Abstract
Exome and whole genome sequencing (ES/WGS) offer potential advantages over traditional approaches to diagnostic genetic testing. Consequently, use of ES/WGS in clinical settings is rapidly becoming commonplace. Yet there are myriad moral, ethical, and perhaps legal implications attached to the use of ES and health care professionals and institutions will need to consider these implications in the context of the varied practices and policies of ES service providers. We developed “core elements” of content and procedures for informed consent, data sharing, and results management and a quantitative scale to assess the extent to which research protocols met the standards established by these core elements. We then used these tools to evaluate the practices and policies of each of the 6 U.S. CLIA-certified labs offering clinical ES. Approaches toward informed consent, data sharing, and results return vary widely among ES providers as do the overall potential merits and disadvantages of each, and more importantly, the balance between the two.
Keywords: exome sequencing, whole genome sequencing, informed consent, data sharing, return of results, ethical implications
INTRODUCTION
Exome and whole genome sequencing (ES/WGS) have emerged as powerful tools for identifying novel disease genes [Lupski et al. 2010; Ng et al. 2010a; Ng et al. 2010b]. ES/WGS offer substantial advantages when compared to traditional diagnostic genetic testing approaches that interrogate a single or subset of known genes, which can be expensive in aggregate as genes are analyzed in a step-wise manner. As a result, ES/WGS have quickly been transitioned from use as a research tool to use in clinical service labs as an alternative to targeted genetic testing. Several widely publicized examples in which these tools have been used to facilitate a clinical diagnosis have further engendered enthusiasm for adopting ES/WGS into clinical practice [Bainbridge et al. 2011; Welch et al. 2011; Worthey et al. 2011]. We are similarly enthused, but such swift translation of ES/WGS into clinical settings raises important questions such as: (1) under what circumstances is ES/WGS likely to be of greatest clinical benefit, (2) how can informed consent for ES/WGS effectively be obtained, (3) what are reasonable options to offer for sharing individual ES/WGS (iES/WGS) results, (4) how should iES/WGS results be managed for providers and families, and (5) how can providers’ and families’ expectations about risk profiling using ES/WGS be managed. While these questions frame the discussion, there are little empirical data about how to best answer them. This is a major obstacle to integrating ES/WGS into clinical care.
We assessed the informed consent, data sharing, and results management practices and policies of the six commercial labs that currently provide ES testing for clinical service. A comparison study such as this one is intended to inform the clinical genetics community of the similarities and differences among exome sequencing providers and to help genetics professionals make an informed decision about which service is best suited to their needs. No service provider stands out as “best” or “worst” nor do we make any judgment about which provider should or should not be used.
MATERIALS AND METHODS
Over the past several years, our group has participated in several dozen research studies including the NHLBI Exome Sequencing Project [Tennessen et al. 2012] and the University of Washington Center for Mendelian Genomics [Bamshad et al. 2012] in which we perform ES/WGS. In the course of these studies, we were required to review approaches and documents used for obtaining informed consent, informing individuals of options for data sharing, and managing results return across a broad range of research settings (e.g., investigation of both Mendelian and complex traits in pediatric and adult study populations). The human genetics community, patient advocacy groups, as well as private and public funding sources have broadly recognized these three areas as important. Moreover, research of these areas has been widely supported by National Institutes of Health, the focus of numerous national and international symposia, and discussed in hundreds of publications in the scientific literature. Based on this review and our experience, we developed policies and language via an iterative process with investigators, ethicists, and regulatory bodies (e.g., institutional review boards). These evolved into “core elements” (Table I) of content and procedures for informed consent (n=10), data sharing (n=3), and results management (n=7). Subsequently, we devised a quantitative scale to assess the extent to which research protocols met the standards established by these core elements (Appendix Tables I-III). A score of zero reflected the absence of a core element, whereas a maximum score represented a core element that had all the content and procedures considered ideal. Because the complexity of core elements varied, so did the maximum score for each core element.
Table I.
Core elements
| Informed Consent | Result types returned |
| Potential risks | |
| Re-analysis of data | |
| Explanation of ES | |
| Patient preferences related to secondary results | |
| Discussion of secondary results | |
| Informed consent and noted discrepancies | |
| Discussion of sharing of de-identified data | |
| Data disposition policy | |
| Limitations of ES | |
|
| |
| Data Sharing | Data sharing options for ordering providers / institutions |
| Deposition into public databases | |
| Education and research purposes | |
|
| |
| Results Management | Updating of result interpretation |
| Range of results returned | |
| Patient preferences related to secondary results, and time constraints | |
| Patients accessing their results | |
| Report construction: methods | |
| Informational/counseling resources available to patients | |
| Report construction: results / interpretation | |
We applied these assessment tools to the informed consent, data sharing, and results management policies and protocols for each of the six U.S.-based CLIA-certified labs currently offering clinical ES for service: Ambry Genetics Corporation, ARUP Molecular Genetics Laboratory, Baylor College of Medicine’s Whole Genome Sequencing Laboratory, Emory Molecular Genetics Laboratory, GeneDx, and UCLA’s Diagnostic Molecular Pathology Laboratory. Although additional labs offer ES/WGS, these services are provided only for researchers or in-house care-providers and therefore were excluded from our analysis. Assessment was based on review of all documents available from each service provider (e.g., requisitions, informed consent documents, test information sheets, and sample results reports). Since none of the service providers had written data sharing or results management policies, we obtained such information via direct correspondence with the lab representative assigned to provide clients with additional information. Tests of Readability were performed using Microsoft Word’s word count and Flesch Reading Ease (FRE [Flesch 1948]) statistics. The higher the word count, the lengthier the document, and the higher the FRE score, the easier the comprehensibility of the document. FRE scores below the range of 60-70 is easily understood by 13-15 year old students, and scores within the range of 0-30 is best understood by university graduates. Additionally, we summarized information on practical considerations of ES such as cost, turn around time, sample requirements, and methodology (Table II). Scoring was completed by face-to-face consensus review and discussion. Scores were summed for each service provider, and normalized to range from 0-1. Principal components analysis and tests of correlation were performed in R.
