Abstract
Introduction
Osteogenesis imperfecta (OI), Ehlers-Danlos syndrome (EDS), and osteopetrosis (OPT)are collectively common inherited skeletal diseases. Evaluation of subjects with these conditions often includes molecular testing which has important counseling, therapeutic and sometimes legal implications. Since several different genes have been implicated in these conditions, Sanger sequencing of each gene can be a prohibitively expensive and time consuming way to reach a molecular diagnosis.
Methods
In order to circumvent these problems, we have designed and tested a NGS platform that would allow simultaneous sequencing on a single diagnostic platform of different genes implicated in OI, OPT, EDS, and other inherited conditions leading to low or high bone mineral density. We used a liquid-phase probe library that captures 602 exons (~100 kb) of 34 selected genes and have applied it to test clinical samples from patients with bone disorders.
Results
NGS of the captured exons by Illumina HiSeq2000 resulted in an average coverage of over 900X. The platform was successfully validated by identifying mutations in 6 patients with known mutations. Moreover, in 4 patients with OI or OPT without a prior molecular diagnosis, the assay was able to detect the causative mutations.
Conclusions
In conclusion, our NGS panel provides a fast and accurate method to arrive at a molecular diagnosis in most patients with inherited high or low bone mineral density disorders.
Introduction
Skeletal dysplasias are heritable disorders that can affect the growth, development, and homeostasis of bone and cartilage. Although individual forms of skeletal dysplasias are relatively rare, collectively, skeletal dysplasias have an incidence of 1:5,000 births [1]. Due to the large number and overlapping phenotypic features in these disorders, arriving at a specific molecular diagnosis in those affected can be a challenging process[1, 2]. A recent study concerning the nosology of genetic skeletal disorders lists a total of 226 unique genes that are implicated in 456 well-characterized skeletal dysplasias grouped in 40 distinct groups[3].
An accurate molecular diagnosis can provide the basis for counseling regarding prognosis and reproductive options. An accurate genetic diagnosis has been shown to provide psychological benefits to patients and their families[4]. This is especially relevant in the prenatal context where diagnostic modalities for definitive diagnoses are even more limited. Postnatally, an accurate diagnosis may help initiate appropriate therapeutic interventions in some conditions, like bisphosphonate therapy in osteogenesis imperfecta (OI) and bone-marrow transplantation for some forms of osteopetrosis (OPT). Hence, improvements in the accuracy, speed, and cost of the diagnosis will have significant impact on clinical care of patients. Given the extensive phenotypic and genetic heterogeneity of many forms of skeletal dysplasias, reaching an appropriate diagnosis on a molecular basis often involves Sanger sequencing of multiple genes in a sequential manner which, as expected, is time-consuming, costly, and challenging. Recently, van Dijk et al., reported best practice guidelines for the laboratory diagnosis for OI, but this was mostly based on Sanger sequencing of relevant genes [5]. Exponential growth in the fields of high-throughput capture and next-generation sequencing has made NGS one of the most promising techniques for molecular diagnoses [6–8]. While whole exome sequencing allows for simultaneous interrogation of thousands of genes, the average coverage achieved is below what can be achieved with targeted sequencing, and there are thus more exons with coverage too low for accurate base calling. For example, at 30X average coverage (lower than what is typically done in clinical exome laboratories), less than 80% of disease-causing mutation sites from the Human Genome Mutation Database showed genotype sensitivity (defined as >10× coverage and a Phred software base-calling quality score >30)[9]. Hence, targeted NGS panels which have higher coverage for a select group of genes continue to be useful for diagnosis when a specific group of conditions is suspected.
In order to facilitate sequencing of multiple genes causing some of the common forms of heritable skeletal disorders including genetic forms of low- and high-bone mineral density, we have designed and validated a novel testing panel on a single diagnostic platform. The panel includes 21 genes implicated in low bone mineral density (LBMD)diseases, including 9 genes for OI, and 14 genes implicated in high bone mineral density (HBMD)diseases including various forms of OPT. TNFRSF11A is included in both categories since different mutations can either give osteopetrosis, expansile osteolysis or Paget disease of the bone.
