Abstract
The 3′-untranslated region (UTR) is known to be a critical regulator of post-transcriptional events that determine the gene expression at the RNA level. The gene CCN1 is one of the classical members of the matricellular CCN family and is involved in a number of biological processes during mammalian development. In the present study, the 600-bp 3′-UTR of CCN1 was functionally characterized. Reporter gene analysis revealed that the entire 3′-UTR profoundly repressed gene expression in cis in different types of the cells, to which both the proximal and distal-halves of the 3′-UTR segments contributed almost equally. Deletion analysis of the 3′-UTR indicated a distinct functional element in the proximal half, whereas a putative target for microRNA-181s was predicted in silico in the distal half. Of note, the repressive RNA element in the proximal half was shown to be capable of forming a stable secondary structure. However, unexpectedly, a reporter construct with a tandem repeat of the predicted miR-181 targets failed to respond to miR-181a. In addition, the other major structured element predicted in the distal half was similarly characterized. To our surprise, the second element rather enhanced the reporter gene expression in cis. These results indicate the involvement of multiple regulatory elements in the CCN1 3′-UTR and suggest the complexity of the miRNA action as well as the 3′-UTR-mediated gene regulation.
Keywords: CCN family, CCN1, Cyr61, miRNA, Post-transcriptional regulation, 3′-UTR
Introduction
The CCN family is a group of proteins with novel structure and multiple functionalities. This family consists of six members in vertebrates, and the family name represents the original name of three classical members: cysteine-rich 61 (Cyr61), connective tissue growth factor (CTGF), and nephroblastoma-overexpressed gene product (NOV)(Perbal and Takigawa 2005; Jun and Lau 2011; Kubota and Takigawa 2007a, b, 2011; Rachfal and Brigstock 2005). All of the members are composed of conserved protein modules connected in tandem. Following the amino-terminal signal peptide for secretion, insulin-like growth factor-binding protein-like module (IGFBP), the von Willebrand factor type C repeat (VWC), the thrombospondin (TSP) type 1 repeat, and the C-terminal cystine knot (CT) except for CCN5 are lined up (Holbourn et al. 2008). Generally, CCN proteins are known to exert multiple functions, which are based on the multiple interactions with a vast number of molecular counterparts via these four conserved modules, which are highly interactive with a variety of ligands (Jun and Lau 2011; Kubota and Takigawa 2007a, b, 2011; Perbal and Takigawa 2005; Rachfal and Brigstock 2005). Indeed, CCN1, the most classical member of the family, plays critical roles in several developmental events, including chondrogenesis and angiogenesis (Jun and Lau 2011; Mo et al. 2002). Indeed, the Ccn1-null mice suffer embryonic death due to chorioallantonic fusion failure or placental vascular insufficiency, suggesting the important roles of CCN1 in vessel formation (Mo et al. 2002). CCN1 also plays an important role in cell proliferation by activating several growth factors, regulating the cell cycle, and participating in matrix remodeling (Jun and Lau 2010, 2011; Si et al. 2006). Besides, CCN1 not only mediates physiological processes, but is also associated with pathological ones. For example, involvement of this molecule in different types of malignancies was earlier indicated (Jun and Lau 2011; Ohgawara et al. 2011; O’Kelly et al. 2008). Interestingly, CCN1 protein acts either as a positive regulator or growth inhibitor of cancer cells (Jun and Lau 2011; Leask 2010). Also, CCN1 levels have been reported to be significantly higher in metastatic lesions than in primary pancreatic tumors (Holloway et al. 2005). Recently, a novel role of CCN1 in inflammation-induced apoptosis was uncovered as well (Bai et al. 2010). As a background of such functions, interaction with integrins, proteoglycans, and other extracellular matrix molecules and its biological significance are widely recognized (Jun and Lau 2011). As such, the property of CCN1 protein is being uncovered, and it is currently regarded as a matricellular protein with diverse functions (Jun and Lau 2011; Bai et al. 2010). Nevertheless, the regulatory mechanism of CCN1 gene expression at the post-transcriptional level mostly remains to be investigated.
In view of genetic structure, the mRNAs of all of the members except for CCN6 are commonly characterized by the retention of the 3′-untranslated regions (UTRs) of significant lengths. Recently, the profound functional significance of the 3′-UTR in post-transcriptional gene regulation has been attracting the interest of a vast number of scientists (Merritt et al. 2008). According to recent reports, several RNA-binding proteins bind to 3′-UTR region of target mRNA, which regulates gene expression at the posttranscriptional level (Ishimaru et al. 2010; Zhang et al. 2010). For example, expression of the p63 gene, a member of the p53 tumor suppressor family, is regulated via mRNA stability by an RNA-binding protein, RNPC1 (Zhang et al. 2010). Interestingly, RNPC1 is a target of the p53 family, which represses p63 gene expression by accelerating RNA degradation under the interaction with the 3′-UTR. As such, the 3′-UTR of an mRNA is known to determine the stability, localization, and translation of the mRNA. Moreover, functional characterization of miRNA, a class of the non-coding RNAs that control gene expression, revealed that its target is mostly located in the 3′-UTR region of target mRNAs, further indicating the critical importance of the 3′-UTR in post-transcriptional regulatory events (Didiano and Hobert 2008; Lee et al. 2009; Sandberg et al. 2008).
Concerning the CCN family, the best-characterized 3′-UTR among the members is that of CCN2 (1,19). It has been recognized that CCN2 gene expression is regulated at multiple steps such as transcriptional, posttranscriptional, and translational stages. Initial studies reported the repressive effect of the 3′-UTR on the expression of CCN2. Subsequently, the cis-acting element of structure-anchored repression (CAESAR) in the 3′-UTR was identified as an RNA element regulating mRNA stability and translation efficiency (Kubota et al. 2000; Kubota and Takigawa 2007a, b, 2011). Furthermore, a nucleocytoplasmic shuttling trans-factor, nucleophosmin (NPM/B23), was found to actually bind to another repressive cis-element in the 3′-UTR of the CCN2 mRNA, playing an important role in the posttranscriptional regulation of CCN2 during endochondral ossification (Mukudai et al. 2005, 2008). Finally, miR-18a has been reported to target the CCN2 mRNA at its 3′-UTR, regulating chondrocytic phenotype (Ohgawara et al. 2009). It should be noted that the miR-18a target region and NPM-binding sites are in close proximity, suggesting common machinery shared by these two regulatory systems (Jacobsen et al. 2010). In contrast to CCN2, until today, no reports have described the post-transcriptional regulatory function of the 3′-UTR of CCN1. Therefore in this study, we analyzed the functional properties of the CCN1 3′-UTR and found an element that mediate repressive gene regulation therein.
Materials and methods
Cell culture
Human cervical carcinoma HeLa, human kidney-derived 293T and human chondrocytic HCS-2/8 (Takigawa et al. 1989) cell lines were cultured in Dulbecco’s modified Eagle’s minimum essential medium (D-MEM) supplemented with 10 % fetal bovine serum (FBS). Chicken normal embryonic fibroblasts were isolated from a 10-day-old whole chicken embryo. Those cells were maintained in high-glucose D-MEM supplemented with 10 % FBS. All of the cells were incubated in humidified air containing 5 % CO2 at 37 °C.
