitochondrial localization [12].
‘ Candidatus Liberibacter asiaticus’ is a Gram-negative, fastidious alpha-Proteobacterium, causing huanglongbing (HLB), a devastating disease of citrus worldwide. HLB causes rapid decline and shortens the life span of infected trees [13]. Having a greatly reduced genome of approximately 1.23 Mb, Las bacteria reside in phloem sieve cells of infected citrus plants and are transmitted by the citrus psyllids, Diaphorina citri [14,15]. Intriguingly, even with such a small genome size, the Las psy62 genome contains multiple prophage-related regions, and two were identified as prophages/temperate phages, which occupy ca. one-sixteenth of the entire Las genome [15,16]. Within these prophage regions, two hypothetical hypervariable proteins (HyvI and HyvII) were identified that contained multiple, nearly-identical, leucine-rich repeats (LRRs). The diversity and plasticity of these two genes may have implications for how these intracellular bacteria adapt to their host ecological niches [17].
Prophages in many bacterial genomes are associated with bacterial pathogenicity and biofilm formation [18]. In the present study, we discovered that these two hypervariable proteins encoded by Las prophages are novel autotransporters (redesignated as LasAI and LasAII). We determined that LasAI and LasAII are polar and surface localized in bacteria in addition to being targeted to the mitochondria when expressed in plant cells. Previously, we demonstrated that the Las bacterium may act as an “energy parasite” by encoding a functional ATP translocase for direct ATP/ADP importation from their host cells [19]. Together these findings may lead us to understand how these intracellular bacteria modulate their host energy biosyntheses during their pathogenesis.
Results
Characteristics of unique autotransporters, lasA I and lasA II, in Las
The lasA I and lasA II, previously reported as hyv I and hyvII, are located in two prophage regions in the Las Psy62 genome [17]. The 2760 bp lasA I encodes a 919 amino-acid hypothetical protein with a predicted molecular mass of 103.5 kDa. LasAI contains 12 full nearly identical leucine-rich repeats (LRRs) and 4 partial LRRs. The repeated sequence of the motifs is LEQIDLSKLEQIDLSEMAVLTQKMNIIDGIVNNLATQTKDVGRK. The 12 full repeats within the lasA I gene share 93-100% identity at the nucleic acid level and 84-100% identity at the protein level. lasA II contains 1026 bp and encodes a 341 amino-acid hypothetical protein with a predicted molecular mass of 38.9 kDa. LasAII has only one partial LRR. The translocator domains of LasAI and LasAII share 80% identity at the amino acid level while the passenger domains share 50% identity at the amino acid level. Using the SignalP signal peptide prediction software, no signal sequence was identified in LasAI or LasAII and little information about the function of LasAI or LasAII was obtained from a BLAST search of the NCBI protein database. The LasAI and LasAII passenger domains share low level (about 25%) amino acid sequence similarities with the LRR protein of Colwellia psychrerythraea 34H (GenBank accession number: AAZ26055) and the cell wall associated biofilm protein of Staphylococcus epidermidis (ZP_06614153). Surprisingly, the passenger domains of LasAI and LasAII share similar LRR repeat structures with the Toll-like receptors (TLRs) that function as sentinels of the innate immune system by binding a variety of ligands, including lipopolysaccharide, flagellin and dsRNA, through a LRR ligand-binding domain [20]. LasAI and LasAII translocator domains were predicted to contain ten and twelve β-stranded secondary structures respectively by the YASPIN Secondary Structure Prediction program. However, the 3D structure predicted by the I-TASSER program did not form the typical β-barrel structure, which was reported for the translocator domain of the autotransporter NalP from Neisseria meningitidis [3,21]. Despite the absence of typical signal peptides and no significant sequence homology with other autotransporters at the amino acid level, sequence analyses predicted that LasAI and LasAII possess architectural features of the autotransporter family, including passenger domains with large repeated sequences that form coiled-coils and translocator domains containing β-stranded structures.
LasAI is an outer membrane protein and non-cleaved from cells
The full length gene lasA I was cloned into the pET102D-TOPO vector and protein expression was induced in E. coli BL21 (DE) cells (Invitrogen, Carlsbad, CA). A protein of the expected size for LasAI was shown on SDS-PAGE and confirmed by Western blot (Figure 1). LasAI was purified under hybrid conditions, and the elution fractions contained two bands detected by SDS-PAGE. The 120 kDa protein band including the 16 kDa fusion tag was verified by Western blot with an antibody against LasAI (N terminus, one full repeat and amino acids from part of the translocator domain). No signal was detected for the 40 kDa protein on the same Western blot (Figure 1B).
Figure 1. Expression and outer membrane localization of a novel autotransporter protein, LasAI, of ‘ Candidatus Liberibacter asiaticus’ (Las).
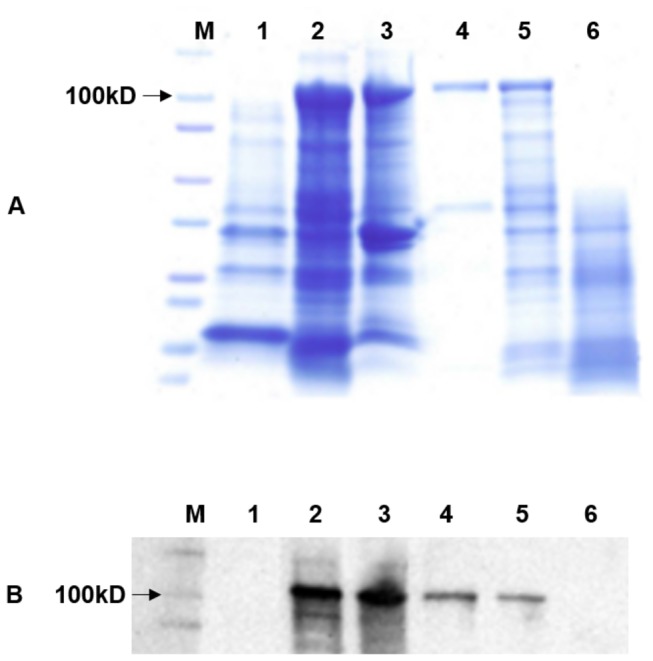
SDS-PAGE (A) and Western blot (B) analysis of E. coli containing the pET102-lasA I construct. M: A molecular mass marker; lane 1: Whole-cell lysate from E. coli BL21 containing plasmid pET102 alone; lane 2: Whole-cell lysate from E. coli BL21 containing recombinant plasmid pET102-lasA I, lane 3: outer membrane protein, lane 4: purified LasAI from whole-cell pellet, lane 5: LasAI protein without proteinase K treatment, Lane 6: LasAI with proteinase K treatment.