Table II.
Practical Considerations
| Ambry | ARUP | Baylor | Emory | GeneDx | UCLA | |
|---|---|---|---|---|---|---|
| Name of test | Clinical Diagnostic Exome™ |
Exome Sequencing with Symptom- Guided Analysis |
Whole Exome Sequencing |
EmExome: Clinical Whole Exome Sequencing |
XomeDx | Clinical Exome Sequencing |
| Began offering | 09/2011 | 04/2012 | 10/2011 | 06/2012 | 01/2012 | 01/2012 |
| Turn around time (weeks) |
8-16 | 12-16 | 15 | 15 | 12-16 | 11-12 |
| Cost | $7,900 | $8,000-10,000 | $7,000 | $9,000 | $9,000 | $4,500-6,500 |
| Method (strategy) | Individual ES, Trio ES |
Individual ES | Individual ES | Individual ES | Trio ES | Individual ES, Trio ES |
| Method (exome capture) |
Agilent SureSelect |
Agilent SureSelect, NimbleGen SeqCap |
NimbleGen (custom designed) VCRome 2.1 |
NimbleGen SeqCap |
Agilent SureSelect |
Agilent SureSelect |
| Coverage: (mean depth of coverage) |
90-100× | >100× | >100× | 100× | 100-120× | >100× |
| Coverage (%target bases covered at ≥10×) |
90% | 95% | >95% | 96% | 90-95% | 95% |
| Variant confirmation | + (only primary) |
+ (only primary) |
+ (primary, some secondary results) |
+ (primary, all secondary results) |
+ (only primary) |
+ (only primary) |
ES: exome sequencing
RESULTS
Informed Consent
Core element content
The median score for informed consent was 0.55 with a range of 0.26 (Emory) to 0.64 (Baylor) indicating substantial variation across the ten core elements assessed (Fig 1A). Emory consistently scored the lowest with two exceptions (patient preferences related to secondary results; and discussion of sharing of de-identified data) while Baylor scored highest for four core elements (re-analysis of data; explanation of ES; discussion of secondary results; and informed consent and noted discrepancies), each of which was fairly variable among service providers. Scores for four or five of the service providers were identical for four core elements (potential risks; secondary findings; data disposition policy; and limitations of ES).
Figure 1. Distribution of scores for core elements assessed (A,C,D) and readability of informed consent documents (B).
Each ES provider is represented by a symbol of a different shape: hexagon: Ambry; circle: ARUP; star: Baylor; pentagon: Emory; square: GeneDx; triangle: UCLA.
If Emory and Baylor are excluded from the analysis, most of the variation among service providers was limited to two core elements (re-analysis of data; and patient preferences related to secondary results). Baylor and GeneDx offer reanalysis of iES data and reporting to a healthcare provider on an annual basis. ARUP and UCLA offer updates of the primary result while Ambry, like Emory, offers no option to reanalyze ES data. All service providers require an unspecified fee to re-analyze ES data.
In the informed consent documents, Ambry, Baylor and Emory offer return of secondary results, but the extent to which patient preferences related to secondary results are considered was highly variable. Baylor reports certain secondary results per routine disclosure, whereas Ambry and Emory consider patient preference. Ambry’s preferences options are categorized by a combination of potential utility (carrier status of recessive disorders, predisposition to later-onset disease, predisposition to increase cancer risk, and early-onset disease) and/or actionability. For minors, Ambry offers return of only secondary results associated with early-onset disease. While UCLA does not routinely report secondary ES results, their informed consent document indicates that provider requests for secondary results may be considered. ARUP and GeneDx do not currently offer secondary ES results and therefore also do not consider patient preferences.
Three core elements (result types returned; sharing of data; and data disposition policy) varied modestly across service providers. All service providers to some extent report iES results binned by functional annotation. Emory limits reporting to only suspected causal variants. All other service providers report pathogenic variants and variants of unknown significance (VUS), while ARUP and GeneDx also explain that non-pathogenic variants will be identified, but not reported. In addition, Ambry, ARUP and UCLA inform patients that the interpretation of results is likely to change in the future.
Four providers (Ambry, ARUP, Baylor, and UCLA) explicitly discussed data sharing in their consent documents while two (Emory and GeneDx) did not. Both ARUP and Baylor provide information about what and where data are shared. Baylor indicates only aggregate data will be shared, while ARUP suggests individual-level genotype data will be deposited in “national or international databases.” Neither provides an opt-out option. Ambry and UCLA include somewhat vague statements about what and how data could be shared (e.g., “data will be used for the purpose of research, test validation, and/or education”—Ambry; “we will use your results to improve [the test] by comparing your data to others”—UCLA). However, both Ambry and UCLA provide an option to opt out.
Four service providers do not mention their data disposition policies (e.g. how and whether data will be retained and/or accessible) in the consent document. ARUP and GeneDx indicate that data will be saved for at least five years and one year, respectively, after a report is issued. No service provider includes information on how data are stored or destroyed, and what safeguards are in place to ensure data security.
Document
Median word count among the six informed consent documents was 1,154 and ranged from 724 (Emory) to 3,429 words (Ambry); the median readability score (Flesch Reading Ease) among the six informed consent documents was 40 and ranged from 24 (ARUP) to 49 (GeneDx) (Figure 1B). Word count was independent of readability (R2=0.012), and neither word count nor Flesch Reading Ease scores were correlated with the total informed consent score (R2=0.073 and 0.001, respectively). Baylor’s informed consent document was shorter in length, easier to read, and yet received the highest overall score in our assessment. Accordingly, an informed consent document can contain the core elements we assessed and need not be lengthy or difficult to read.All six service providers require the signature of a clinician, genetic counselor or health care provider for the submission of a sample. Five providers recommend, and only Emory requires, some form of genetic counseling in their consent document (Appendix Table IV).