Plasmid constructs
All of the reporter constructs for the evaluation of the repressive effect of the CCN 3′-UTR and its fragments were constructed by inserting them into a pGL3-L(+) or pGL3-L(-) parental vector (Kubota et al. 2000) at the multiple cloning sites located immediately downstream of the firefly luciferase gene. The original luciferase-CCN1 chimeric constructs, pGL3-CyrUTRS and pGL3-CyrUTRA (Fig. 1), were constructed by subcloning a human CCN1 3′-UTR cDNA that had been amplified by PCR with primers Cyr61UTRS (5′-TTC TGC AGG GAC TAA ATG CTA CCT G-3′) and Cyr61UTRA (5′-GGC TTA AGG TAA ATT ATT TCT TTA TAA ATG-3′) between unique Pst I and Afl II sites in pGL3-L(+) and pGL-3L(-), respectively. To construct pGL3-CyrUTRS-XN (Fig. 2), the Xba I-Nru I fragment of pGL3-CyrUTRS, which contained the entire 3′-UTR, was transferred between unique Nhe I and Sma I sites on the same backbone. Next, the 3′-UTR was split into proximal and distal halves at a unique Blp I site, and each fragment was similarly built in pGL3-L(+) to yield pGL3-CyrBE or pGL3-CyrFS (Fig. 2). All of the deletion mutants of pGL3-CyrBE were constructed by subcloning PCR-derived fragments of the 3′-UTR between unique Xba I and Eco RV sites in pGL3-L(+) (Fig. 3). For this objective, three oligonucleotide primers containing human CCN1 cDNA sequences were prepared. The nucleotide sequence of these primers were the following: CY270A (5′-GAG ATA TCA GCA GTA ACA TGT GCT CCA A-3′), CY160A (5′-GAG ATA TCA AGA ATG ACA AGG CTT-3′), and CY165S (5′-TTT CTA GAT TAA GGT ATT TCG AAA GGC C-3′). In addition, two primers corresponding to a sense sequence of the downstream end of the luciferase gene (LUCS: 5′-CTC GAC GCA AGA AAA ATC AG-3′) and an antisense sequence immediately downstream of the multiple cloning site (GLMCSA: 5′-TGT CCA AAC TCA TCA ATG TAT C-3′) in pGL3-L(+) were also designed and synthesized. PCR amplification was carried out with combinations of LUCS/CY270A, LUCS/CY160A, CY165S/GLMCSA, and CY165S/CY270A; and the amplicons were inserted into pGL3-L(+) to yield pGL3-CyrBEXEP, BEXE, BEXENB and BEMI, respectively, after having been double-digested with Xba I and EcoRV. For the construction of pGL3-FSMI, the structured segment was amplified from pGL3-CyrUTRS with primers CyrLattS (5′- GCT CTA GAT TCC ATC CCT TCC TG -3′) and CyrLattA (5′- GGA ATT CCC CTC CCT CCC -3′). The amplicon was then digested by Xba I and EcoRI and was subcloned between the corresponding sites in pGL3-L(+). A reporter gene construct for miR-181, pGLS-181DS, was constructed by inserting a synthetic double strand DNA by utilizing the same Xba I and EcoRI sites in the same parental plasmid, as illustrated in Fig. 4. Nucleotide sequence of the inserted DNA segment containing the predicted miR-181a targets is also given therein. Integrity of all of the plasmids was confirmed by DNA sequencing.
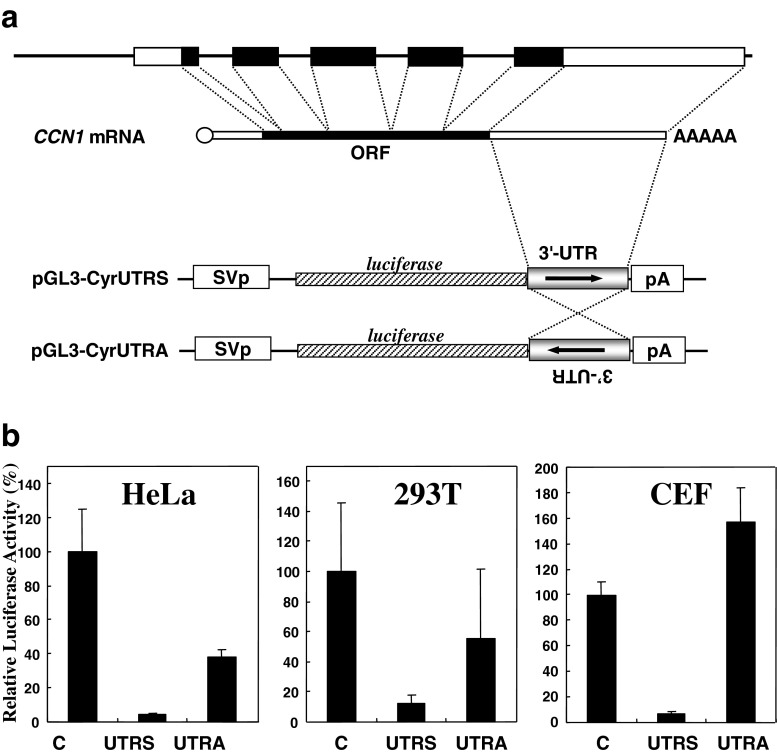
Fig. 1.