Patients and Methods
Human Subjects
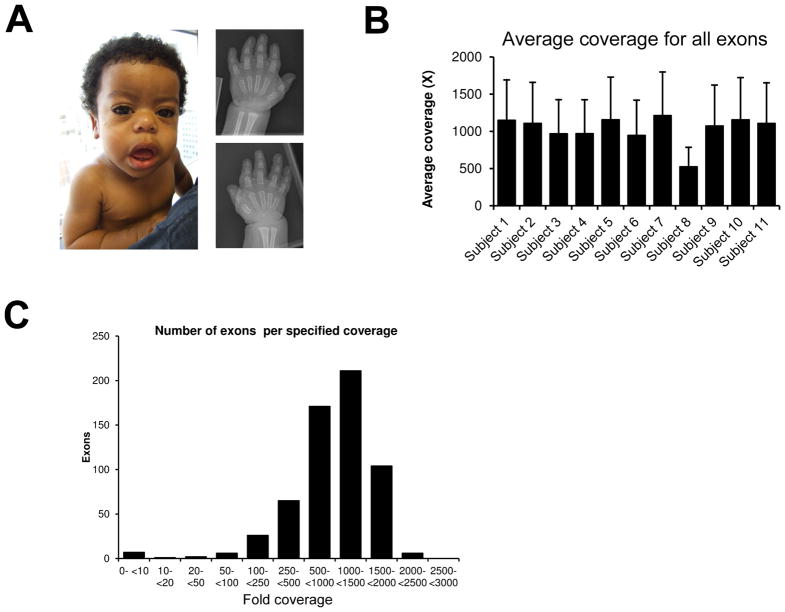
Ten subjects were enrolled in the study. The study procedures were approved by the Institutional Review Board of the involved institutions. Informed consent was obtained from each subject. Subjects 1 through 4 had recessive forms of OI and have been previously described [10–13]. Subjects 5, 7, 8 and 9 had a diagnosis of OI, and subject 10 had osteopetrosis Subject 11 with dysosteosclerosis was described in detail previously [14–16]. Subject 6 has geleophysic dysplasia based on the presence of classical clinical findings which we will describe here (see fig. 1A). He had short stature, mild coarsening of facial features, a prominent forehead, a depressed nasal bridge, broad base nose, long philtrum, midface hypoplasia, prominent lips, a short neck, a chest with muscular appearance, shortened long limbs, short phalanges and metacarpals, limitation of elbow extension, and brachydactyly (Fig. 1A). He also presented with inguinal hernias, thickened skin, a narrow larynx with tracheomalacia, nystagmus, and mild developmental delay. Echocardiogram showed mitral valve leaflet thickening, short chordae and mild concentric left ventricular hypertrophy.
Figure 1.
A) Subject 6 with a diagnosis of geleophysic dysplasia. Photograph showing classical clinical features and hand radiograph demonstrating generalized shortening of phalanges and metacarpals. B) The average coverage per patient for all exons was above 400X. C) Distribution of coverage per exon by the NGS panel.
The mutations had been described in subjects 1 through 4 as reported previously [10–13]. The COL1A1 mutation in subject 5 was identified in a separate laboratory. Subject 6 had Sanger sequencing to exclude ADAMTSL2 mutations [17], then exome sequencing was performed and identified a mutation in FBN1 before that mutation was published to be associated with geleophysic dysplasia [18]. These subjects served as positive controls and were used to validate the accuracy of the panel testing. Subjects 7, 8 and 9 did not have a known molecular diagnosis for their OI as was the case with subject 10 with OPT. These samples were assayed as test samples to assess the sensitivity and accuracy of our platform.