For five service providers (Ambry, ARUP, Emory, GeneDx, and UCLA), we identified one or more discrepancies between the information included in their informed consent document, and either their data sharing or results management policies as obtained from correspondence with representatives from each lab (Appendix Table V). However, these discrepancies were difficult to assess objectively because written policy documents describing data sharing and results management were not made available to us upon request.
Data Sharing
Core element content
The median score across the three core elements assessed for data sharing was 0.62 with a range of 0.31 (Baylor) to 0.77 (Emory) (Figure 1C). Baylor scored the lowest for each element while Emory scored highest for two core elements (deposition into public databases; and education and research purposes). Scores for four of the service providers were identical for each of two core elements (deposition into public databases; and education and research purposes). Most variable among ES providers were options for data sharing offered to ordering providers / institutions. Ambry and Baylor limit what is shared with providers to the primary and secondary results included in the results report, while all other providers will share .vcf files, upon request. ARUP and GeneDx will also return BAM files if supplied with hard drives from requesting providers.
All service providers propose to use their ES results for educational and research purposes. Baylor and UCLA intend to publish ES results in aggregate. Opting out is not an option. Ambry, ARUP, and Emory indicate they plan to use de-identified data for “education purposes” and do provide an opt-out option.
All service providers have a data deposition policy. Emory indicated they would submit “clinical information and test results in HIPAA-compliant de-identified public databases” and provide patients with an option to opt-out. Ambry, ARUP, GeneDx, and UCLA each expressed interest and/or future plans to submit de-identified data into public-access databases (e.g., dbSNP) and indicated they would ensure their informed consent document reflected their deposition plans. Each intended to provide patients with an option to opt-out. None of these service providers explained the type or scope of data (aggregate results, individual level genotypes) that they intended to deposit in such databases. Baylor had no plans to submit de-identified data into public databases.
Results Management Policy
Core element content
The median score for results management was 0.55 with a range of 0.35 (ARUP) to 0.68 (Emory) (Figure 1D). For two core elements (information / counseling resources available to patients; and report construction: result / interpretation) the score for each service provider was identical. ARUP consistently scored the lowest and Emory the highest for four of five remaining core elements assessed. Among these core elements, most of the variation among service providers was limited to range of results returned, and patient preferences related to secondary results.
Aside from returning the primary iES result, five service providers (Ambry, Baylor, Emory, GeneDx, and UCLA) will return secondary results either through routine disclosure or driven by patient preferences. Each of these five service providers, except Ambry, returns “medically actionable” secondary iES results. Ambry, Baylor, and Emory offer return of secondary iES results in categories other than “medically actionable” and patients can opt for return among these categories (Table III). GeneDx also allows patients to select among limited preferences for return. ARUP does not offer secondary iES results.
Table III.
Categories of secondary results
| Ambry | Baylor | Emory | ||
|---|---|---|---|---|
| No opt in/out option | Focused report | Expanded report | ||
| “Medically actionable” | N/A | “Medically actionable” | ||
| Opt in/out option | (no age minimum): Early-onset disease |
|||
| (≥18 years): Carrier status of recessive disorders |
(no age minimum) Carrier status for autosomal recessive conditions (restrictions*) |
(no age minimum) Other carrier status for autosomal recessive conditions |
(≥18 years): Carrier status for autosomal recessive conditions |
|
| (≥18 years): Predisposition to later- onset disease |
(no age minimum) Adult onset conditions (restrictions**) |
(≥18 years): Adult onset dementia disorders, “other” adult onset disorders |
||
| (≥18 years): Predisposition to increase cancer risk |
(≥18 years): Adult onset cancer disorders |
|||
| (no age minimum) Pharmacogenetics |
(no age minimum): Pharmacogenetics |
|||
| Categories not reported |
Pharmacogenetics, Common disease |
Common disease | Common disease | |
restricted to “disorders recommended for reproductive screening by professional societies”
“findings in genes causing adult onset dementia syndromes or other adult onset neurological conditions for which there is presently no prevention or cure” are not reported.
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06), correspondence with lab representative
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12), correspondence with lab representative
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012), correspondence with lab representative
Emory EmExome Informed Consent Document (Rev. 6/2012), correspondence with lab representative
GeneDx XomeDx Informed Consent Document (GeneDx 07/12), correspondence with lab representative
UCLA Informed Consent for Postnatal Clinical Exome Sequencing, correspondence with lab representative
Ambry and Emory require that patients decide whether to receive secondary iES results at a single point in time (i.e. a clinic visit). In contrast, Baylor provides patients with up to six months during which they can receive additional secondary iES results (i.e. an “expanded” report). Each service provider reports results only to the ordering providers; none of the service providers allow patients direct access to their results.
Three core elements (updating of result interpretation; patients accessing their results; reports construction: methods) varied modestly across service providers. Each service provider offers an update of its iES result interpretation, although whether the service provider or the ordering provider is responsible for prompting an update varies. ARUP, GeneDx, and UCLA put the onus on the ordering provider (i.e., they recommend that ordering clinicians periodically query them to determine whether the original results interpretation has changed). Ambry and Baylor proactively contact providers when a VUS is reclassified as a pathogenic variant. Emory offers a unique publicly available online variant classification tool, “EmVClass,” that provides status updates on all variants that have been observed by them. If a previously identified variant has been reclassified, this tool allows ordering providers to submit an online request for an updated report.
DISCUSSION
ES/WGS are powerful albeit imperfect new tools to facilitate diagnosis and genetic risk profiling. Unlike a targeted genetic test that generates the same putative single result regardless of the laboratory performing the test, the scope of results available from ES are highly dependent on the service provider. Moreover, varied consenting practices and data sharing policies minimally impact decisions about whether to perform targeted testing much less which lab to choose to do the test. Accordingly, which lab performs a test makes little if any difference for a healthcare provider or institution. In contrast, there are myriad moral, ethical, and perhaps legal implications attached to the use of ES/WGS and healthcare providers and institutions will need to consider these implications in the context of the varied practices and policies of ES service providers [Tabor et al. 2011].