It has been shown that autotransporter passenger domains are transported to the cell surface and most of them are processed, thus releasing the passenger domain into the culture’s supernatant. To determine the subcellular localization of LasAI and its passenger domain, outer membrane proteins, surface-associated proteins and secreted proteins in culture supernatant were isolated. As shown in Figure 1, a 120 kDa protein was detected in outer membrane fraction by SDS-PAGE gel and confirmed by western blot, which suggests LasAI containing both passenger domain and translocator domain in these fractions. In contrast no LasAI protein was detected from the culture supernatant using the anti-LasAI antibody (Figure S1A), which suggests that even though the LasAI protein contains the signal information required for cell pole localization in E. coli, its passenger domain was not cleaved and released into the culture’s supernatant in E. coli. The surface-associated protein was isolated from cell pellets and a Western blot was performed using an anti-LasAI antibody. No specific binding signal was observed (Figure S1A). Taken together, the results indicate that the LasAI is an outer membrane protein and its passenger domain was not cleaved and was still tightly associated with the translocator domain in E. coli.
Polar localization of LasAI and LasAII
Several autotransporters from a variety of rod-shaped pathogenic bacteria are polar-localized in the bacterial [11]. We examined the localization of LasAI and LasAII by constructing GFP fusion proteins. The expression of GFP and GFP fusion proteins was detected by Western blot with an anti-GFP antibody (Figure S1B). When GFP was fused with the translocator domain of lasA I (pET102-gfp-lasA I-TD), or the full length lasA II gene (pET102-gfp-lasA II), the expression of GFP was observed at the cell poles of E. coli by confocal laser scanning microscopy (CLSM) (Figure 2D–I). In the control panel, the expression of GFP (pET102-gfp) without the fusion partner was observed in the whole cell, which indicates GFP itself is not directed to bacterial cell poles (Figure 2A–C).
Figure 2. Polar localization of Las autotransporters LasAI and LasAII from ‘ Candidatus Liberibacter asiaticus’ (Las) in E. coli .
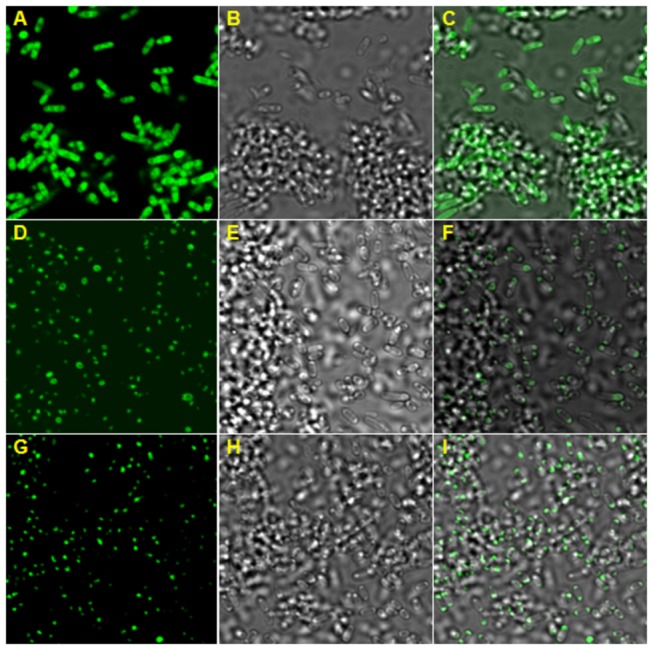
A, D, G: GFP expression detected by confocal laser scanning microscopy (CLSM) in E. coli containing recombinant plasmids pET102-gfp, pET102-gfp-lasA I-TD and pET102-gfp-lasA II, respectively. A, D, G: 505 nm LP filter; B, E, H: differential interference contrast (DIC) of bacterial cells; C, F, I: FITC-DIC merged.
Exportation of the LasAI and LasAII passenger domains by the translocator domains
Although the typical N-terminal signal sequence found in most autotransporters was not identified in LasAI, our results showed that the passenger domain of the LasAI protein is localized at the E. coli cell surface. Immuno-dot blot results showed strong signals indicating LasAI passenger domain is transported out of the bacterial cells when expressed in E. coli, and no signal was observed in the control strain of E. coli (Figure 3). Proteinase K-treated E. coli containing the LasAI constructs did not bind the LasAI antibody, indicating that the passenger domain of LasAI was degraded on the surface of the bacterial cells. LasAI degradation by proteinase K was confirmed by SDS-PAGE and Western blot. Proteinase K-treated E. coli cells expressing LasAI contained no signal while the untreated control cells contained the full 120 kDa protein (Figure 1). Proteinase K has no ability to cross the bacterial membrane and only digest the surface protein of intact bacteria. This confirmed that the passenger domain of LasAI was digested on the surface of the bacterial cells.
Figure 3. Surface location in E. coli of the autotransporter LasAI as determined by immuno-dot blot analysis.
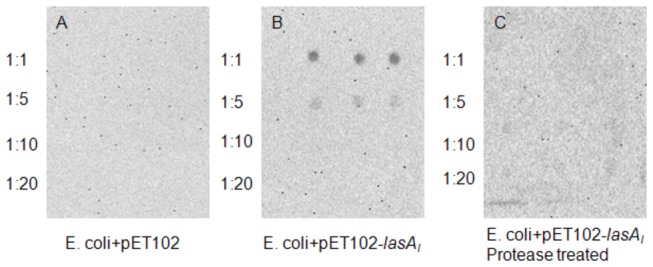
Serial dilutions of E. coli cells were deposited onto a nitrocellulose membrane. The presence of LasAI was detected with anti-LasAI antibody. A: E. coli containing control plasmid pET102; B: IPTG-induced E. coli containing recombinant plasmid pET102-lasA I; C: IPTG induced E. coli containing recombinant plasmid pET102-lasA I treated with Proteinase K.
The LasAI translocator domain not only exports its native passenger domain but also the LasAI-GFP fusion protein to the cell surface. When GFP alone was expressed, the GFP protein stayed inside the cells and no GFP binding signal was detected with an anti-GFP antibody. However, when GFP was fused with the translocator domain of lasA I, or the full length lasA II gene, the GFP proteins were detected on intact bacterial surfaces with an anti-GFP antibody (Figure 4). After proteinase K digestion, immuno-dot blot results showed no GFP signal (data not shown). However, when the whole-cell lysate was treated with proteinase K and analyzed by SDS-PAGE, a slight reduction in the intensity of the fusion proteins was observed with Coomassie blue staining (data not shown). This indicated that not all fusion proteins expressed in E. coli are exposed on the surface of the bacteria and that the translocation of the GFP fusion protein is not as efficient as with the native LasAI passenger domain.