Process to select genes
The bone mineral density diagnostic platform was divided into two panels based on the bone mineral density: the LBMD panel and the HBMD panel. We included forms of skeletal dysplasia commonly encountered in clinical practice that present with alterations in bone mineral density along with recurrent fractures. Only genes that had been proven to cause disease in these disorders were included in the panel. Genes such as COL1A1, COL1A2, CRTAP, FKBP10, LEPRE1, PLOD2, PPIB, SERPINF1, SERPINH1, and SP7 which are all implicated as the cause of various forms of OI were included in the LBMD panel [3, 5, 10, 19]. Genes associated with generalized or localized high bone density disorders such as CA2, CLCN7, OSTM1, and TCIRG1 were included in the high bone mineral density panel [3, 20]. Genes leading to Ehlers-Danlos syndrome (EDS) and Paget disease were also included. Some of the genes have pseudogenes or paralogs, and thus the homology of the exons with other sequences could make next-generation sequencing problematic, both at the hybridization step and alignment step. This was the case for LRP5 (13 paralogs, 23 coding exons), SERPINH1 (15 paralogs, 3 coding exons), and PLEKHM1 (5 paralogs, 11 coding exons). These genes were excluded from the hybridization capture library, and they are rarely involved in the diseases targeted by our panel. Thus, the low bone mineral density panel includes 21 genes associated with low bone mineral density phenotypes (see Table 1 for the list and inheritance patterns for the associated disorders). The high bone mineral density panel includes 14 genes associated with high bone mineral density phenotypes (see Table 2 for the list and inheritance patterns for the associated disorders).
Table 1.
Low bone mineral density panel genes and conditions
| Gene | Disease |
|---|---|
| COL1A1 | Osteogenesis imperfecta, types I, II, III and IV (AD); Ehlers-Danlos syndrome, type VIIA (AD) |
| COL1A2 | Osteogenesis imperfecta, types II, III and IV (AD); Ehlers-Danlos syndrome, type VIIB (AD) and cardiac valvular form (AR) |
| CRTAP | Osteogenesis imperfecta, types II, III, IV and VII (AR) |
| LEPRE1 | Osteogenesis imperfecta, types II, III and VIII (AR) |
| PPIB | Osteogenesis imperfecta, types II, III, IV and IX (AR forms) |
| FKBP10 | Osteogenesis imperfecta types III and VI (AR forms); Bruck syndrome (AR) |
| SERPINF1 | Osteogenesis imperfecta, types III and VI (AR) |
| SP7 | Osteogenesis imperfecta, types IV and XI (AR) |
| PLOD2 | Bruck syndrome 2 (AR) |
| PLOD3 | Bone Fragility with Contractures, Arterial rupture and Deafness (AR) |
| COL3A1 | Ehlers-Danlos syndrome, type IV (AD) |
| COL5A1 | Ehlers-Danlos syndrome, types I and II (AD) |
| COL5A2 | Ehlers-Danlos syndrome, types I and II (AD) |
| B4GALT7 | Ehlers-Danlos syndrome, Progeroid Form (AR) |
| SLC39A13 | Ehlers-Danlos syndrome, Spondylocheirodysplastic form (AR) |
| ALPL | Hypophosphatasia (AR, AD), Odontohypophosphatasia (AD) |
| SLC34A1 | Hypophosphatemic nephrolithiasis/osteoporosis 1 (AD); Fanconirenotubular syndrome 2 (AR) |
| SLC9A3R1 | Hypophosphatemic nephrolithiasis/osteoporosis 2 (AD) |
| FBN1 | Marfan, MASS, Stiff skin, Shprintzen-Goldberg and Weill-Marchesani syndromes, Acromicric and Geleophysicdysplasias (all AD) |
| TNFRSF11A | Paget disease of bone (AD); Osteolysis, familial expansile (AD) |
| TNFRSF11B | Paget disease, juvenile (AR) |
OI types are classified according to the Sillence classification[26]. SERPINH1 can cause OI but needs to be added by Sanger sequencing. AD= autosomal dominant; AR= autosomal recessive.
Table 2.
High bone mineral density panel genes and conditions
| Gene | Disease |
|---|---|
| TCIRG1 | Osteopetrosis, autosomal recessive 1 (AR) |
| TNFSF11 | Osteopetrosis, autosomal recessive 2 (AR) |
| CA2 | Osteopetrosis, autosomal recessive 3, with renal tubular acidosis (AR) |
| CLCN7 | Osteopetrosis, autosomal dominant 2 (AD); Osteopetrosis, autosomal recessive 4 (AR) |
| OSTM1 | Osteopetrosis, autosomal recessive 5 (AR) |
| TNFRSF11A | Osteopetrosis, autosomal recessive 7 (AR) |
| ANKH | Craniometaphyseal dysplasia (AD form); Chondrocalcinosis 2 (AD) |
| CTSK | Pycnodysostosis (AR) |
| FAM123B | Osteopathiastriata with cranial sclerosis (X-linked dominant) |
| LEMD3 | Buschke-Ollendorff syndrome/ Osteopoikilosis (AD) |
| SOST | Van Buchem disease; Sclerosteosis (AR) |
| TGFB1 | Camurati-Engelmann disease (AD) |
| TYROBP | Nasu-Hakola disease (AR) |
| FAM20C | Raine syndrome (AR) |
AD= autosomal dominant; AR= autosomal recessive.