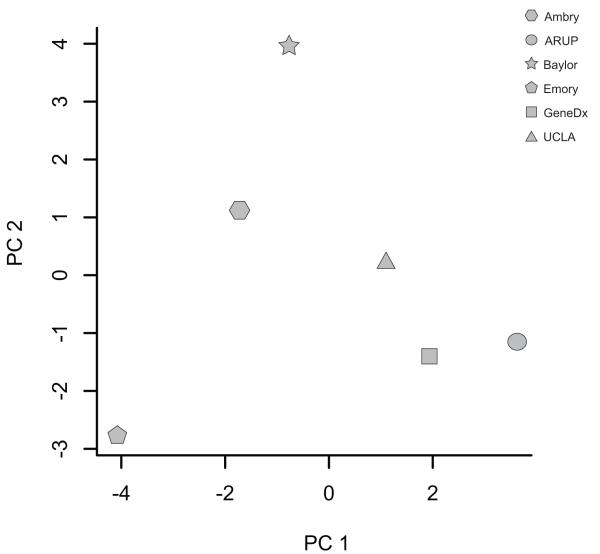
Our analysis of the practices and policies of ES service providers reveals that overall their approaches toward informed consent, data sharing, and results return vary widely (Fig 2). Accordingly, the potential merits and disadvantages of each, and more importantly, the balance between the two differ among ES providers. Such variation is perhaps not surprising at the outset of the integration of new technologies into clinical practice. However, this variation has practical implications for healthcare providers and institutions evaluating, comparing, and ultimately deciding which ES provider to use. The impact is even greater as many institutions look toward the practices of ES providers for guidance as they develop their own policies about the use ES.
Figure 2. Principal component (PC) analysis summarizing variation in core elements among six clinical ES providers.
Forty-four percent of the variance in the scores of core elements assessed for informed consent, data sharing and return of results is explained by PC1, and thirty-two percent of the variance is explained by PC2.
Perhaps the most substantial barrier to the effective use of ES for clinical service is how to maximize the possible benefits and minimize potential harms to patients. The most powerful characteristic of ES is its potential to identify virtually all pathogenic coding variants in an individual or so-called secondary results, in addition to the cause of the phenotype that warranted ordering the test (i.e. the primary result). Yet most healthcare providers and patients have a limited appreciation of the scope of secondary results that might be identified, whether secondary results should be returned, and if so, how to do it effectively and conveniently. The results management policies of the ES service providers do not fully address the challenges of offering secondary results for return, and vary considerably in their approach to allowing individuals access to these results. Five of the six providers offer secondary results, but differ substantially in which results are offered, whether patient preferences are considered, and what options exist to reanalyze ES data for secondary results.
We think that healthcare providers and medical institutions have an ethical and fiduciary obligation to offer patients access to clinically important and valid secondary iES results. Some individuals will opt to receive some or all such results while others will choose to not learn about secondary results, particularly if they are mainly concerned and focused on the diagnosis and treatment of a primary condition. Nevertheless, many clinicians and ethicists have expressed concern that even offering patients this option will unnecessarily exacerbate anxiety and stress, and create opportunities for misunderstanding [Christenhusz et al., 2012; Sharp, 2011]. We, and others have agreed, that withholding information about the potential to identify secondary results or the option to receive such results violates the autonomy of the individual and the requirements of fully informed consent [Townsend et al., 2012]. More importantly, the extent to which individuals want secondary results, whether potential benefits and harms of offering secondary results are realized, and the long-term outcome of providing secondary results needs to be empirically tested to enable development of evidence-based policies.
The consensus of the field on what secondary iES results should be offered in clinical care, or even in research, is elusive and evolving [Biesecker et al., 2012]. The American College of Medical Genetics and Genomics is developing guidelines on a minimum set of results that must be offered for return, although it is unclear if this will be useful in all circumstances and how effectively it could be implemented by ES providers, caregivers, or explained to patients [Levenson, 2012]. A recent study of a convenience sample of “experts” showed some agreement on conditions for which results should be returned, but only took into consideration the context of adults vs. children, and did not consider which individual variants should be returned [Green et al., 2012]. More importantly, as the life circumstances and clinical condition/risks of an individual change over time, the importance of some iES results may change. For example, if a patient develops a family history of cancer or a physician is choosing between drug regimens, a different set of priorities about iES secondary results may be operating than at the time of initial sequencing. If iES results exist, it is hard to contemplate a sound ethical argument in which iES data or results would not be made available for re-analysis for the benefit of the patient. For all of these reasons, ES service providers will need to provide options for reanalysis and results return.
As consensus on return of secondary iES results builds, an even greater challenge might be how to implement a practical, cost-effective system to update and offer secondary results that respects patient preferences. This begins by refocusing return of iES results from a model based on ordering a test for a discrete clinical purpose to one in which iES results are viewed as a dynamic, sustained resource of information that is available to an individual not only at a single point in time, but over many years and even possibly a lifetime.
We think that iES results management can be best optimized using a self-guided approach that enables individuals to self-select among offered results in a convenient, confidential, and personalized context that is responsive to their value system. This approach respects autonomy, allows individuals to maximize potential benefits of genomic information, minimize potential harms, and preserves their right to an open future to the extent they desire or think is appropriate [Yu et al., 2013]. Given the necessary features and potential challenges that need to be overcome in managing iES results, we think an interactive web-based information system for management of iES results is needed, if not a necessity. Such a system would allow individuals to set their preferences about which results they do and do not want to receive according to their own values and assessments of benefits and risk and these preferences could be modified at their discretion. It would also facilitate the use of accurate standardized educational information about disease-related phenotypes allowing genetics professionals to focus on interpreting results and considering the impact of results on medical management.