a Structures of the gene, mRNA, and reporter constructs of human CCN1. The genomic CCN1 structure is illustrated at the top with schematic representation of the corresponding mRNA with 5′-cap and polyadenyl tail. Solid and open boxes denote open reading frames and untranslated regions, respectively, whereas intronic regions are shown in bold lines. In the lower half of the panel, construction of the original reporter plasmids, pGL3-CyrUTRS and pGL3-CyrUTRA, is displayed. SV40p, pA, and 3′-UTR indicate SV40 promoter, SV40 polyadenylation signal, and the 3′-UTR of CCN1, respectively. b Effect of the cis-linked 3′-UTR at the 3′-end on luciferase gene expression. The parental pGL3-L(+) (C), pGL3-CyrUTRS (UTRS), or pGL3-CyrUTRA (UTRA) was used to transfect the cells along with an internal control vector, pRL-TK. Forty-eight hours later, the cells were harvested for the luciferase assay, and firefly luciferase gene activities were standardized against those of the internal control. The name of the cell line used is shown on each graph. Mean values are shown with error bars of standard deviations
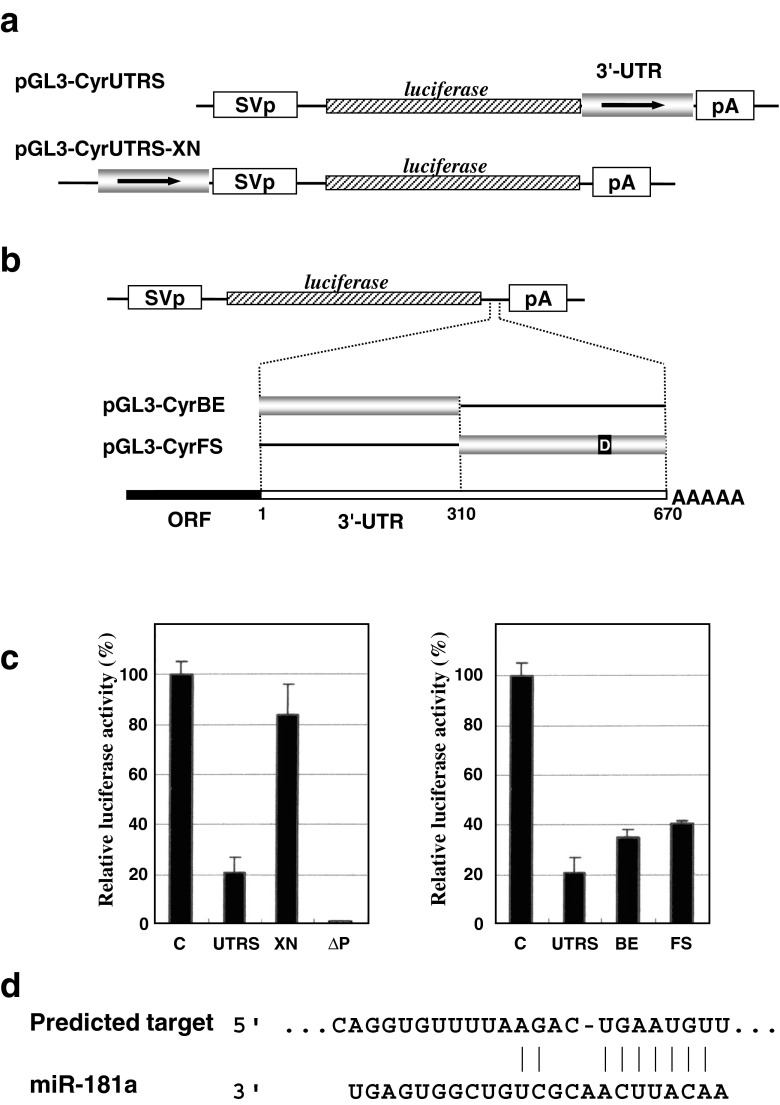
Fig. 2.
a Structure of the plasmids used to evaluate the positional effect on the 3′-UTR function. In pGL3-CyrUTR-XN, the CCN1 3′-UTR was relocated immediately upstream of the SV40 promoter. b Structures of the reporter constructs for the independent functional evaluation of the proximal and distal halves of the 3′-UTR. The junction of the cDNA fragments subcloned is indicated by the nucleotide number counted from the upstream end (1) of the 3′-UTR. The small solid box labeled “D” represents the putative miRNA target predicted in silico, which is detailed in panel d. c Relative luciferase activities from the plasmids illustrated in panels a and b in HeLa cells. The left panel shows the results obtained with the plasmids described in panel a, where ∆P indicates a plasmid without a promoter, pGL3∆P. Similarly, the right panel shows those from the plasmids in panel b. Mean values are shown with error bars of standard deviations. d Putative miRNA target in the CCN1 3′-UTR predicted by Targetscan. The nucleotide sequence of the corresponding region in the 3′-UTR is displayed, together with that of miR-181a forming an RNA duplex
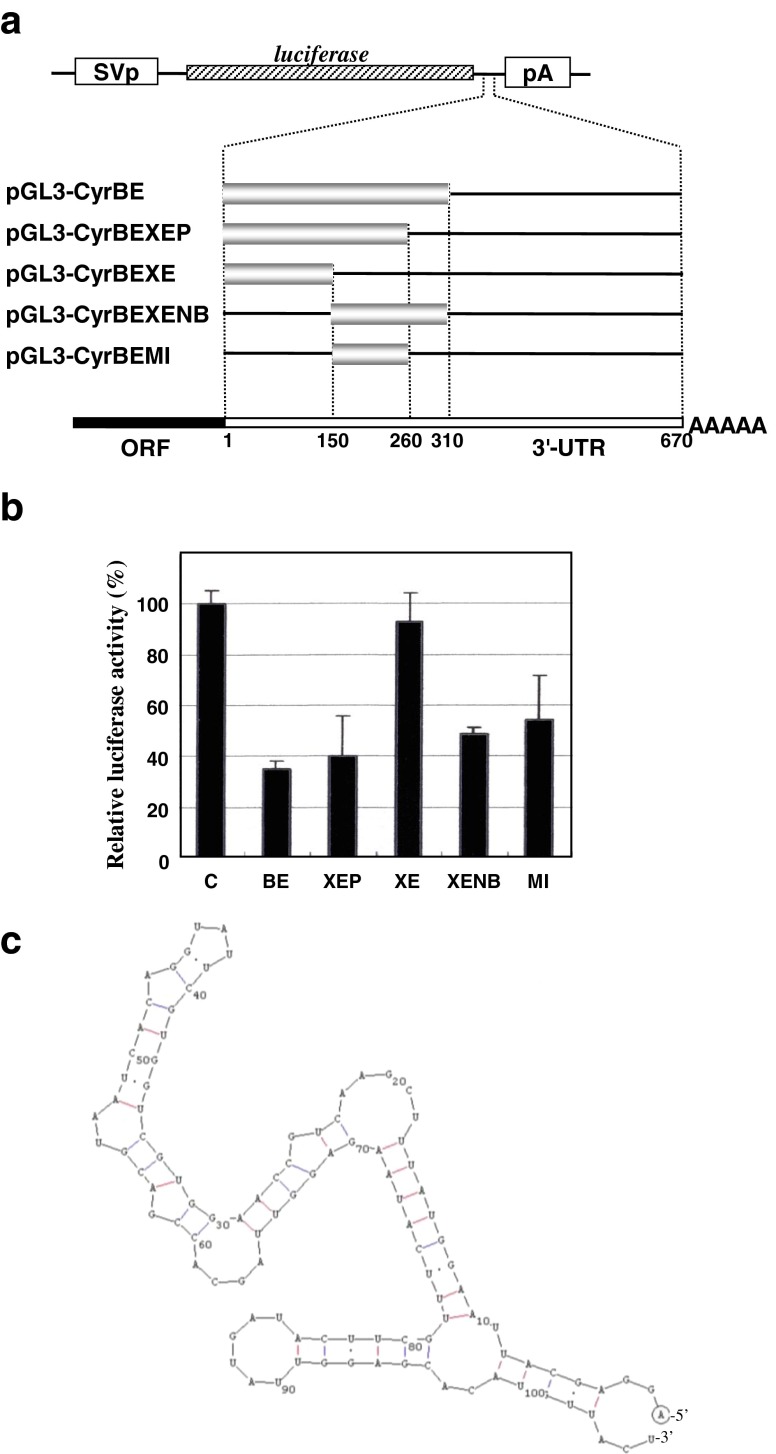
Fig. 3.