Targeted Next-Generation sequencing
A custom in-solution based capture library for the 34 selected genes(602 exons totaling almost 100 kb) was designed and used to enrich the target regions of genes implicated in numerous disorders with LBMD and HBMD. The experimental procedures followed the manufacturer’s recommendations (Roche NimbleGen Inc., Madison, WI, USA). Because of the very high read depth afforded by NGS, several different samples can be pooled in a single lane, or “flow cell” of an Illumina sequencer. The DNA fragments from each patient were tagged with bar coded oligonucleotides. Equimolar amounts of the libraries from 8 indexed samples were pooled and their sequences determined in one lane of a HiSeq2000 (Illumina Inc., San Diego, CA, USA) with single-end, 75 base read length (since bases beyond that are generally less accurately called). An internal identity control system and an external monitoring system were designed and incorporated in the custom library with the specific probes, and sequenced by NGS together with each sample. Raw data in BCL format were converted to qseq files before separating the data according to genetic barcodes (demultiplexing) using CASAVA v1.7 (Illumina Inc., San Diego, CA, USA). Demultiplexed data were further processed by NextGENe software (SoftGenetics, State College, PA, USA) for alignment to the human genome (hg19). An in-house bioinformatics pipeline was used for the variant annotation.
Results
Sequencing and analysis
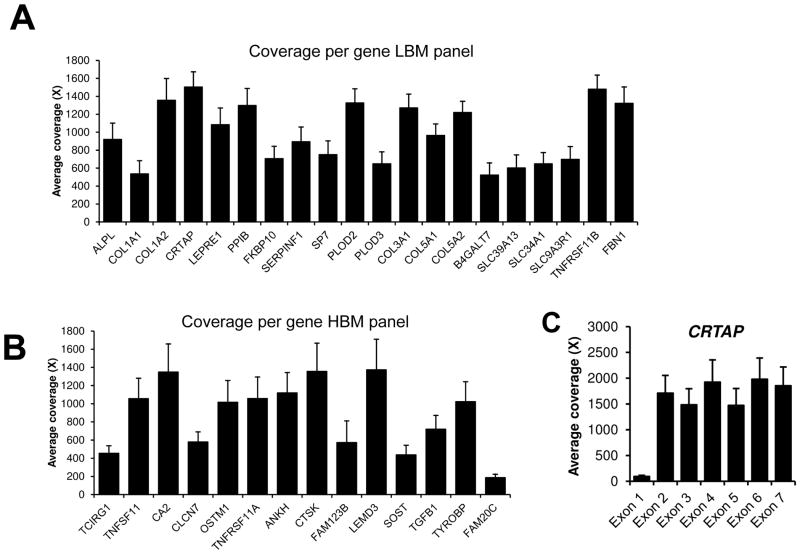
We performed coverage analysis considering the coding sequences and 20 bases inside the introns and obtained an average overall coverage per base 1032X (±191). The coverage was quite consistent between each patient (Figure 1B). Figure 1C demonstrates that the median coverage was in the 1000X–1500X range. The average coverage per gene is given in Figure 2 (here TNFRSF11A is only shown in the HBMD panel in order not to duplicate the data shown). Of the 34 genes, only 3 genes had an average coverage below 500X: FAM20C (187X), SOST (438X) and TCIRG1 (454X). Considering a minimum cutoff of 20X to call a heterozygous change, there were on average 16 exons (range 12–21) with targeted bases below this cutoff per patient. This would mean that to have a complete coverage of all genes, we need to perform 16 Sanger sequencing reactions per patient. The coverage across the exons of each gene can vary greatly, but is usually lower for the first exon. The low coverage in the first exon is illustrated by the exon by exon average coverage of CRTAP in figure 3C. For FAM20C, coverage was insufficient in most exons for most patients. This gene has 2 paralogs (FAM20A and FAM20B), and hybridization capture might not adequately distinguish between the three, contributing to the low coverage.