Our results suggest some ambivalence and confusion on the part of ES providers about the disposition of ES data and what information or options they will provide to patients about data sharing. If patient access to secondary results over time is going to be maximized, ES providers will need to either share the annotated variants with or develop tools for reanalysis for clinicians and/or institutions [Bick and Dimmock, 2011]. Furthermore, it is essential that ES providers’ plans of sharing individual-level data / results be disclosed to patients, including a description of the possible risks of identification and violation of privacy [Presidential Commission for the Study of Bioethical Issues, 2012]. ES providers should also include an opt-out clause for broad data sharing of individual-level data / results to protect patient autonomy. This would parallel current practices in research settings, and would maximize patient autonomy about the use of their data for purely research purposes [Hull et al., 2008; Ludman et al., 2010; Oliver et al., 2012].
There are several important limitations to our study. The core elements that we identified, and the scoring system that we developed are based on our experience and reflect what we consider to be the key priorities and issues in ES. The scale is not validated, and there may be other elements that need to be taken into consideration in evaluating these services, including expertise, experience with ES, and cost. Our scoring was based on conversations with representatives from the ES service providers, without access to written policies, and therefore may reflect the limitations of individual knowledge, the policies themselves or the absence of written policies. Ideally, companies should develop such written policies and make them publically available. We included only the six laboratories that currently offer ES for clinical service. We excluded several laboratories and institutions that only provide clinical ES for in-house providers, and companies that offer direct-to-consumer ES or WGS outside the context of clinical care. We also did not review the informed consent documents or policies of the institutions that are conducting clinical ES in the context of research studies (e.g., the Clinical Sequencing Exploratory Research studies funded by NHGRI [Mjoseth, 2012; NHGRI, 2012]). A broader review could provide a clearer picture of current practices, but will not assist providers, institutions or policy makers in evaluating the ES options available to them.
In summary, the informed consent, data sharing, and results management policies of ES service providers vary widely and are likely to evolve rapidly as technology changes, attitudes toward the use and management of secondary results evolves, and experience with the use of ES for clinical care deepens. Clearly this affords healthcare providers / ES services with options that they may or may not consider advantageous depending on diagnostic intent and institutional context. This is likely of benefit only insofar as healthcare providers and institutions have access to expert input to weigh these options. Many do not. For this reason, there may be benefit to some standardization for best practices among ES providers. Our analysis is, at a minimum, an attempt to further this discussion.
ACKNOWLEDGMENTS
We thank Heather Mefford for review and thoughtful discussion. Our work was supported in part by the National Institutes of Health / National Human Genome Research Institute (1U54HG006493, 5R00HG004316 and 5R01HG006618), the Life Sciences Discovery Fund (2065508 and 0905001), and the Washington Research Foundation.
APPENDIX TABLES
Appendix Table I.
Core Elements for Informed Consent
| Core Elements | Score | Explanation of Score | |
|---|---|---|---|
| Explanation of ES | 0 | No explanation of ES/purpose of test | |
| 1 | Purpose defined as targeted test* | *i.e. solely for diagnosis of specific clinical concern |
|
| 2 | Targeted test, and provides explanation of what a diagnosis may or may not provide* |
*Provides explanation of what a diagnosis may or may not provide= may or may not predict prognosis or influence treatment |
|
| 3 | Targeted test with explanation of what a diagnosis may or may not provide, and as a test that can identify service lab determined medically actionable results |
||
| 4 | Targeted test with explanation of what a diagnosis may or may not provide, and as a test that can identify some secondary results that may be of patient interest |
||
| 5 | Resource of information that has the potential to provide health and life-planning benefits |
||
| Potential risks | 0 | No mention of potential risk | |
| 1 | Mentions ES has risks | ||
| 2 | Describes the potential psychological risks* or privacy risks* associated with ES |
*Psychological risks= e.g. learning genetic information about self and/or family members, potential nonpaternity *Privacy risks= e.g. computer security breaches arising from maintaining data in an electronic format); others potentially having access to results potential consequences of such access |
|
| 3 | Describes the potential psychological and privacy risks associated with ES |
||
| 4 | Describes potential psychological or privacy risks and how such risks would be handled* |
*How such risks would be handled= e.g. genetic counseling; the safeguards in place to handle/minimize these risks: GINA |
|
| 5 | Describes potential psychological and privacy risks associated with ES, and how such risks would be handled |
||
| Discussion of secondary results |
0 | No mention of secondary results | |
| 1 | Mentions secondary results | ||
| 2 | Incomplete explanation of secondary results, and incomplete explanation of IF management |
||
| 3 | Complete explanation of secondary results*, and incomplete explanation of IF management |
*Complete explanation of secondary results= the full range of secondary results along with benefits/risks of why one may want or not want to learn about secondary results |
|
| 4 | Incomplete explanation of secondary results, and complete explanation of secondary results management* |
*Complete explanation of secondary results management= clear what secondary results are/are not returned |
|
| 5 | Complete explanation of secondary results, and complete explanation of secondary results management |
||
| Patient preferences related to secondary results |
0 | No mention of patient preferences | |
| 1 | Mentions patient preferences | ||
| 2 | Allows for patient preferences for some service lab determined secondary results |
||
| 3 | Allows for patient preferences for all service lab determined secondary results |
||
| Result types returned |
0 | No mention of result types* returned | *Results types= pathogenic, variant of unknown significance, nonpathogenic |
| 1 | Mention of result types, and no explanation of what result types are returned |
||
| 2 | Incomplete explanation of result types returned, and no explanation of temporal nature of results* |