Figure 4. Surface localization in E. coli of GFP fusion proteins of LasAI and LasAII by immuno-dot blot.
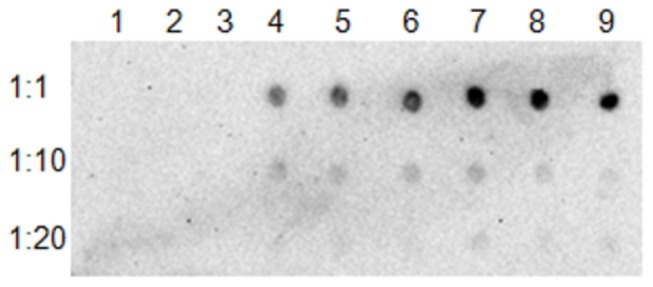
Serial dilutions of bacterial cells were deposited onto a nitrocellulose membrane; GFP was detected with anti-GFP antibody. Lane 1, 2, 3: IPTG induced E. coli containing plasmid pET102-gfp; Lane 4, 5, 6: IPTG-induced E. coli containing plasmid pET102-gfp-lasA I-TD; Lane 7, 8, 9: IPTG induced E. coli containing plasmid pET102-gfp-lasA II.
To further confirm the surface localization of LasAI, an immunofluorescence assay (IFA) was performed. The E. coli cells expressing the LasAI protein were not labeled by anti-LasAI antibody even though the isolated LasAI protein and the E. coli cells expressing the LasAI protein can be detected by Western blot and immuno-dot blot. The absence of surface labeling indicates that the LasAI protein produced in E. coli may not be secreted and folded properly.
LasAI targeting to mitochondria
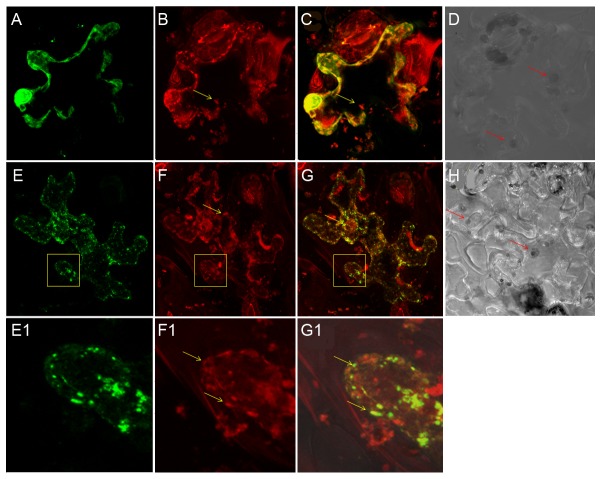
The autotransporter YadA translocator domain was expressed in yeast and imported into the mitochondria, which did not interfere with mitochodrial function [12]. To investigate the potential function and cellular localization of LasAI and LasAII in plant cells and the role of the LasAI and LasAII translocator domains, full-length LasA I, full-length LasA II and the translocator domain of LasA I were cloned into the pGDY vector and transformed into Agrobacterium tumefaciens strain GV 2660. Transient expression results showed no detectable GFP in tobacco plants inoculated with these constructs, except with the pGDY vector alone. Co-inoculation of tobacco leaves with a P19 gene silencing suppressor and pGDY-lasA I-TD construct facilitated GFP expression (data not shown). Only a few cells had detectable GFP in the infiltrated zone when the full length lasA I or lasA II constructs were co-inoculated with the gene silencing suppressor. CLSM and propidium staining results showed that the expression of pGDY-lasA I-TD appeared to localize in the mitochondria. MitoTracker labeling and CLSM confirmed that GFP-lasA I-TD targeted to the mitochondria in tobacco leaves. As shown in Figure 5G and G1, the appearance of yellow mitochondria confirmed the localization of the pGDY-lasA I-TD fusion protein. The autofluorescence of chloroplasts was not observed by MitoTracker detection using a 560nm low-pass filter as shown by differential interference contrast (DIC) (Figure 5 D and H). In contrast, GFP alone in whole cells expressed mainly in the nucleus and did not show yellow mitochondria (Figure 5C). Our results demonstrated that the translocator domain of LasAI contains sufficient structural information for targeting mitochondria. No obvious cell death response was observed in the infiltrated leaf zone; however, mitochondria aggregation was observed when infiltrated with full length lasA I and lasA II constructs (Figure 6A2 and A3). In infiltrated leaves, enlarged mitochondria and morphology change in chloroplast were observed and both of them are detached from cell wall (Figure 6B2). In addition, aggregation and changes in mitochondrial morphology were observed in infected periwinkle (Figure 6C2). Collectively, these results suggest that lasA I and lasA II may affect mitochondria and chloroplast function and manipulate energy production during Las infection.
Figure 5. Mitochondrial localization of the Las autotransporter LasAI.
A-G1: confocal laser scanning micrographs. GFP expression and MitoTracker labeling were detected in tobacco leaves infiltrated with pGDY and pGDY-lasA I-TD plasmids, respectively. A, E: GFP detection with 505-530 nm BP filter; B, F: MitoTracker detection with 560 nm LP filter; C, G: merged scans; D, H: differential interference contrast (DIC) micrographs of tobacco cells with chloroplasts (red arrows). E1, F1, G1: magnifications of yellow boxes in panels E, F and G. Mitochondria (yellow arrows).
Figure 6. Mitochondria aggregation and morphology in plant cells.