Figure 2.
A) Average coverage per gene for the low bone mineral density panel. B) Average coverage per gene for the high bone mineral density panel. C) Coverage per exon for some genes, such as CRTAP here, can vary greatly owing to high GC content of the first exon of some genes.
Variants identified
We validated the sensitivity and accuracy of our platform by assay of DNA samples from 6 patients with known mutations in SERPINF1, CRTAP, FKBP10, COL1A1, and FBN1(Table 3). The assay was able to detect mutations in all 6 samples, except for subject 2, where only one of the two heterozygous mutations was detected. The other mutation was located in exon 1, which has a 72% GC content and had very poor coverage, so Sanger sequencing was necessary to detect this mutation.
Table 3.
Panel validation by identifying known mutations
| Subject and reference | Phenotype | Gene | Genbank mRNA and CCDS ID | Mutation (cDNA) | Mutation (protein) | Zygosity of mutation |
|---|---|---|---|---|---|---|
| 1 [10] | Recessive OI | SERPINF1 | NM_002615.5, CCDS11012 | c.1163_1166dupACTA | p.His389Glnfs*4 | Homozygous |
| 2 [11] | Recessive OI | CRTAP | NM_006371.4, CCDS2657 | c.822_826delAATACinsT and c.278_293dupGCCCG | p.Lys274Asnfs*11 and p.Glu95Glyfs*82 | Compound Heterozygous |
| 3 [12] | Recessive OI | FKBP10 | NM_021939.3, CCDS11409 | c.831dupC | p.Gly278Argfs*17 | Homozygous |
| 4 [13] | Recessive OI | FKBP10 | NM_021939.3, CCDS11409 | c.831dupC | p.Gly278Argfs*17 | Homozygous |
| 5 | OI | COL1A1 | NM_000088.3, CCDS11561 | c.1903G>A | p.Gly635Ser | Heterozygous |
| 6 | Geleophysic dysplasia | FBN1 | NM_000138.4, CCDS32232 | c.5117G>A | p.Cys1706Tyr | Heterozygous |
We detected mutations in COL1A1 and COL1A2 in three patients with OI without a previous molecular diagnosis and a CLCN7 mutation in a patient with OPT (Table 4). All mutations were confirmed by Sanger sequencing. Thus, the panel was successful to reach a molecular diagnosis all 4 patients with OI or OPT and unknown mutations. No mutations in the panel genes were identified for subject 11 who has dysosteosclerosis. Sanger sequencing for SLC29A3 (which we recently demonstrated causes dysosteosclerosis[21]) was also negative; it thus appears that there is more than one gene causing dysosteosclerosis and we are currently investigating this further.
Table 4.
Screening for mutations in individuals not previously tested.
| Subject | Phenotype | Gene | Genbank mRNA and CCDS ID | Mutation (cDNA) | Mutation (protein) | Zygosity of mutation |
|---|---|---|---|---|---|---|
| 7 | OI type IV | COL1A2 | NM_000089.3, CCDS34682 | c.3938T>C | p.Leu1313Pro | Heterozygous |
| 8 | OI type III | COL1A1 | NM_000088.3, CCDS11561 | c.3118G>A | p.Gly1040Ser | Heterozygous |
| 9 | OI | COL1A1 | NM_000088.3, CCDS11561 | c.3505G>A | p.Gly1169Ser | Heterozygous |
| 10 | Osteopetrosis | CLCN7 | NM_001287.5, CCDS32361 | c.856C>T | p.Arg286Trp | Heterozygous |
| 11 | Dysosteosclerosis | No pathogenic mutation identified. This patient also had Sanger sequencing for SLC29A3 [21]. | ||||
Discussion
In this study, we describe a diagnostic panel for the molecular diagnosis of the heritable skeletal disorders that commonly cause low or high bone mineral density. The target genes were enriched by a solution-based oligonucleotide probe hybridization capture method followed by massively parallel sequencing. In total, 34 genes spanning 602 exons were simultaneously sequenced with deep coverage. Using this approach, mutations were identified in three subjects with OI and one with osteopetrosis without prior molecular testing.