Post-transcriptional regulatory element in the proximal half of the CCN1 3′-UTR. a Deletion mutants of the 3′-UTR in the reporter gene constructs were made to locate a functional element in the proximal half. Approximate locations and size of the 3′-UTR subfragments are indicated by nucleotide numbers counted from the upstream end of the 3′-UTR. Abbreviations used are as described in Fig. 1 legend. b Relative luciferase activities from the plasmids illustrated in panel a. Evaluation was performed in HeLa cells with a control experiment (C) using pGL3-L(+). Mean values are shown with error bars of standard deviations. c Predicted secondary structure of the minimal functional segment involved in pGL3-CyrUTR-BEMI. The prediction was performed by use of GENETYX software
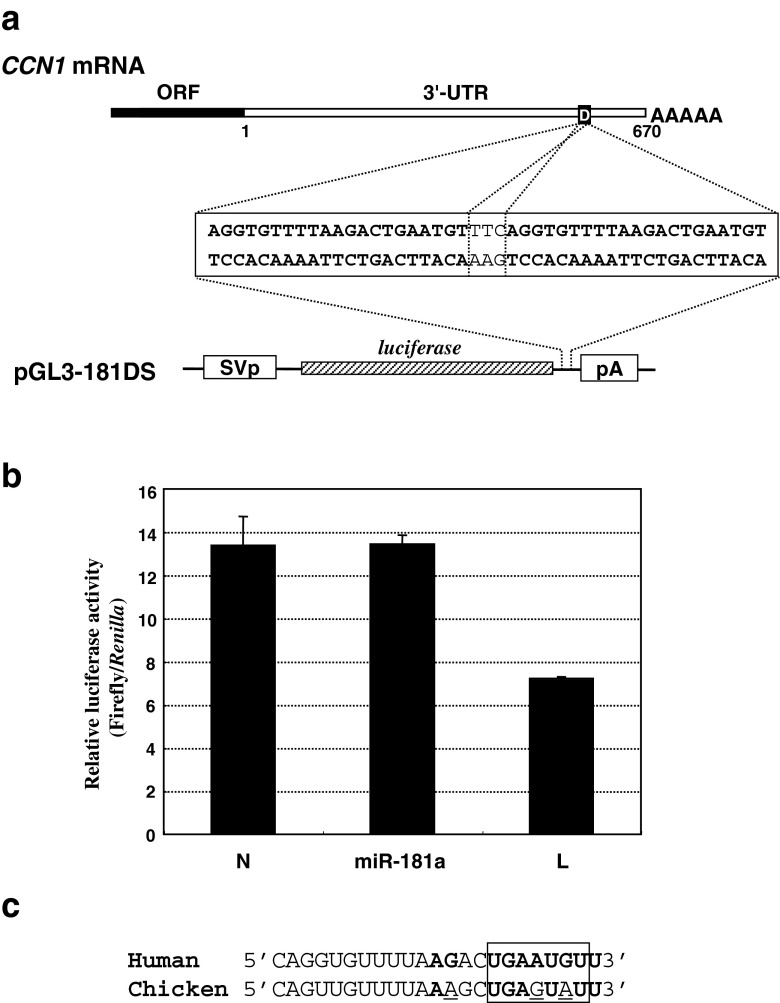
Fig. 4.
Functional evaluation of the predicted miR-181a target sequence in the CCN1 3′-UTR. a Structure of the plasmid used to evaluate the function of the miR-181a target predicted by Targetscan. A double strand DNA containing 2 copies of the miR-181a target sequences predicted in human CCN1 was synthesized and inserted into the parental reporter plasmid to yield pGL3-181DS. The nucleotide sequence of the corresponding region is illustrated in the middle of the panel. b Relative luciferase activities from the plasmid illustrated in a in HCS-2/8 cells in the presence or absence of exogenous miR-181a. The cells were transfected with miR-181a or a control siRNA (N, negative control siRNA; L, control luciferase siRNA). Eight hours later, the cells were further transfected with the reporter plasmid along with an internal control, phR-TK (int-), and were harvested for the luciferase assay after 48 h of incubation. c Absence of the seed element for miR-181 in chicken CCN1. Nucleotide sequences of the predicted miR-181a target in human CCN1 and the correspondent in chicken CCN1 are aligned. Nucleotides complementary to miR-181a are shown in bold cases, whereas those mismatched in chicken CCN1 are underlined. The seed element is indicated by a box
DNA transfection and luciferase assays
Twenty-four hours prior to transfection, cells were seeded in a 12-well tissue culture cluster plate at 80-90 % confluence. For the functional analysis of the 3′-UTR, the cells were co-transfected with 750 ng of a reporter construct and 250 ng of pRL-TK (internal control: Promega, Madison, WI) with the aid of Lipofectamine (Invitrogen, Carlsbad, CA) for HeLa, 293T and CEF or Fugene 6 (Roche, Basel, Switzerland) for HCS-2/8 after a medium change. Forty-eight hours after transfection, the cells were lysed in 250 μl of a passive lysis buffer (Promega), which lysate was directly used for the analysis by the Dual-luciferase assay system (Promega). For miRNA-mediated reporter gene assays, HCS-2/8 cells were seeded in a 6 -well tissue culture plate and were transfected with miR-181a or control siRNA as described in another subsection. After incubated for 8 h, DNA transfection was conducted as described above with 500 ng of pGL3-181DS and 50 ng of phRL-TK (int-) (Promega); and the cells were harvested 48 h later with 500 μl of the lysis buffer. Sequential measurement of firefly and Renilla luciferase activities was performed as described previously (Kubota et al. 2000).
In silico analyses
RNA secondary structure prediction and nucleotide sequence alignment were performed by using a commercial software package, GENETYX ver. 7 (Genetyx Corporation, Tokyo, Japan). For the prediction of miRNA targets in the CCN1 3′-UTR, three on-line devices were employed (Targetscan ver. 4.1., http://www.targetscan.org/; miRanda, http://www.microrna.org/microrna/home.do; DIANA, http://diana.cslab.ece.ntua.gr/microT/).
Synthesis and application of miRNAs
Synthetic RNA oligonucleotides of the sequences 5′-AAC AUU CAA UGC UGU CGG UGA GU-3′ and 5′-ACC ACU GAC CGU UGA CUG UAC were prepared by B-Bridge International Inc (Mountain View, USA). These single-stranded RNAs were annealed to obtain mature-form miR-181a duplex. As a negative and positive controls, Silencer Negative Control siRNA (Ambion, Austin, TX) and Control siRNA luciferase (GL3)(Cosmo Bio, Tokyo, Japan) were used, respectively. Twenty-four hours before transfection, HCS2/8 cells were seeded at 80–90 % confluence, and miR-181a transfection was performed with Lipofectamine 2000 (Invitrogen) at a concentration of 50 nM, according to the manufacturer’s instructions.
Results
Cis-repressive activity of the 3′-UTR of the human CCN1 gene
In the case of another classical CCN family member, CCN2, it is widely recognized that post-transcriptional repressive regulation by the 3′-UTR plays a crucial role in cell growth and differentiation (Kubota and Takigawa 2007a, b; Mukudai et al. 2005, 2008). Since both CCN1 and CCN2 were reported to similarly respond to the same stimuli (Moritani et al. 2005), we suspected that similar repressive regulation would be mediated by the CCN1 3′-UTR as well. To examine this hypothesis, we employed a conventional reporter gene assay system. The cDNA of the CCN1 3′-UTR was amplified by RT-PCR and was subcloned into a luciferase expression vector between the downstream end of the luciferase gene and polyadenylation signal in a sense direction, as described in Fig. 1a, to construct pGL3-CyrUTRS. Comparative analysis of the luciferase activity from this plasmid with that from the parental control in a few cell lines revealed even a stronger cis-repressive activity against gene expression than that of CCN2 (Kubota et al. 2000). This effect was orientation dependent, as indicated by the results obtained with another plasmid containing the UTR in an anti-sense direction (pGL3-CyrUTRA: Fig. 1a), suggesting its property as a post-transcriptional regulatory element. Of note, the strong repression by the 3′-UTR was observed not only in human cell lines, but also in chicken normal fibroblasts, which indicates that this regulation is fundamental and universal among vertebrates (Fig. 1b).