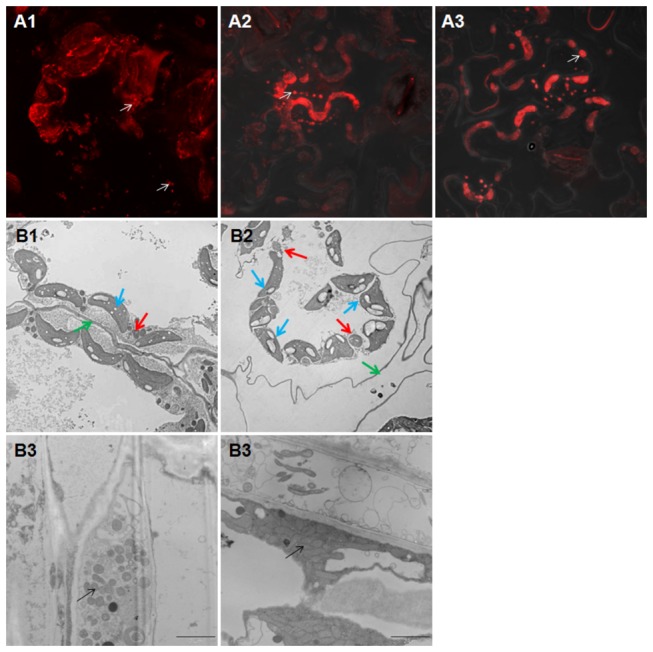
A1-A3: confocal laser scanning micrographs. MitoTracker labeling was detected in tobacco leaves infiltrated with pGDY (A1), pGDY-lasA I (A2) and pGDY-lasA II (A3). Arrows indicate normal and aggregated mitochondria respectively. B1-B4: transmission electron microscopy micrographs. B1: normal mitochondria (red arrow), chloroplast (blue arrow) and cell wall (green arrow) from infiltrated pGDY. B2: enlarged mitochondria (red arrow), abnormal chloroplast (blue arrow) and detached cell wall (green arrow) from infiltrated pGDY-lasA I. B3: normal mitochondria (arrow) from healthy periwinkle. B4: aggregated abnormal mitochondria (arrow) from Las-infected periwinkle.
Discussion
We previously reported the genetic diversity and characteristics of two hypervariable proteins (HyvI and HyvII) from the Psy62 Las genome and global Las isolates that contain up to 12 nearly identical tandem repeats [17]. In the present study, we discovered that LasAI and LasAII are two novel autotransporters. Most known autotransporters are virulence proteins in animal and human pathogens [2]. Typically autotransporters contain an N-terminal signal peptide, a passenger domain and a C-terminal translocator domain. However, no typical signal peptide was predicted in LasAI or LasAII, and while the amino acid sequences of LasAI and LasAII translocator domains contained predicted β-stranded structures, they shared no homology with translocator domains from other autotransporters. We propose that these proteins are new members of the autotransporter family because the translocator domains of LasAI and LasAII not only deliver their native passenger domains, but also exported GFP fusion proteins (GFP-LasAI-TD and GFP-LasAII) onto the bacterial cell surface. These findings reveal that LasAI and LasAII are unique autotransporters and the T5SS may play an important role in Las pathogenesis. Furthermore, using transient gene expression in tobacco leaves, we demonstrated that LasAI contains sufficient structural information for targeting the host mitochondria as do other members of the autotransporter family [12].
Secreted proteins play a central role in the interactions of bacteria and their hosts. Gram-negative bacteria have evolved several specialized secretion systems to deliver effectors into their hosts, such as the Type Ш secretion system (T3SS) and the Type IV secretion system (T4SS). As an intracellular bacterium, Las does not have a T3SS or a T4SS, but may use the Sec secretion system [15,22]. Among the known bacterial secretion systems, the autotransporter or T5SS is the simplest pathway. Since the first report in the 1980s, the autotransporter family has been continuously expanding. Most of the characterized T5SS secreted proteins contribute to the virulence of animal or human pathogens [2]. The relatively few autotransporters reported from plant pathogens include the adhesins HecA/HecB of Erwinia chrysanthemii [23] and the XatA of Xylella fastidiosa which is important for virulence and is also associated with bacterial autoaggregation and biofilm formation [24]. In addition, the EstA autotransporter was reported as a member of the esterase family from the rice root colonizing and beneficial bacterium, Pseudomonas stutzeri A15 [25].
The amino acid sequences of autotransporters are highly divergent except for the conserved translocator domain. LasAI and LasAII proteins are unique autotransporters because they share no homology with any other members of the autotransporter family. Only coiled-coil domains were predicted and no signal peptides or anchor domains were identified in the LasAI and LasAII proteins when using the Trimeric Autotransporter Adhesins (TAAs) domain annotation tool [26]. Since the translocator domains of LasAI and LasAII have the ability to export their native passenger domains and GFP fusion proteins to the E. coli cell surface, the C-terminal translocator domains of LasAI and LasAII may form β-barrel structures through which the passenger domain can pass [2]. However, using the I-TASSER program to predict the 3D structures of LasAI and LasAII, the β-barrel structures of these translocator domains are atypical [21]. In other translocator domains, such as that of NalP from Neisseria meningtidis , the crystal structure contains a 12-stranded β-barrel with a hydrophilic pore filled by an N-terminal α-helix [3]. It is not surprising that the predicted structures of the LasAI and LasAII translocator domains are different from other translocators as there are no conserved amino acids between the Las translocator domains and the known translocator domains. The crystallized structures of LasAI and LasAII will be important for understanding the passenger domain export mechanism.
LasAI and LasAII localize at bacterial poles as do other reported autotransporter members, including IcsA and SepA of Shigella flexneri, AIDA-I of diffusely adherent E. coli and BrkA of Bordetella pertussis [11]. It has been shown that NalP from spherically shaped N. meningitides and BrkA from B. pertussis localize at the pole of E. coli, suggesting that autotransporters contain information required for polar localization [11]. It is interesting to note that in the IcsA protein of S. flexneri two regions within the passenger domain were involved in pole targeting [27]. In contrast, the LasAI translocator domain alone has the ability to target the bacterial poles. Further investigation should identify the region(s) essential for LasAI and LasAII to localize to the bacterial poles.
Typically T5aSS autotransporters exposed at the cell surface are proteolytically cleaved at the junction of the passenger domain and the outer-membrane embedded translocation domain [2]. Although LasAI and LasAII were surface-localized autotransporters, LasAI was present in greater amounts in whole-cell lysates and outer membrane but undetectable in the culture supernatant, indicating either that itis inefficient cleavage in E. coli or that the cleaved passenger domain remains tightly associated with the translocator domain. The passenger domains of T5cSS, such as Hia from H . influenza , are usually not cleaved and stay tightly associated with the cells [4]. In E. coli, BrkA is proteolytically cleaved at the bacterial surface, and the extracellular domain, though cleaved, remains tightly associated with the translocator domain [11]. Further efforts to confirm the LasAI and LasAII surface localization failed by IFA, although production of LasAI and GFP fusion proteins in E. coli was confirmed by immuno-dot blot and Western blot. The exported native passenger domains could not bind the primary antibody, indicating that they may not fold properly on the cell surface of E. coli. This was also observed with the IcsA of S. flexneri, which can be labeled at the surface of wild type S. flexneri but cannot be labeled in the E. coli cells expressing IcsA [28]. Once a pure Las culture is obtained, it will be possible to confirm the LasAI and LasAII surface localization by IFA and determine whether passenger domains are cleaved.