Poor coverage was observed in genes having paralogs. The presence of paralogs can negatively impact hybridization and alignment. Another problematic area is the first exon because of the high GC content. This is an inherent problem of NGS; the presence of GC-rich regions prevents proper hybridization resulting in low capture and coverage of that sequence [22, 23]. Sanger sequencing of the problematic areas is one of the solutions available for the areas of low coverage.
Another solution could be an alternative enrichment approach such as microdroplet PCR. This method is reported to be more efficient in overcoming problems associated with high GC content and is also able to resolve paralogs [24]. However, a comparison of the different target enrichment platforms by Valencia et al. demonstrated that both the solution phase-based method and the droplet PCR-based methods had problems with regions of high GC content and had similar overall coverage [25]. Droplet PCR was better able to distinguish between paralogs while the capture-based enrichment had a smaller number of missed exons [25]. Thus, both methods seem to be comparable in terms of performance for molecular diagnostic panels, each with different pitfalls.
In cases for which a mutation cannot be identified with a NGS platform and/or Sanger sequencing, alternative methods including Multiplex Ligation-dependent Probe Amplification (MPLA) and array Comparative Genomic Hybridization (aCGH)to detect whole exon or gene deletions or duplications may be useful. Although NGS read coverage can be used to detect whole exon deletion or duplication, MLPA and aCGH remain gold-standard techniques to demonstrate whole exon or gene deletion or duplication. Exome sequencing to find novel disease genes or variant phenotypes of an unsuspected disease may be useful in cases where mutations are not found. In addition, biochemical studies such as collagen modification studies for OI, and specific gene expression studies when mutations in non-coding regions are suspected can be performed.
An appropriate diagnostic paradigm should balance cost, sensitivity, specificity, and nature of subsequent downstream sequencing. The finding of high bone mineral density should lead to testing of a collection of genes using a NGS approach. The finding of low bone mineral density, however, could direct a staged testing approach. For example, if dominantly inherited OI is strongly under consideration, targeted COL1A1 and COL1A2 testing could be considered first as these genes account for over 85% of OI cases. If negative, then the LBMD panel could be considered. Alternatively, the LBMD panel could be a first step if an alternative clinical cause of LBMD is more likely such as in the presence of clinical symptoms of Marfan syndrome or Ehlers Danlos syndrome. Similarly, the LBMD panel would be a viable first testing option if a recessive pattern of inheritance of OI is likely making type I collagen gene mutation less likely. Ultimately, as whole exome capture coverage increases with decreasing cost, it is likely that this approach will eventually supersede targeted approaches. This reflects the dynamic nature of testing paradigms given rapidly evolving technologies.
In summary, we present a study demonstrating that our diagnostic platform can successfully be used in a clinical setting for the accurate and rapid molecular diagnosis of patients with disorders of low and high bone mineral density. Future directions would include incorporation of additional genes to the existing panel and the design of new panels that target other causes of skeletal dysplasia.
Acknowledgments
We thank Alyssa Tran for patient enrollment and Yuqing Chen for technical assistance. Research funding includes NIH grant PO1 HD22657 and the Baylor College of Medicine BCM Intellectual and Developmental Disabilities Research Center (HD024064). PC is supported by a CIHR clinician-scientist training award. SNSC is supported by fellowship grants from the Osteogenesis Imperfecta Foundation and the National Urea Cycle Disorders Foundation.
Footnotes
Statement regarding potential conflict of interest/disclosure: Baylor College of Medicine, more specifically the Molecular Genetics Laboratories of which V.W.Z., H.C. and L.-J.C.W. are members, plans to eventually offer an iteration of this panel on a commercial basis. Baylor College of Medicine is a non-profit institution.