Function and potential of the 3′-UTR as a post-transcriptional regulatory machinery
Next, to further estimate the post-transcriptional property of the 3′-UTR-mediated regulation of CCN1, we constructed another reporter plasmid, in which the 3′-UTR was translocated to the upstream site of the promoter driving the luciferase gene and evaluated its effect. The result of luciferase assay showed no significant effect of the 3′-UTR at this location outside of the transcribed area (Fig. 2, panels a and c). This finding further supports the idea that the 3′-UTR exerted its effect after transcription.
In the middle of the 3′-UTR, a unique Blp I site is located. Taking advantage of this structure, we split the 3′-UTR into two pieces and evaluated the functionality of each fragment under the same experimental conditions. Interestingly, both fragments were equally functional to repress gene expression in cis, displaying approximately half of the effect by the full-length UTR (Fig. 2 panels, b and c). These results indicate independent functional elements in proximal and distal halves, which show additive effects when they work together.
Among the post-transcriptional RNA regulatory elements found in 3′-UTRs, miRNA targets are known to be predictable in silico. Thus, initially by using Targetscan we searched for the miRNAs that might target the CCN1 3′-UTR. Unexpectedly, only one miRNA target was predicted. This target was also predicted as one of two putative miRNA targets by DIANA, or one of 12 targets by miRanda. Of note, involvement of a miR-155 target in the ones predicted by miRanda also supported a previous finding that miR-155 actually targeted CCN1 (Zhang et al. 2010). According to these predictions, the CCN1 mRNA might be under the regulation of miR-181 members miR-181a, b, c and d. This putative miR-181 target was located in the distal half of the 3′-UTR (Fig. 2d).
A major regulatory element revealed by the deletion analysis of the proximal-half fragment of the 3′-UTR
Since no miRNA target was predicted in the proximal half of the 3′-UTR, we subsequently tried to localize a functional element experimentally by deletion analysis. Several deletion mutants of the proximal-half of the 3′-UTR were connected to the luciferase in the sense direction, and then the effect was evaluated by performing the luciferase assay. Removal of the downstream 50 bases did not affect the repressive activity, whereas further deletion up to 160 bases completely abolished it (Fig. 3). Upstream deletion analysis showed that the 5′- quarter region was not functional and was dispensable for the repressive regulation (Fig. 3). Based on these results, we defined a minimal 110-base functional element in the second quarter area of the 3′-UTR. It should be noted that in silico prediction revealed a stable RNA secondary structure of the element, suggesting its property as a post-transcriptional genetic element.
Functional evaluation of the possible regulation by miR-181 via the 3′-UTR
Since an miR-181 target was predicted in silico, we examined whether or not the predicted target sequence in CCN1 mRNA 3′-UTR was actually functional by performing a reporter gene assay. In our previous study, we observed only a modest repressive effect of chemically synthesized miR-181a duplex on the reporter gene construct containing a single copy of the predicted target (Sumiyoshi et al. 2013). Here, to verify the functionality of the predicted target, another reporter gene construct, pGL3-181DS, with two copies of the predicted target was newly prepared, and the responsiveness against miR-181a was further examined. For this evaluation, human chondrocytic