*Temporal nature of results= how result classification/meaning may change in the future |
|
| 3 | Incomplete explanation of result types returned, and explanation of temporal nature of results |
||
| 4 | Complete explanation of result types* returned, and no explanation of temporal nature of results |
*Complete explanation of results types= discusses all 3 categories |
|
| 5 | Complete explanation of result types returned, and explanation of temporal nature of results |
||
| Limitations of ES | 0 | No limitations of ES addressed | |
| 1 | Mentions ES has limitations | ||
| 2 | Incomplete explanation of technical and analytical limitations of ES addressed |
||
| 3 | Complete explanation of technical and analytical limitation of ES* so that expectations of what ES can/cannot do is clear, as well as explaining the implications of the absence of finding |
*Complete explanation of technical limitations= e.g. entire exome not targeted; even what is targeted, not all genes are adequately covered to allow for confident variant calls; even if adequately covered, certain parts of genes (exon 1, GC rich regions), are problematic; not all mutations are in the exome; not all types of mutations, even if within an exon, are detected by ES (e.g. triplet repeat expansions, large deletions); analytical limitations= filtering approach (assumed mode of inheritance, reduced penetrance, genetic heterogeneity, variant identified in gene whose function is not known) |
|
| Re-analysis of data | 0 | No mention | |
| 1 | Does not offer re-analysis of any exome data | ||
| 2 | Offers re-analysis, but no explanation offered | ||
| 3 | Offers re-analysis/updates related only to primary result and provides updated information to ordering provider |
||
| 4 | Offers re-analysis/updates related to any result, and provides updated information to ordering provider at any time |
||
| 5 | Offers re-analysis related to any result, and provides patient access to updated information at any time |
||
| Data disposition policy |
0 | No mention of data disposition policy | |
| 1 | Mention of data disposition policy | ||
| 2 | Incomplete explanation of data disposition policy, and no mention of safeguards in place to ensure security of data |
||
| 3 | Complete explanation of data disposition policy*, no mention of safeguards in place to ensure security of data |
*Complete explanation of data disposition policy= length of storage, what/where data are stored, storage system is HIPAA-compliant, how data is destroyed after retention period ends |
|
| 4 | Incomplete explanation of data disposition policy, and mentions safeguards in place to ensure security of data |
||
| 5 | Complete explanation of data disposition policy, and safeguards in place to ensure security of data storage |
||
| Discussion of sharing de-identified data |
0 | No mention of data sharing | |
| 1 | Mention of data sharing, where/with whom de-identified data would be shared, and no opt-out option |
||
| 2 | Mention of data sharing, no mention of where/with whom de-identified data would be shared, and opt out option |
||
| 3 | Mention of data sharing, where/with whom de-identified data would be shared, and opt-out option |
||
| Informed consent and noted discrepancies |
0 | ≥3 discrepancies noted between informed consent document and policies (data sharing, results management) |
|
| 1 | 2 discrepancies noted between informed consent document and policies (data sharing, results management) |
||
| 2 | 1 discrepancy noted between informed consent document and policies (data sharing, results management) |
||
| 3 | 0 discrepancies noted between informed consent document and policies (data sharing, results management) |
||
ES: exome sequencing; GINA: Genetic Information Nondiscrimination Act of 2008; HIPAA: Health Insurance Portability and Accountability Act of 1996
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06)
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12)
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012)
Emory EmExome Informed Consent Document (Rev. 6/2012)
GeneDx XomeDx Informed Consent Document (GeneDx 07/12)
UCLA Informed Consent for Postnatal Clinical Exome Sequencing
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06)
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12)
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012)
Emory EmExome Informed Consent Document (Rev. 6/2012)
GeneDx XomeDx Informed Consent Document (GeneDx 07/12)
UCLA Informed Consent for Postnatal Clinical Exome Sequencing
Appendix Table II.
Core Elements for Data Sharing
| Core Elements | Score | Explanation of Score | |
|---|---|---|---|
| Deposition into public databases |
0 | No data deposition policy | |
| 1 | Policy is to not deposit data into databases | ||
| 2 | Policy is to deposit data into databases, and no other details provided |
||
| 3 | Policy specifies any 1 of the following: [type of data*, volume of data*, with or without phenotypic information, which databases*], opt-in/out option for patients |
*Type of data= sequence vs. variant level *Volume of data= entire vs. partial sequence; all vs. some variants *Databases= by name or category (e.g. restricted vs. private) |
|
| 4 | Policy specifies any 2 of the following: [type of data*, volume of data*, with or without phenotypic information, which databases*], opt-in/out option for patients |
||
| 5 | Policy specifies any 3 of the following: [type of data*, volume of data*, with or without phenotypic information, which databases*], opt-in/out option for patients |
||
| 6 | Policy specifies all 4 of the following: [type of data*, volume of data*, with or without phenotypic information, which databases*], opt-in/out option for patients |
||
| Education and research purposes |
0 | No policy regarding data use for educational/research purposes |
|
| 1 | Data used for educational/research purposes, and no opt out option |
||
| 2 | Data used for educational/research purposes, and opt- out option |
||
| Data sharing options for ordering provider/ institution |
0 | No sharing with provider/institution at all | |
| 1 | Sharing of primary result only | ||
| 2 | Sharing of all primary and secondary results as determined by service lab |
||
| 3 | Sharing fully annotated variant list | ||
| 4 | Sharing .vcf files | ||
| 5 | Sharing .vcf and BAM files | ||
vcf: variant call format; BAM: compressed binary version of a Sequence Alignment/Map format
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06), correspondence with lab representative
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12), correspondence with lab representative
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012), correspondence with lab representative
Emory EmExome Informed Consent Document (Rev. 6/2012), correspondence with lab representative
GeneDxXomeDx Informed Consent Document (GeneDx 07/12), correspondence with lab representative
UCLA Informed Consent for Postnatal Clinical Exome Sequencing, correspondence with lab representative
Appendix Table III.