To understand the function of these novel autotransporters, lasA I and lasA II were constructed for Agrobacterium -mediated transient expression. Because no GFP expression was detected with the infiltrations containing our constructs, we used the gene silencing suppressor p19 to enhance the ectopic expression in plant leaves since post-transcriptional gene silencing (PTGS) is reported as a general feature in Agrobacterium -mediated transient expression [29–31]. As expected, strong GFP expression was observed when the lasA I translocater domain (pGDY-lasA I-TD) was co-infiltrated with the p19 construct; while no GFP was detected when infiltrated with the lasA I translocater domain (pGDY-lasA I-TD) alone. However, only a few GFP expressing cells were detected from the leaves co-inoculated with p19 and the full gene constructs of either lasA I or lasA II (pGDY-lasA I and pGDY-lasA II, respectively). This result may be due to GFP-LasAI and GFP-LasAII fusion proteins improperly folded in the plant cells. Surprisingly, we observed the mitochondria aggregation in these infiltrated cells even though there was no detectable GFP expression. We speculate that LasAI and LasAII may have the ability to self-cleave and produce functional subunits targeted to the mitochondria similar to the autotransporter VacA from Helicobacter pylori [32]. Further investigations are underway to confirm this hypothesis. Collectively, these results suggest lasA I and lasA II from a Las prophage/phage (bacterial virus) can act as inducers of PTGS in plant cells. To the best of our knowledge, this is the first evidence of bacterial prophage/phage gene inducing PTGS in plants. Further characterization of the PTGS conferred by LasAI and LasAII may shed light on the evolution and adaptation of the Las bacterium.
With the aid of the gene silencing suppressor p19, we revealed that the LasAI translocator domain-GFP fusion protein targeted to the mitochondria of tobacco leaf cells. Using the YASPIN secondary structure program, LasAI and LasAII were predicted to contain at least ten β-stranded structures in the translocator domain. It is worth noting that four β-strands in the YadA autotranspoter of Yersinia are sufficient for its mitochondrial localization in yeast [12]. Furthermore, several bacterial proteins without typical N-terminal signal sequences also target mitochondria [33], indicating that the lack of signal peptides in LasAI and LasAII is not exceptional.
Proteins containing tandem repeats are associated with diverse functions, and the variable numbers of tandem repeats affect the pathogenicity or antigenicity of several human and animal pathogens [34]. Deletions or insertions of these repeats within the lasA I and lasA II genes were reported in samples of distinct geographical origins and a single origin [17]. It is interesting that the tandem repeats of the LasAI and LasAII passenger domains contain characteristics of the LRR family of proteins. The LRR proteins are important for immune responses, adhesion, invasion, signal transduction, and DNA/RNA processing [35]. The LRR motif of these proteins forms a “horseshoe-shaped” molecule that provides a versatile scaffold for protein–protein interactions [36]. Several LRR proteins have been shown to be located on the cell surface and play a role in surface adherence and aggregation [37]. Compared to the tobacco cells expressing the GFP-LasAI translocator domain, which did not affect mitochondrial morphology, we observed mitochondrial aggregation in cells infiltrated with full length lasA I or lasA II. This phenomenon could be explained if the translocator domain was integrated into the mitochondrial outer membrane with the LRR passenger domain facing the cytosol, thus causing mitochondrial aggregation. By transmission electron microscopy, mitochondrial aggregation was observed in Las infected periwinkle, which agrees with our observation that mitochondrial aggregation is caused by LasAI and LasAII expression in tobacco. Most of the reported LRR proteins contain an N-terminal signal peptide for secretion across the bacterial membrane and a C-terminal membrane attachment region followed by a hydrophobic transmembrane [37]. In contrast, LasAI and LasAII lack the classical signal sequence but the translocator domain can export the LRR passenger domain across bacterial membranes. AdpC, a LRR protein lacking a signal peptide, was also reported to be located on the outer membrane surface when it was expressed in a heterologous E. coli host [38]. Further investigation into whether LasAI and LasAII passenger domains target mitochondria are critical to understanding the functions of these proteins.
In conclusion, ‘Ca. Liberibacter asiaticus’ is an obligate, intracellular bacterium with a significantly reduced genome. We are the first to demonstrate that Las encodes two novel autotransporters (LasAI and LasAII) that target mitochondria when expressed in plant cells. Although the functions of these effectors remain to be elucidated, we hypothesize that Las encodes these autotransporters to modulate energy biosynthesis since Las may directly import ATP/ADP from the cytosol of host cell for its energy and biosynthesis [19]. On the other hand, these proteins may serve as suppressors for plant immune responses since Las encodes a functional flagellin that induces PAMP-triggered immunity in tobacco leaves [39]. Future work will focus on the functional elucidation of LasAI and LasAII, including an investigation into whether LasAI and LasAII passenger domains target mitochondria, identification of the eukaryotic binding partners, and characterization of protein structures. These studies will lead to a better understanding of Las pathogenesis, and thereby yield a better control strategy for HLB.
Materials and Methods
Bacterial strains, plants and cultivation
Strains and plasmids used in this study are listed in Table 1. Escherichia coli Top10 (Invitrogen, Carlsbad, CA) was used as a host for plasmid construction and E. coli BL21 (DE3) cells (Invitrogen, Carlsbad, CA) for recombinant protein expression. E. coli was grown in Luria-Bertani (LB) medium at 37°C. Agrobacterium tumefaciens strain GV2660 was cultured at 28°C in LB and used to mediate transient expression in the leaves of Nicotiana benthamiana . Antibiotics were used at the following concentrations: carbencilin, 50 µg/mL; kanamycin, 50 µg/mL.