HCS-2/8 cells were employed, since CCN1 production was repressed by exogenous miR-181a in these cells (Sumiyoshi et al. 2013). Nevertheless, co-transfection of a synthetic miR-181a duplex at the same concentration with pGL3-181DS yielded no significant effect on luciferase gene expression, compared with the results obtained with the control double-stranded RNA. Under the same condition, an siRNA against the luciferase mRNA efficiently repressed the reporter gene expression from the same plasmid (Fig. 4). These results indicate that the predicted miR-181 target in the distal half of the 3′-UTR does not mediate the post-transcriptional repressive effect of miR-181a on CCN1. The fact that the predicted target is not conserved between the CCN1 genes in chicken cells (Fig. 4c), in which the 3′-UTR also acts repressive in cis, supports this conclusion.
Functional property of the second major structured element that was predicted in the distal half of the 3′-UTR
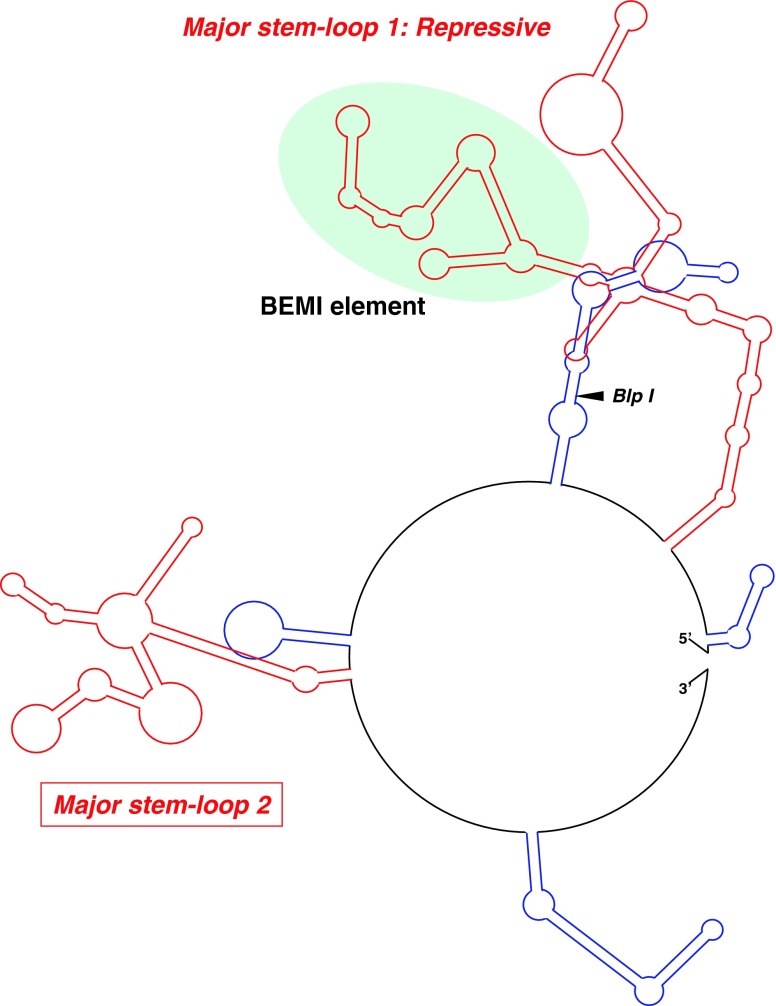
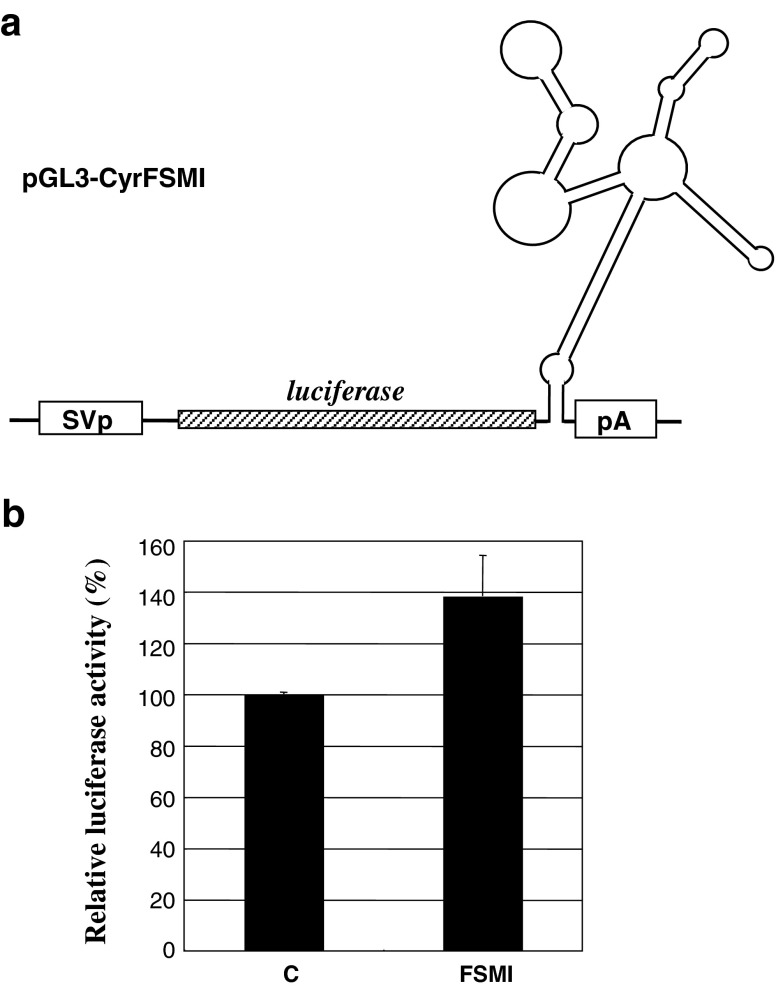
Based on the fact that the functional element minimized by a series of deletion analyses perfectly corresponded to the structured element predicted in silico, we searched for another one as a candidate for the repressive element in the distal half. In silico prediction of the entire CCN1 3′-UTR indeed revealed the repressive element as a part of the major stem loop in the proximal half (Fig. 5: in red). According to this result, the second major stem loop was located in the distal half as well. Thus, suspecting a repressive function therein, we constructed and functionally evaluated another reporter plasmid with the second major stem loop immediately downstream of the luciferase coding region. Unexpectedly, this plasmid, entitled pGL3-CyrFSMI, displayed rather enhancing effect on the luciferase gene expression in cis in HeLa cells (Fig. 6). Except for these two structures, only minor stem loops were predicted in addition (Fig. 5: blue lines).
Fig. 5.
a Distribution of putative structured elements in the 3′-UTR of human CCN1 mRNA. Two major stem-loops and 4 minor ones are represented by red and blue lines, respectively. Functional evaluation firmly indicated the repressive regulatory function of the major stem-loop 1 in the proximal half, which involved the minimal functional segment (BEMI)
Fig. 6.
Functional characterization of the major stem-loop 2 predicted in the distal half of the 3′-UTR. a Structure of the reporter construct. A cDNA corresponding to the major stem-loop 2 illustrated in Fig. 5 was isolated by PCR and was subcloned into the same parental vector at the downstream of the luciferase gene. b Relative luciferase activity in the cell lysate of HeLa cells transfected with the plasmid shown in panel a as represented by the percentage against the control. The parental pGL-3L(+) was utilized for the control experiment
Structural requirement for the RNA cis-repressive elements
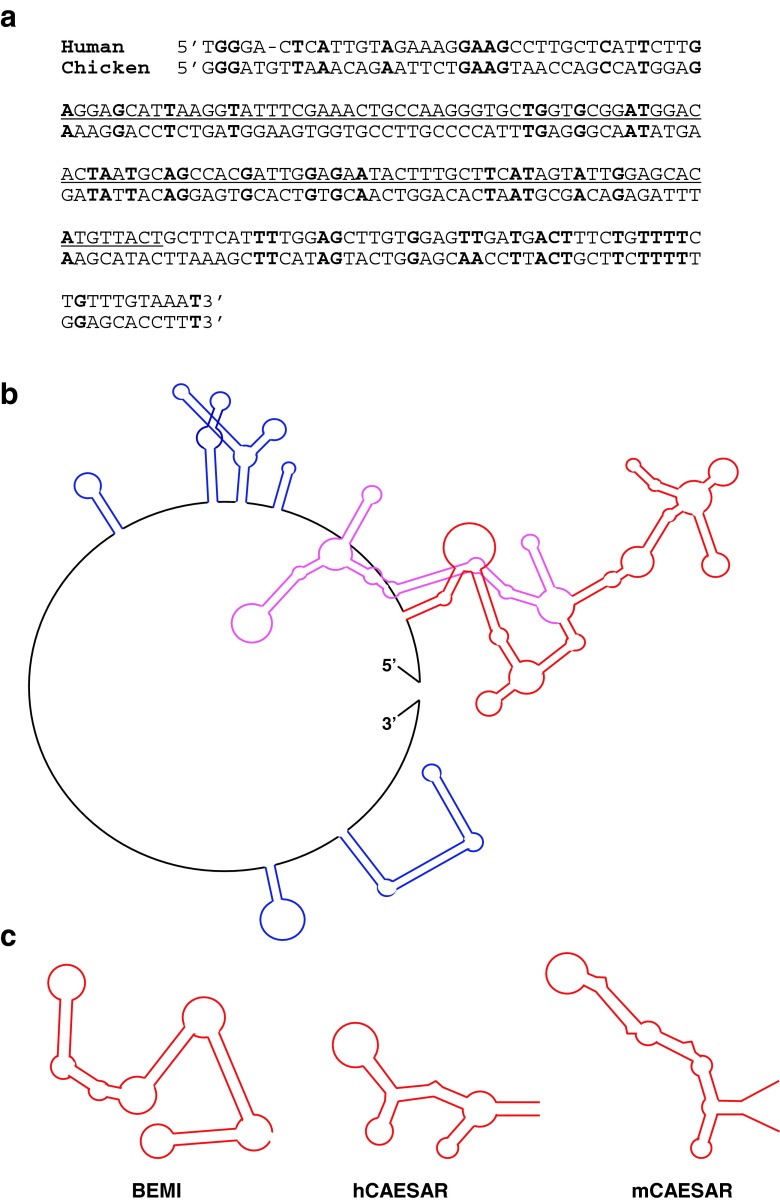
According the result shown in Fig. 1, the 3′-UTR in human CCN1 exerted the repressive effect in chicken cells as well as human cells. Therefore, under a hypothesis that similar regulatory system with a structured element may exist also in the 3′-UTR of chicken CCN1, we initially analyzed the corresponding nucleotide sequence therein. However, in silico alignment of the human and chicken 3′-UTRs revealed no significant homology at all. Surprisingly, the percentage of the matched nucleotides in the BEMI region was 22 %, which was even less than that expected between random sequences (25 %), indicating no conservation during the course of evolution (Fig. 7a).