Core Elements for Results Management Policy
| Core Elements | Score | Explanation of Score | |
|---|---|---|---|
| Range of results returned | 0 | No result report offered for return | |
| 1 | Only primary result returned | ||
| 2 | Primary result and medically actionable results as determined by service lab routinely returned |
||
| 3 | Primary result and medically actionable results as determined by service lab guided by patient preference returned |
||
| 4 | Primary result and restricted categories of secondary results as determined by service lab guided by patient preference returned |
||
| 5 | Primary result and non-restricted categories of secondary results guided by patient preference returned |
||
| Patients related to secondary results, and time constraints |
0 | Does not allow for patient preferences | |
| 1 | Allows for patient preferences for some secondary results as determined by service lab, and one time to decide |
||
| 2 | Allows for patient preferences for all secondary results as determined by service lab, and one time to decide |
||
| 3 | Allows for patient preferences for some secondary results as determined by service lab, and period of time to decide |
||
| 4 | Allows for patient preferences for all secondary results as determined by service lab, and period of time to decide |
||
| 5 | Allows for patient preferences for all secondary results (beyond that offered by service lab), and ability to change preferences over time |
||
| Updating of result interpretation |
0 | No updating of results offered | |
| 1 | Offers to update results, but no details provided | ||
| 2 | Recommends/requires provider request if results interpretation has changed |
||
| 3 | Contacts provider if new information related only to primary finding is discovered |
||
| 4 | Contacts provider if new information related to any result reported is discovered |
||
| 5 | Provides web interface for patients/providers to see up to date information about any given result |
||
| Patients accessing their results |
0 | Patients cannot access any result | |
| 1 | Patients can only access primary result through their ordering provider/EMR |
||
| 2 | Patients can access primary and company determined medically actionable findings through ordering provider/EMR |
||
| 3 | Patients can access primary and restricted range of secondary results through their ordering provider and/or EMR |
||
| 4 | Patients can access all results through ordering provider and/or EMR |
||
| 5 | Patients given independent ability to access their results at their discretion and over time |
||
| Informational/counseling resources available to patients |
0 | No informational/counseling resources available to patients |
|
| 1 | Results report is the only resource provided to patients through an intermediary (i.e. clinician) |
||
| 2 | Results report and service lab provided online patient focused brochures/information sheets on ES |
||
| 3 | Results report and service lab recommends in pre- and post-test genetic counseling |
||
| 4 | Results report, service lab recommends pre- and post-test genetic counseling, and provides online patient focused resources |
||
| 5 | Results report, service lab offers telemedicine pre- and post-test genetic counseling and online educational resources |
||
| Report construction (results, interpretation) |
0 | No report returned to provider | |
| 1 | Results section and interpretation of report does not follow any recommendations as recommended by ACMG (ACMG recommendations for standards for interpretation and reporting of sequence variation)* |
*II. Reporting of Sequence Variations: Recommendations 1-6; A. Use of standardized terminology and established databases for reporting sequence variants: Recommendations 1-2, B. Limitations of sequence-based testing should be reported: Recommendations 1-5 |
|
| 2 | Results section and interpretation of report follows some recommendations as recommended by ACMG |
||
| 3 | Results section and interpretation of report follows all* recommendations as recommended by ACMG |
*Results section follows all ACMG recommendations= all recommendations in II (1-6), A (1-2), B (1-6) |
|
| Report construction (methods) |
0 | No methods section included in results report | |
| 1 | Methods section has incomplete description of technical aspects |
||
| 2 | Methods section has incomplete description of technical aspects, and lab offers to provide genes/regions of exome with poor to no coverage |
||
| 3 | Complete explanation of the technical aspects*, lab offers to provide gene/regions of exome with poor to no coverage |
*Complete explanation of technical aspects= exome capture kit used, sequencing platform used, variant calling/annotating/filtering, report-specific coverage/quality metrics |
|
EMR: electronic medical record; ES: exome sequencing
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06), correspondence with lab representative
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12), correspondence with lab representative
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012), correspondence with lab representative
Emory EmExome Informed Consent Document (Rev. 6/2012), correspondence with lab representative
GeneDxXomeDx Informed Consent Document (GeneDx 07/12), correspondence with lab representative
UCLA Informed Consent for Postnatal Clinical Exome Sequencing, correspondence with lab representative
Appendix Table IV.
Submitting samples and genetic counseling
| Who can submit a sample? | Is genetic counseling required? | |
|---|---|---|
| Ambry | Requires “Clinician (geneticist/genetic counselor)” signature | “Post-test counseling and interpretation: I understand the importance of seeking genetic counseling post-testing due to the nuances. It is HIGHLY recommended that you seek genetic counseling to specifically address the risks related to any findings from this test.” |
| ARUP | Requires “Ordering health care provider” signature | “Genetic counseling is recommended prior to, as well as following, this complex testing.” |
| Baylor | Requires “Physician/Counselor” signature | “Due to the complex nature of the WES testing it is recommended that families seek genetic counseling in conjunction with testing.” |
| Emory | Requires “Physician/Counselor/Clinician” signature | “Given the complexity of the EmExome, genetic counseling by a trained medical geneticist or genetic counselor is required prior to and after undergoing this testing.” |
| GeneDx | Requires “Physician/Counselor” signature | “Because of the complexity of the test, and the type of information that might result from the test, it is important that you have genetic counseling both before and after the test is done.” |
| UCLA | Requires “Physician/Genetic Counselor” signature | “We recommend counseling with a clinical geneticist or genetic counselor.” |
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06)
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12)
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012)
Emory EmExome Informed Consent Document (Rev. 6/2012)
GeneDxXomeDx Informed Consent Document (GeneDx 07/12)
UCLA Informed Consent for Postnatal Clinical Exome Sequencing
Appendix Table V.