Table 1. Strains and plasmids used in this study.
| Strains or plasmids | Description | Source |
|---|---|---|
| Escherichia coli | ||
| TOPO10 | Chemically competent cells for cloning | Invitrogen |
| BL20(DE3) | Chemically competent cells for protein expression | Invitrogen |
| Plasmids in Escherichia coli | ||
| pCR 2.1 | Cloning vector, Ampr, Kmr | Invitrogen |
| pET102D-TOPO | Expression vector, Cmr | Invitrogen |
| pET102-gfp | pET102D carrying gfp, Cmr | This study |
| pET102-lasA I | pET102D carrying lasA I, Cmr | This study |
| pET102-gfp-lasA I -TD | pET102D carrying gfp and lasA I translocator domain, Cmr | This study |
| pET102-gfp-lasA II | pET102D carrying gfp and lasA II, Cmr | This study |
| Agrobacterium tumeficiences | ||
| GV2660 | Strain for transient expression in the plant | 40 |
| Plasmids in Agrobacterium tumeficiences | ||
| pGDY | GFP transient expression vector, Kmr | 40 |
| pGDY-lasA I | pGDY carrying lasA I, Kmr | This study |
| pGDY-lasA I -TD | pGDY carrying lasA I translocator domain, Kmr | This study |
| pGDY-lasA II | pGDY carrying lasA II, Kmr | This study |
| p19 | Gene suppressor, Kmr | 31 |
N . benthamiana seeds were stored at 4°C for 2 days prior to germination. Subsequently the seeds were germinated in chambers programmed for cycles of 16 h light and 8 h dark at 26°C. The seedlings were then transferred into FaFard 4P mix soil in plastic containers and grown in controlled greenhouse conditions.
Plasmid construction
For Agrobacterium -mediated transient expression, the full length lasA I gene, lasA II gene and translocator domain (TD) of lasA I were amplified using genomic DNA from infected plants using the primers listed in Table 2. The respective PCR products were cloned into a binary vector, pGDY, in which gene expression was under control of a CaMV 35S promoter [40], generating pGDY-lasA I, pGDY-lasA II and pGDY-lasA I-TD. The recombinant plasmids were verified by sequencing and then transformed into A. tumefaciens GV2660 by electroporation.
Table 2. Primers used in this study.
| Primer | Sequence |
|---|---|
| Primers for transient expression | |
| lasAI -F | 5’- GCGA G A T C A*ATTAGAAAAGTAAACATGG-3’ |
| lasA -R | 5’- ATTG C T G A G*TTAGTCATCAAAATTAATAAC-3’ |
| lasAII -F | 5’- ATGAGATCTA G A T C T*GAGGACACTAGAAGG-3’ |
| lasAI -TD-F | 5’- ATGA G A T C T*GAGGACACTAGAAGG-3’ |
| Primers for protein expression | |
| GFP-pET-F | 5’- CACCATGGTGAGCAAGGGCGAGGA-3’ |
| GFP-pET-R | 5’- TTATCTAGATCCGGTGGATCC-3’ |
| lasAI -pET-F | 5’- CACCATGATTAGAAAAGTAAACAT-3’ |
| lasAI -pET-R | 5’- ATAGTCATCAAAATTAATAACTTC-3’ |
* Restriction enzyme sites are in italics and underlined
For protein expression, the GFP-pET-F and GFP-pET-R primers were used for PCR amplification of either GFP alone, the N-terminal GFP fusion with the translocator domain of lasA I, or the N-terminal GFP fusion with the full length lasA II from pGDY, pGDY-lasA I-TD and pGDY-lasA II plasmids, respectively (Table 2). Full length lasA I was amplified from infected plant DNA with primers lasAI-pET-F and lasAI-pET-R (Table 2). PCR products for each gene were ligated into the pET102/D-TOPO vector and transformed into E. coli TOP10 cells according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Selected clones were cultivated at 37°C in LB broth for plasmid isolation and sequence verification. The clones were designated as pET102-gfp, pET102-lasA I, pET102-gfp-lasA I-TD and pET102-gfp-lasA II.
Purification of LasAI
The pET102-lasA I consensus clone was transformed into E. coli BL21 (DE3) expression cells (Invitrogen, Carlsbad, CA). LasAI expression was induced and the protein purified using ProBond™ purification system under hybrid conditions according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The purified protein was separated using one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. Membrane transfer was performed by iBlot according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Western blotting was performed with a primary antibody against purified partial LasAI (N-terminus, one repeat and part of the translocator domain) from an immunized mouse (ProMab Biotechnologies, Richmond, CA). Goat anti-mouse HRP-conjugated antibody was used as the secondary antibody and detected by chemiluminescence following the manufacturer’s instructions (Life Technologies, Carlsbad, CA).
Protein fraction preparation and Western blot
Whole-cell protein lysates, culture supernatant protein (CS), outer membrane protein (OM) and surface associated protein (AD) were prepared from E. coli BL21 containing plasmid pET102-lasA I as previously described [11,41–43]. For the CS fraction, culture supernatants were filtered through 0.22 µM-pore size filters and concentrated approximately 100-fold by passage through Amicon centrifuge tubes with a molecular mass limit of 50 KDa (Millipore, Billerica, MA). The final pellet was dissolved in SDS-PAGE buffer. The outer membrane protein was isolated on the basis of sarkosyl insolubility [43]. Briefly the cells were collected and broken down by sonication. Total membrane proteins were separated by ultracentrifugation at 28000 rpm for 1hr at 4°C. To obtain the outer membrane protein, the pellet was suspended in 20mM Tris buffer (pH7.4) containing 0.5% sarkosyl and centrifuged again at 28000 rpm for 1hr at 4°C. The final pellet was dissolve in SDS-PAGE buffer. To obtain the AD fraction, the protein secreted but remaining bound to the cell surface, and the cell pellets were suspended in PBS and N-hexadecane was added. The suspensions were vortexed and centrifuged, and the liquid phase was filtered. The proteins were precipitated by acetone and dissolved in SDS-PAGE buffer. Proteins were separated on SDS-PAGE and detected by Western blot as described above.
Immuno-dot blot and proteinase K treatment
Immuno-dot blotting was performed as described previously [39]. The protein expression from bacterial cells containing different constructs was induced as described above. Bacterial cultures were centrifuged at 2,000 g for 10 min at 4 C°, washed three times with PBS and adjusted to a final concentration of 0.45 as determined by measurements of the optical density at 660 nm. Three microliters of each serial dilution (1:1, 1:5, 1:10 and 1:20) was spotted onto a nitrocellulose membrane in three replicates. The membrane was air dried and the Western blot procedure was performed as described above. The proteinase K treatment of intact cells was performed in PBS with 10 mM MgCl2 [44] and then detected by immuno-dot blot. The proteins, either untreated or treated with proteinase K, were recovered for SDS-PAGE analysis and Western blotting.