Fig. 7.
Structural characteristics of cis-repressive elements in the 3′-UTR of the CCN family member genes. a Nucleotide sequence alignment of the 3′-UTR segment around the human BEMI element with its corresponding region in chicken CCN1 3′-UTR. Nucleotides matched between the two are shown in bold letters, whereas the BEMI element is underlined. b Predicted secondary structure of the 3′-UTR in chicken CCN1 mRNA. The major and minor stem-loops are represented in red and blue, respectively. The substructure in the major loop (purple) indicates a putative repressive element. c Comparison of the secondary structures of functional cis-repressive elements identified in the 3′-UTRs of CCN family member genes. In addition to the BEMI element found in this study, human and murine cis-acting elements of structure-anchored repression (CAESARs) are illustrated (Kubota et al. 2000; Kondo et al. 2000)
Next, in silico prediction of the secondary structure was performed with the chicken 3′-UTR as well. As shown in Fig. 7b, only one major structured element was predicted in the proximal half, whereas no major one was in the distal half. Interestingly, a minor structure that is quite close to the one in the human 3′-UTR was predicted at a similar location. Together with the results of reporter gene assays, this estimation further strengthen the importance of the RNA secondary structure in the regulation of gene expression by the CCN1 3′-UTR.
Finally, in order to find out a common structural feature shared among functional RNA repressive elements, predicted secondary structures of the cis-acting repressive elements involved in the 3′-UTRs of CCN family members were analyzed. As shown in Fig. 7c, the cis-acting elements of structure-anchored repression (CAESARs) of human CCN2 (Kubota et al. 2000) and mouse Ccn2 (Kondo et al. 2000) and the BEMI element were compared. These functional elements are commonly characterized by the retention of multiple-bulged major loop accompanied by a minor loop near the bottom, which may be required for the repressive regulation (Fig. 7c). Of note, such a structure is predicted in the chicken CCN1 as well (Fig. 7b: purple lines).
Discussion
The profound role of the 3′-UTR as a post-transcriptional regulator in tissue and organ development has been uncovered by a number of recent studies. Indeed, one such study indicates that the 3′-UTR-mediated post-transcriptional gene regulation is predominant, rather than the promoter-mediated transcriptional control, in conducting the germ line development in nematodes (Merritt et al. 2008). The critical importance of the 3′-UTR has been widely recognized also in mammalian cells. In neuronal cells, the 3′-UTR is known to regulate gene expression by determining the localization of certain mRNAs (Andreassi and Riccio 2009). Moreover, our recent studies revealed the novel function of the post-transcriptional elements in the CCN2 3′-UTR, which regulate the CCN2 gene expression by controlling the mRNA stability and translational efficacy (Kubota et al. 2000; Mukudai et al. 2005, 2008). Additionally, in a variety of malignancies, the anti-apoptotic Bcl-2 protein is overexpressed, especially in leukemia. Nucleolin is one of the proteins that bind to the bcl-2 mRNA 3′-UTR region, thereby increasing its half-life by regulating its stability (Ishimaru et al. 2010). As a new aspect, it has been reported that the 3′-UTR of a particular mRNA has the ability to regulate miRNA functions (Sandberg et al. 2008). These cumulative findings firmly indicate how important the 3′-UTR is in post-transcriptional regulation. In this study, we investigated the functional properties of the CCN1 3′-UTR as a post-transcriptional regulatory machinery. First, the strong repressive effect of the human CCN1 3′-UTR on gene expression was found not only in human cells, but also in chicken cells, indicating that the regulatory system has been evolutionally conserved. Also, the orientation and position dependence of its function indicates that the 3′-UTR mediates the repressive effect at the RNA level. Given that life was acquired through the molecular evolution of RNA under the interaction with peptide counterparts, such regulation is fundamental and thus may have been conserved over a long period of time.
As a result of our deletion analysis, the involvement of multiple regulatory elements was indicated to exist in the CCN1 3′-UTR. One of the elements was located in the proximal half of the 3′-UTR and was predicted to form a stable secondary structure, whereas the other in the distal half remains unidentified. The predicted secondary structure of the minimal functional segment in the proximal half was characterized by a distinct stem-loop with a length of 110 bases. The requirement of RNA secondary structure for the function of post-transcriptional elements was strongly indicated in a number of previous studies. For example, critical effects of the secondary structure and nucleotide length of the 3′-UTR of hepatitis C virus (HCV) mRNA on efficient HCV replication were indicated (Murayama et al. 2010). In this regulatory machinery, the generation of mRNAs with a shorter 3′-UTR caused by early polyadenylation results in more efficient RNA replication, for which the RNA structure is responsible. As another example, a structural change in the 3′-UTR of Hip2, which activates T lymphocytes, alters gene expression during CD4+ T lymphocyte differentiation (Lin et al. 2009). In these regulatory systems, involvement of an RNA-binding cofactor that specifically interacts with the structured target has been strongly suspected. In the case of chicken CCN2, a specific binding counterpart of an RNA element, nucleophosmin (NPM), controls the fate of CCN2 mRNA, which results in the regulation of chondrocyte differentiation (Mukudai et al. 2005, 2008). Therefore, also with this 110-base segment of the CCN1 mRNA, certain RNA-binding proteins are suspected to specifically interact. For understanding the precise regulation system via this element, further detailed examinations are currently underway.
Recently, the 3′-UTR has become quite commonly observed to be a target of miRNA. Being interested in this issue, we suspected the presence of several miRNA targets that might have supported the repressive effect of the distal half of the CCN1 3′-UTR on gene expression. However unexpectedly, in silico analysis with Targetscan predicted only a single miRNA target therein. Furthermore, functional evaluation revealed that the predicted target was irresponsive to miR-181a, indicating that this nucleotide sequence does not mediate the miR-181-associated repressive regulation.
As such, we subsequently searched for structured RNA segments that could mediate the repressive regulation in the distal half of the 3′-UTR, based on the criterion obtained through the identification of the BEMI element in the proximal half. This strategy was supported by the fact that the formation of BEMI element was predicted also in the context of full length 3′-UTR. As a consequence of the secondary structure prediction of the full-length 3′-UTR, only one major structured element was indicated in the distal half. Reporter gene analysis revealed an enhancing effect of this element on gene expression in cis. This outcome was for us unexpected, but was acceptable, since it represented that secondary structure formation was the structural basis for RNA segments to gain regulatory function. Nevertheless, it remains unclear that which portion(s) in the distal half of the 3′-UTR is responsible for the gene repressive effect. According to our in silico prediction, one minor stem loop may be present in the distal half. Considering that the 84-bases long CAESAR element in the CCN2 3′-UTR mediates distinct repressive regulation on gene expression (Kubota et al. 2000), such a minor stem-loop can as well. Including this issue, more comprehensive analysis to illustrate the entire network and molecular mechanism of the 3′-UTR-mediated gene regulation around CCN1 is currently in progress.