Informed consent document and noted discrepancies
| Stated in informed consent document | Policy in which discrepancy found | ||
|---|---|---|---|
| Ambry | “Only those variants identified as “candidate mutations” are reviewed by a medical director and can be placed into a patient’s chart….I understand that all candidate variants* will be provided.” |
Data sharing: | The candidate variant list is only returned when requested, and when the patient and provide sign a separate consent which states that “any information gleaned is strictly for research purposes and shall not be used for clinical decision-making purposes and any additional results shall not be made available to the patient.” |
| ARUP | “Deidentified information from individuals undergoing exome sequencing will be provided to national or international databases to enhance knowledge of the human genome.” |
Data sharing: | Current policy is to not do so. |
| Emory | Reporting for children (<18 years of age): “changes in genes that have been reported to be diagnostic or possibly diagnostic.” NOTE: no secondary results are offered as options for minors |
Results management: |
Pharmacogenetics variants are offered for return to all patients, regardless of patient age. |
| Categories of secondary results offered for return: “Carrier status for autosomal recessive conditions, pharmacogenetic variants, adult onset cancer disorders, adult onset dementia disorders, other adult onset disorders” NOTE: no mention of term “medically actionable |
Results management: |
Routinely report “medically actionable” secondary results. |
|
| GeneDx | “GeneDx will not release raw data to patients or physicians.” |
Data sharing: | Offer to return .vcf and BAM files to requesting providers |
| “Variations in disease-associated genes that are not related to the disease under investigation in the patient/family will not be reported.” |
Results management: |
May report “medically actionable” secondary results. |
|
| UCLA | “Clinically significant mutations known to be associated with a genetic disease that the symptoms are not evident at this time and may or may not develop in the future (such as cancer risk or Alzheimer’s disease risk) will not be reported here.” NOTE: no mention of term “medically actionable |
Results management: |
Routinely report “medically actionable” secondary results. |
candidate variants defined as “any gene alteration which results from bioinformatics filtering based on inheritance pattern and, upon medical director review, cannot be ruled out as possibly associated with the patient’s clinical phenotype under evaluation.
Source:
Ambry Exome Patient Consent Form (F0912-02-011p-PTM-06), correspondence with lab representative
ARUP Informed Consent for Exome Sequencing with Symptom-Guided Analysis (ARUP Rev.1 5/12), correspondence with lab representative
Baylor Whole Exome Sequencing Requisition - Information and Consent for Testing (Last Updated: 9/24/2012), correspondence with lab representative
Emory EmExome Informed Consent Document (Rev. 6/2012), correspondence with lab representative
GeneDxXomeDx Informed Consent Document (GeneDx 07/12), correspondence with lab representative
UCLA Informed Consent for Postnatal Clinical Exome Sequencing, correspondence with lab representative
REFERENCES
- Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3(87):87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, Mane S, Nickerson DA. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A(7):1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick D, Dimmock D. Whole exome and whole genome sequencing. Curr Opin Pediatr. 2011;23(6):594–600. doi: 10.1097/MOP.0b013e32834b20ec. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: are we ready? Nat Rev Genet. 2012;13(11):818–24. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz GM, Devriendt K, Vermeesch J, Dierickx K. Why genomics shouldn’t get too personal: in favor of filters: Re: invited comment by Holly K. Tabor et al. in American Journal of Medical Genetics Part A Volume 155. Am J Med Genet A. 2012;158A(10):2641–2. doi: 10.1002/ajmg.a.35547. author reply 2643-2644. [DOI] [PubMed] [Google Scholar]
- Flesch R. A new readability yardstick. J Appl Psychol. 1948;32(3):221–233. doi: 10.1037/h0057532. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Berry GT, Biesecker LG, Dimmock DP, Evans JP, Grody WW, Hegde MR, Kalia S, Korf BR, Krantz I, McGuire AL, Miller DT, Murray MF, Nussbaum RL, Plon SE, Rehm HL, Jacob HJ. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14(4):405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull SC, Sharp RR, Botkin JR, Brown M, Hughes M, Sugarman J, Schwinn D, Sankar P, Bolcic-Jankovic D, Clarridge BR, Wilfond BS. Patients’ views on identifiability of samples and informed consent for genetic research. Am J Bioeth. 2008;8(10):62–70. doi: 10.1080/15265160802478404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson D. The tricky matter of secondary genomic findings: ACMG plans to issue recommendations. Am J Med Genet A. 2012;158A(7):ix–x. doi: 10.1002/ajmg.a.35521. [DOI] [PubMed] [Google Scholar]
- Ludman EJ, Fullerton SM, Spangler L, Trinidad SB, Fujii MM, Jarvik GP, Larson EB, Burke W. Glad you asked: participants’ opinions of re-consent for dbGap data submission. J Empir Res Hum Res Ethics. 2010;5(3):9–16. doi: 10.1525/jer.2010.5.3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjoseth J. NHGRI funds return of results studies, forms expert consortum. 2012. [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010a;42(9):790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010b;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHGRI . Clinical sequencing exploratory research. 2012. [Google Scholar]
- Oliver JM, Slashinski MJ, Wang T, Kelly PA, Hilsenbeck SG, McGuire AL. Balancing the risks and benefits of genomic data sharing: genome research participants’ perspectives. Public Health Genomics. 2012;15(2):106–114. doi: 10.1159/000334718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presidential Commission for the Study of Bioethical Issues . Privacy and Progress in Whole Genome Sequencing. 2012. [Google Scholar]
- Sharp RR. Downsizing genomic medicine: approaching the ethical complexity of whole-genome sequencing by starting small. Genet Med. 2011;13(3):191–194. doi: 10.1097/GIM.0b013e31820f603f. [DOI] [PubMed] [Google Scholar]
- Tabor HK, Berkman BE, Hull SC, Bamshad MJ. Genomics really gets personal: how exome and whole genome sequencing challenge the ethical framework of human genetics research. Am J Med Genet A. 2011;155A(12):2916–2924. doi: 10.1002/ajmg.a.34357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend A, Adam S, Birch PH, Lohn Z, Rousseau F, Friedman JM. “I want to know what’s in Pandora’s box”: Comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet A. 2012 doi: 10.1002/ajmg.a.35554. [DOI] [PubMed] [Google Scholar]
- Welch JS, Westervelt P, Ding L, Larson DE, Klco JM, Kulkarni S, Wallis J, Chen K, Payton JE, Fulton RS, Veizer J, Schmidt H, Vickery TL, Heath S, Watson MA, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Ley TJ, Wilson RK. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305(15):1577–1584. doi: 10.1001/jama.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]