Transient gene expression in N benthamiana
The A. tumefaciens GV2660 strain containing different constructs was cultured over night in 2 mL LB medium with the addition of antibiotics, and the following day 50 µL of the cell cultures were inoculated into new 5 mL LB aliquots containing the same antibiotics. These cultures grew until the OD600 value reached approximately 1.0. The cultures were then centrifuged for 10 min at 3000 g, and re-suspended in 5 mL of Agromix (10 mM MgCl2, 10 mM MES, and 100 µM acetosyringone). After the suspension stood at room temperature for at least 3 h, the OD600 value was adjusted to different optical densities with Agromix. The P19 suppressor was also cultured and treated in the same way [31]. The final cell suspension with an OD600 of 1.0 was mixed with the p19 suppressor and infiltrated into 4 week-old N . benthamiana leaves with a 1 mL needleless syringe. The experiments were performed with ten independent replicates.
After two days of infiltration, the infiltrated zone was excised and the epidermal layers were peeled for mitochondrial staining with MitoTracker Red CMXRos according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA) and imaged using a confocal laser scanning microscope (CLSM), Zeiss LSM 510. GFP was detected with a 505-530 nm BP filter and MitoTracker was detected with a 560 nm LP filter.
Polar localization by confocal microscope
The E. coli BL21 (DE3) cells containing pET102-gfp, pET102-gfp-lasA I-TD and pET102-gfp-lasA II constructs were cultured and induced as described above. The collected cells were mounted in antifade solution (Invitrogen, Carlsbad, CA) for CLSM. Images were taken with a 505 nm LP filter and 40Χ or 63Χ (oil) objectives.
Transmission electron microscopy (TEM)
Midribs were sampled from healthy and Las-infected periwinkle. The infiltrated zone was sliced from tobacco. The samples were fixed and sectioned for TEM micrographs as described previously [44].
Supporting Information
A: Western blot of E. coli containing the pET102-lasAI construct. Lane1: whole-cell lysate; lane 2: culture supernatant; lane 3: cell associated protein. B: Western blot of E. coli containing GFP and GFP fusion proteins. Lane 1: E. coli containing plasmid pET102-gfp; lane 2: E. coli containing plasmid pET102-gfp-lasA I-TD; lane 3: E. coli containing plasmid pET102-gfp-lasA II.
(TIF)
Acknowledgments
We are thankful to Scott Adkins and Carrie Vanderspool for providing N . benthamiana plants, and Lesley Benyon and Cheryl Vahling-Armstrong for critical reviews of the manuscript. We also thank Dr. Michael Goodin from the University of Kentucky for providing the pGDY vector and Dr. József Burgyán from the Agricultural Biotechnology Center, Hungary and Dr. Rima Menassa from Agriculture and Agri-Food Canada for providing the p19 suppressor. Special thanks go to Dr. Byung-Ho Kang, University of Florida for providing electron microscopy of mitochondria from infected and non-infected plant cells.
Funding Statement
Funding for this work was provided by the Florida Citrus Advanced Technology Program award 162 and 310. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Henderson IR, Navarro-Garcia F, Nataro JP (1998) The great escape: structure and function of the autotransporter proteins. Trends Microbiol 6: 370–378. doi:10.1016/S0966-842X(98)01318-3. PubMed: 9778731. [DOI] [PubMed] [Google Scholar]
- 2. Scott-Tucker A, Henderson IR (2009) In Bacterial secreted proteins: Secretory mechanisms and role in pathogensis, Wooldridge K. Nottingham, UK: pp. 139-157. [Google Scholar]
- 3. Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J et al. (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J 23: 1257–1266. doi:10.1038/sj.emboj.7600148. PubMed: 15014442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. St Geme JW, Cutter D (2000) The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J Bacteriol 182: 6005–6013. doi:10.1128/JB.182.21.6005-6013.2000. PubMed: 11029419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M et al. (2004) The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J 23: 701–711. doi:10.1038/sj.emboj.7600100. PubMed: 14765110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laarmann S, Cutter D, Juehne T, Barenkamp SJ, St Geme JW (2002) The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol Microbiol 46: 731–743. doi:10.1046/j.1365-2958.2002.03189.x. PubMed: 12410830. [DOI] [PubMed] [Google Scholar]
- 7. Hendrixson DR, de la Morena ML, Stathopoulos C, St Geme JW (1997) Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol Microbiol 26(3): 505-518. doi:10.1046/j.1365-2958.1997.5921965.x. PubMed: 9402021. [DOI] [PubMed] [Google Scholar]
- 8. Surana NK, Cutter D, Barenkamp SJ, St Geme JW (2004) The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J Biol Chem 279: 14679–14685. doi:10.1074/jbc.M311496200. PubMed: 14726537. [DOI] [PubMed] [Google Scholar]
- 9. Roggenkamp A, Ham JH, Deng WL, Doyle JJ, Collmer A (2003) Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J Bacteriol 185: 3735–3744. doi:10.1128/JB.185.13.3735-3744.2003. PubMed: 12813066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim DSH, Chao Y, Saier MH (2007) Protein-translocating trimeric autotransporters of Gram-Negative bacteria. J Bacteriol 188: 5655–5667. PubMed: 16885434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jain S, van Ulsen P, Benz I, Schmidt MA, Fernandez R et al. (2006) Polar localization of the autotransporter family of large bacterial virulence proteins. J Bacteriol 188: 4841–4850. doi:10.1128/JB.00326-06. PubMed: 16788193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Müller JEN, Papic D, Ulrich T, Grin I, Schütz M et al. (2011) Mitochondria can recognize and assemble fragments of a β-barrel structure. Mol Biol Cell 22: 1638-1647. doi:10.1091/mbc.E10-12-0943. PubMed: 21460184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottwald TR (2010) Current epidemiological understanding of citrus huanglongbing. Annu Rev Phytopathol 48: 119–139. doi:10.1146/annurev-phyto-073009-114418. PubMed: 20415578. [DOI] [PubMed] [Google Scholar]
- 14. Jagoueix S, Bove JM, Garnier M (1994) The phloem-limited bacterium of greening disease of citrus is a member of the subdivision of the proteobacteria. Int J Syst Bacteriol 44: 379-386. doi:10.1099/00207713-44-3-379. PubMed: 7520729. [DOI] [PubMed] [Google Scholar]
- 15. Duan YP, Zhou LJ, Hall DG, Li WB, Doddapaneni H et al. (2009) Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact 22: 1011-1020. doi:10.1094/MPMI-22-8-1011. PubMed: 19589076. [DOI] [PubMed] [Google Scholar]
- 16. Zhang SJ, Flores-Cruz Z, Zhou LJ, Kang BH, Fleites LA et al. (2011) 2011) Ca. Liberibacter asiaticus carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant. Mol Plant Microbe Interact 24: 458-468. doi:10.1094/MPMI-11-10-0256. PubMed: 21190436. [DOI] [PubMed] [Google Scholar]