Acknowledgments
This study was supported by the program Grants-in-aid for Scientific Research (S) [to M.T.], (B) [to M.T.] and (C) [to S.K.] from Japan Society for the Promotion of Science and by a grant from the Terumo Life Science Foundation [to S.K.]. We thank Ms. Yoko Tada and Ms. Eri Yashiro for their secretarial assistance.
Contributor Information
Satoshi Kubota, Phone: +81-86-2356645, FAX: +81-86-2356649, Email: kubota1@md.okayama-u.ac.jp.
Masaharu Takigawa, Phone: +81-86-2356645, FAX: +81-86-2356649, Email: takigawa@md.okayama-u.ac.jp.
References
- Andreassi S, Riccio A. To localize or not to localize: mRNA fate is in 3'UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Bai T, Chen CC, Lau LF. Matricellular protein CCN1 activates a proinflammatory genetic program in murine macrophages. J Immunol. 2010;184:3223–3232. doi: 10.4049/jimmunol.0902792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3' UTR. RNA. 2008;14:1297–1316. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbourn KP, Acharya KR, Perbal B. The CCN family of proteins: structure-function relationships. Trends Biochem Sci. 2008;33:461–473. doi: 10.1016/j.tibs.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway SE, Beck AW, Girard L, Jaber MR, Barnett CC, Jr, Brekken RA, Fleming JB. Increased expression of Cyr61 (CCN1) identified in peritoneal metastases from human pancreatic cancer. J Am Coll Surg. 2005;200:371–377. doi: 10.1016/j.jamcollsurg.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl-2 mRNA by nucleolin and A + U-rich element-binding factor 1 (AUF1) J Biol Chem. 2010;285:27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1–10. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol. 2010;12:676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011;10:945–963. doi: 10.1038/nrd3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Kubota S, Eguchi T, Hattori T, Nakanishi T, Sugahara T, Takigawa M. Characterization of a mouse ctgf 3'-UTR segment that mediates repressive regulation of gene expression. Biochem Biophys Res Commun. 2000;11:119–124. doi: 10.1006/bbrc.2000.3780. [DOI] [PubMed] [Google Scholar]
- Kubota S, Kondo S, Eguchi T, Hattori T, Nakanishi T, Pomerantz RJ, Takigawa M. Identification of an RNA element that confers post-transcriptional repression of connective tissue growth factor/hypertrophic chondrocyte specific 24 (ctgf/hcs24) gene: similarities to retroviral RNA-protein interactions. Oncogene. 2000;19:4773–4786. doi: 10.1038/sj.onc.1203835. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takigawa M. Role of CCN2/CTGF/Hcs24 in bone growth. Int Rev Cytol. 2007;147:1–41. doi: 10.1016/S0074-7696(07)57001-4. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takigawa M. CCN family proteins and angiogenesis: from embryo to adulthood. Angiogenesis. 2007;10:1–11. doi: 10.1007/s10456-006-9058-5. [DOI] [PubMed] [Google Scholar]
- Kubota S, Takigawa M. The role of CCN2 in cartilage and bone development. J Cell Commun Signal. 2011;5:209–217. doi: 10.1007/s12079-011-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A. A sticky situation: CCN1 promotes both proliferation and apoptosis of cancer cells. J Cell Commun Signal. 2010;4:71–72. doi: 10.1007/s12079-009-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3'-untranslated region (3'UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. CYR61 (CCN1) is essential for placental development and vascular integrity. Mol Cell Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritani NH, Kubota S, Sugahara T, Takigawa M. Comparable response of ccn1 with ccn2 genes upon arthritis: An in vitro evaluation with a human chondrocytic cell line stimulated by a set of cytokines. Cell Commun Signal. 2005;3:6. doi: 10.1186/1478-811X-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3' UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukudai Y, Kubota S, Eguchi T, Kondo S, Nakao K, Takigawa M. Regulation of chicken ccn2 gene by interaction between RNA cis-element and putative trans-factor during differentiation of chondrocytes. J Biol Chem. 2005;280:3136–3177. doi: 10.1074/jbc.M411632200. [DOI] [PubMed] [Google Scholar]
- Mukudai Y, Kubota S, Kawaki H, Kondo S, Eguchi T, Sumiyoshi K, Ohgawara T, Shimo T, Takigawa M. Posttranscriptional regulation of chicken ccn2 gene expression by nucleophosmin/B23 during chondrocyte differentiation. Mol Cell Biol. 2008;28:6134–6147. doi: 10.1128/MCB.00495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Weng L, Date T, Akazawa D, Tian X, Suzuki T, Kato T, Tanaka Y, Mizokami M, Wakita T, Toyoda T. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS Pathog. 2010;6:e1000885. doi: 10.1371/journal.ppat.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgawara T, Kubota S, Kawaki H, Kondo S, Eguchi T, Kurio N, Aoyama E, Sasaki A, Takigawa M. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583:1006–1010. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Ohgawara T, Kubota S, Kawaki H, Kurio N, Abd El Kader T, Hoshijima M, Janune D, Shimo T, Perbal B, Sasaki A, Takigawa M. Association of the metastatic phenotype with CCN family members among breast and oral cancer cells. J Cell Commun Signal. 2011;5:291–299. doi: 10.1007/s12079-011-0133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kelly J, Chung A, Lemp N, Chumakova K, Yin D, Wang HJ, Said J, Gui D, Miller CW, Karlan BY, Koeffler HP. Functional domains of CCN1 (Cyr61) regulate breast cancer progression. Int J Oncol. 2008;33:59–67. [PubMed] [Google Scholar]
- Perbal B, Takigawa M. CCN Protein -A new family of cell growth and differentiation regulators- London: Imperial College Press; 2005. pp. 1–311. [Google Scholar]
- Rachfal AW, Brigstock DR. Structural and functional properties of CCN proteins. Vitam Horm. 2005;70:69–103. doi: 10.1016/S0083-6729(05)70003-0. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si W, Kang Q, Luu HH, Park JK, Luo Q, Song WX, Jiang W, Luo X, Li X, Yin H, Montag AG, Haydon RC, He TC. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Mol Cell Biol. 2006;26:2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Abd El Kader T, Nishida T, Ikeda N, Shimo T, Yamashiro T, Takigawa M (2013) Novel role of miR-181a in cartilage metabolism. J Cell Biochem. doi:10.1002/jcb.24556 [DOI] [PubMed]
- Takigawa M, Tajima K, Pan HO, Enomoto M, Kinoshita A, Suzuki F, Takano Y, Mori Y. Establishment of a clonal human chondrosarcoma cell line with cartilage phenotypes. Cancer Res. 1989;49:3996–4002. [PubMed] [Google Scholar]
- Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci USA. 2010;107:9614–9619. doi: 10.1073/pnas.0912594107. [DOI] [PMC free article] [PubMed] [Google Scholar]