References
- 1.Krakow D, Rimoin DL. The skeletal dysplasias. Genet Med. 2010;12:327–341. doi: 10.1097/GIM.0b013e3181daae9b. [DOI] [PubMed] [Google Scholar]
- 2.Makitie O. Molecular defects causing skeletal dysplasias. Endocr Dev. 2011;21:78–84. doi: 10.1159/000328131. [DOI] [PubMed] [Google Scholar]
- 3.Warman ML, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graungaard AH, Skov L. Why do we need a diagnosis? A qualitative study of parents’ experiences, coping and needs, when the newborn child is severely disabled. Child Care Health Dev. 2007;33:296–307. doi: 10.1111/j.1365-2214.2006.00666.x. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk FS, Byers PH, Dalgleish R, Malfait F, Maugeri A, Rohrbach M, Symoens S, Sistermans EA, Pals G. EMQN best practice guidelines for the laboratory diagnosis of osteogenesis imperfecta. Eur J Hum Genet. 2012;20:11–19. doi: 10.1038/ejhg.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku CS, Cooper DN, Polychronakos C, Naidoo N, Wu M, Soong R. Exome sequencing: dual role as a discovery and diagnostic tool. Ann Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Cui H, Zhang VW, Wing L-JC. Next Generation Sequencing Panel for molecular diagnosis of Glycogen Storage Diseases; ACMG annual meeting; Charlotte, NC. 2012. [Google Scholar]
- 8.Tian X, Cui H, Tang S, Wang J, Zhang VW, Wong L-JC, Xia Tian HC, 1, Tang Sha, 1, Wang Jing, 1, WeiZhang Victor, 1, Wong Lee-Jun. Simultaneous sequence analysis of 24 genes responsible for metabolic myopathies. ACMG annual meeting; Charlotte, NC. 2012. [Google Scholar]
- 9.Asan, Xu Y, Jiang H, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homan EP, Rauch F, Grafe I, et al. Mutations in SERPINF1 cause osteogenesis imperfecta type VI. J Bone Miner Res. 2011;26:2798–2803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldridge D, Schwarze U, Morello R, et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008;29:1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley BP, Malfait F, Bonafe L, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011;26:666–672. doi: 10.1002/jbmr.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alanay Y, Avaygan H, Camacho N, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houston CS, Gerrard JW, Ives EJ. Dysosteosclerosis. AJR Am J Roentgenol. 1978;130:988–991. doi: 10.2214/ajr.130.5.988. [DOI] [PubMed] [Google Scholar]
- 15.Lemire EG, Wiebe S. Clinical and radiologic findings in an adult male with dysosteosclerosis. Am J Med Genet A. 2008;146A:474–478. doi: 10.1002/ajmg.a.32182. [DOI] [PubMed] [Google Scholar]
- 16.Packota GV, Shiffman J, Hall JM. Osteomyelitis of the mandible in a patient with dysosteosclerosis. Report of a case. Oral Surg Oral Med Oral Pathol. 1991;71:144–147. doi: 10.1016/0030-4220(91)90455-l. [DOI] [PubMed] [Google Scholar]
- 17.Le Goff C, Morice-Picard F, Dagoneau N, et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat Genet. 2008;40:1119–1123. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Goff C, Mahaut C, Wang LW, et al. Mutations in the TGFbeta binding-protein-like domain 5 of FBN1 are responsible for acromicric and geleophysic dysplasias. Am J Hum Genet. 2011;89:7–14. doi: 10.1016/j.ajhg.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morello R, Bertin TK, Chen Y, et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Cohen MM., Jr The new bone biology: pathologic, molecular, and clinical correlates. Am J Med Genet A. 2006;140:2646–2706. doi: 10.1002/ajmg.a.31368. [DOI] [PubMed] [Google Scholar]
- 21.Campeau PM, Lu JT, Sule G, et al. Whole Exome Sequencing Identifies Mutations in the Nucleoside Transporter Gene SLC29A3 in Dysosteosclerosis, a Form of Osteopetrosis. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulonen AM, Ellonen P, Almusa H, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huentelman MJ. Targeted next-generation sequencing: microdroplet PCR approach for variant detection in research and clinical samples. Expert Rev Mol Diagn. 2011;11:347–349. doi: 10.1586/erm.11.23. [DOI] [PubMed] [Google Scholar]
- 25.Valencia CA, Rhodenizer D, Bhide S, Chin E, Littlejohn MR, Keong LM, Rutkowski A, Bonnemann C, Hegde M. Assessment of target enrichment platforms using massively parallel sequencing for the mutation detection for congenital muscular dystrophy. J Mol Diagn. 2012;14:233–246. doi: 10.1016/j.jmoldx.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dijk FS, Pals G, Van Rijn RR, Nikkels PG, Cobben JM. Classification of Osteogenesis Imperfecta revisited. Eur J Med Genet. 2010;53:1–5. doi: 10.1016/j.ejmg.2009.10.007. [DOI] [PubMed] [Google Scholar]