- 17. Zhou LJ, Powell CA, Hoffman MT, Li WB, Fan G et al. (2011) Diversity and plasticity of the intracellular plant pathogen and insect symbiont "Candidatus Liberibacter asiaticus" as revealed by hypervariable prophage genes with intragenic tandem repeats. Appl Environ Microbiol 77: 6663-6673. doi:10.1128/AEM.05111-11. PubMed: 21784907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyd EF, Brüssow H (2002) Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol 10: 521–529. doi:10.1016/S0966-842X(02)02459-9. PubMed: 12419617. [DOI] [PubMed] [Google Scholar]
- 19. Vahling CM, Duan Y, Lin H (2010) Characterization of an ATP translocase identified in the destructive plant pathogen "Candidatus Liberibacter asiaticus". J Bacteriol 192: 834–840. doi:10.1128/JB.01279-09. PubMed: 19948801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L, Botos I, Wang Y, Leonard JN, Shiloach J et al. (2008) Toll-like receptor 3 (TLR3) recognizes double-stranded RNA (dsRNA), a molecular signature of most viruses, and triggers inflammatory responses that prevent viral spread. Science 320: 379-381. doi:10.1126/science.1155406. PubMed: 18420935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. doi:10.1186/1471-2105-9-40. PubMed: 18215316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akula N, Zheng H, Han FQ, Wang N (2011) Discovery of novel SecA inhibitors of Candidatus Liberibacter asiaticus by structure based design. Bioorg Med Chem Lett 21: 4183-4188. doi:10.1016/j.bmcl.2011.05.086. PubMed: 21684161. [DOI] [PubMed] [Google Scholar]
- 23. Rojas CM, Ham JH, Deng WL, Doyle JJ, Collmer A (2004) HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc Natl Acad Sci USA 99: 13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto A, Huston SL, Killiny N, Igo MM (2010) XatA, an AT-1 autotransporter important for the virulence of Xylella fastidiosa Temecula1. Microbiologyopen 1: 33-45. PubMed: 22950010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicolay T, Devleeschouwer K, Vanderleyden J, Spaepen S (2012) Characterization of esterase A, a Pseudomonas stutzeri A15 autotransporter. Appl Environ Microbiol 78: 2533-2542. doi:10.1128/AEM.07690-11. PubMed: 22307303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szczesny P, Lupas A (2008) Domain annotation of trimeric autotransporter adhesions-data. Bioinformatics 24: 1251-1256. doi:10.1093/bioinformatics/btn118. PubMed: 18397894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Charles M, Pérez M, Kobil JH, Goldberg MB (2001) Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio . Proc Natl Acad Sci USA 98: 9871-9876. doi:10.1073/pnas.171310498. PubMed: 11481451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldberg MB, Bârzu O, Parsot C, Sansonetti PJ (1993) Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol 175: 2189–2196. PubMed: 8468279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hammond SM, Caudy AA, Hannon GJ (2001) Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet 2: 110–119. doi:10.1038/35052556. PubMed: 11253050. [DOI] [PubMed] [Google Scholar]
- 30. Johansen LK, Carrington JC (2001) Silencing on the spot induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126: 930-938. doi:10.1104/pp.126.3.930. PubMed: 11457942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C et al. (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080. doi:10.1093/emboj/cdf312. PubMed: 12065420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Domańska G, Motz C, Meinecke M, Harsman A, Papatheodorou P et al. (2010) Helicobacter pylori VacA Toxin/Subunit p34: Targeting of an Anion Channel to the Inner Mitochondrial Membrane. PLOS Pathog 6:e1000878. doi:10.1371/journal.ppat. 1000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rudel T, Kepp O, Kozjak-Pavlovic V (2010) Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat Rev 8: 693-705. doi:10.1038/nrmicro2421. PubMed: 20818415. [DOI] [PubMed] [Google Scholar]
- 34. Ikegami A, Honma K, Sharma A, Kuramitsu HK (2004) Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola . Infect Immun 72: 4619-4627. doi:10.1128/IAI.72.8.4619-4627.2004. PubMed: 15271922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kobe B, Deisenhofer J J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19: 415–421. doi:10.1016/0968-0004(94)90090-6. PubMed: 7817399. [DOI] [PubMed] [Google Scholar]
- 36. Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP et al. (2002) Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111: 825–836. doi:10.1016/S0092-8674(02)01136-4. PubMed: 12526809. [DOI] [PubMed] [Google Scholar]
- 37. Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D et al. (2008) Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythiae leucine-rich repeat protein BspA. Infect Immun 76: 198–205. doi:10.1128/IAI.01139-07. PubMed: 17967853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iyer D, Anaya-Bergman C, Jones K, Yanamandra S, Sengupta D et al. (2010) AdpC is a Prevotella intermedia 17 Leucine-Rich Repeat Internalin-Like Protein. Infect Immun 78: 2385–2396. doi:10.1128/IAI.00510-09. PubMed: 20308299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zou H, Gowda S, Zhou LJ, Hajeri S, Chen GY et al. (2012) The destructive citrus pathogen, ‘Candidatus Liberibacter asiaticus’ encodes a functional flagellin characteristic of a pathogen-associated molecular pattern. PLOS ONE 7(9): e46447. doi:10.1371/journal.pone.0046447. PubMed: 23029520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383. doi:10.1046/j.1365-313X.2002.01360.x. PubMed: 12164816. [DOI] [PubMed] [Google Scholar]
- 41. Caldwell RB, Lattemann CT (2004) Simple and reliable method to precipitate protein from bacterial culture supernatant. Appl Environ Microbiol 70: 610-612. doi:10.1128/AEM.70.1.610-612.2004. PubMed: 14711696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carroll JA (2010) Methods of identifying membrane proteins in spirochetes. Curr Protoc Microbiol 16: 12C PubMed: 20131222. [DOI] [PubMed] [Google Scholar]
- 43. Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO (1986) Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol 24: 330–332. PubMed: 3489731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koh EL, Zhou LJ, Williams DS, Park J, Ding N et al. (2011) Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus”. Protoplasma 249: 687-697. PubMed: 21874517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Western blot of E. coli containing the pET102-lasAI construct. Lane1: whole-cell lysate; lane 2: culture supernatant; lane 3: cell associated protein. B: Western blot of E. coli containing GFP and GFP fusion proteins. Lane 1: E. coli containing plasmid pET102-gfp; lane 2: E. coli containing plasmid pET102-gfp-lasA I-TD; lane 3: E. coli containing plasmid pET102-gfp-lasA II.
(TIF)