Abstract
Purpose
Previous studies have demonstrated the prognostic importance of the immune microenvironment in follicular lymphoma (FL). To investigate the molecular mechanisms during which tumor-infiltrating T cells (TILs) are altered in the FL microenvironment, we studied highly purified CD4 and CD8 TILs from lymph node biopsies at diagnosis in treatment-naive patients with FL compared with reactive tonsils and the peripheral blood of healthy donors.
Patients and Methods
Gene expression profiling of highly purified CD4 and CD8 TILs was performed on the Affymetrix platform. Diagnostic tissue microarrays from an independent patient set (n = 172) were used to verify protein expression and analyze any impact of TIL-expressed genes on outcome. Time-lapse imaging was used to assess T-cell motility.
Results
The most upregulated genes in both CD4 and CD8 TILs were PMCH, ETV1, and TNFRSF9. PMCH is not expressed in peripheral blood T cells, but expression is highly induced on culture with FL. Both CD4 and CD8 TILs from patients with FL have significantly impaired motility compared with those of healthy TILs from reactive tonsils and this can be induced on healthy T cells by FL cells. During multivariate analysis, a model incorporating the number and location of T cells expressing PMCH, NAMPT, and ETV1 showed prognostic significance for overall survival and for time to transformation.
Conclusion
We showed altered gene expression in TILs in FL and demonstrated that altering the immune microenvironment in FL affects overall survival and time to transformation in this disease.
INTRODUCTION
Follicular lymphoma (FL) is an indolent disease with a very heterogeneous clinical course.1 Though some patients with FL live decades without treatment, others have rapid disease progression and short survival, and 30% of patients undergo histologic transformation to diffuse, large B-cell lymphoma (DLBCL).2 The Follicular Lymphoma International Prognostic Index3 stratifies patient risk but has limitations. Recent studies have demonstrated frequent mutations in FL, particularly inactivating mutations of the MLL2 gene4 that likely drive FL development. Prognostic biomarkers based on disease biology that can accurately predict clinical behavior and aid in disease management are needed.
Cancer cells induce distinctive microenvironments conducive to cancer cell growth. The immune microenvironment plays an important role in FL outcome, and the genes and proteins expressed by infiltrating T cells and macrophages are among the most important predictors of outcome.5–10 The mechanisms through which the immune microenvironment affects FL outcome are poorly understood. Tumor cells alter cytolytic T cells11 and recruitment of protumor macrophages.12,13 CD4 and CD8 tumor-infiltrating lymphocytes (TILs) in FL have impaired function and suppressed recruitment of critical signaling proteins to the immunologic synapse.14 However, these studies shed little light on how FL cells alter the immune environment heterogeneously. To attempt to examine the mechanisms through which FL TILs affect outcome, we analyzed global gene expression profiles of highly purified CD4 and CD8 TILs from FL compared with reactive tonsil. We show altered gene expression in FL TILs and demonstrate that the number and location of TILs with altered expression of PMCH, NAMPT, and ETV1 affects overall survival (OS) and time to transformation (TT).
PATIENTS AND METHODS
Ethical Considerations and Samples
Ethical approval for the study was obtained from the East London and The City Health Authority Local Research Ethics Committee 3. Cryopreserved cell suspensions were obtained from the tissue bank of St Bartholomew's Hospital. All patients consented to storage and use of specimens for research purposes. Lymph nodes (LN) and cryopreserved single-cell suspensions were obtained from previously untreated FL patients and reactive tonsils were used as controls. See Data Supplement for further details.
Gene Expression Profiling
The Affymetrix Expression GeneChip (Affymetrix, Santa Clara, CA) protocol one-cycle procedure with Affymetrix Human Genome U133 plus 2.0 GeneChips were used with 1 μg total RNA as starting material. GeneChip data were analyzed using Partek and R software (R Project for Statistical Computing, Wien, Austria) by two independent experts with 99% similarity in the extracted results. Pathway analysis was performed using Ingenuity software.
Tissue Microarrays
Representative Tissue Microarrays (TMAs) were prepared as triplicate 1-mm diameter cores as previously described9 and in the final validation set of 172 patients at St Bartholomew's Hospital from FL LN biopsies obtained at diagnosis for whom clinical outcome was available. Reactive follicular hyperplasia lymph node samples (n = 12) were arrayed as controls. TMAs were constructed and immunohistochemistry (IHC) staining and analysis was performed.9 Suitable Abs for paraffin-embedded sections were available for pro-melanin-concentrating hormone (PMCH), PMCH variant 1 (ETV1), nicotinamide phosphoribosyltransferase (NAMPT), and CD200, and their expression was studied in more detail. Stained slides were evaluated for percentage of positive cells and expression (mean intensity) via a computerized image analysis system (Ariol, Applied Imaging Molecular Devices, Sunnyvale, CA); pathologist-trained visual parameters were employed, including location relative to the neoplastic follicle: interfollicular, intrafollicular, and total core area. Expert histopathologist analyses (A.M. and M.C.) independently validated the IHC automated analysis.
Time-Lapse Imaging
Random movement of CD4 and CD8 TILs (105 lymphocytes) were assessed on intercellular adhesion molecule–1 coated μ-Slides VI ibiTreat 80,606 (ibidi, Martinsried, Germany) slides.15 Images were taken with a Nikon BioStation IM microscope using a 20× objective lens at 20-second intervals for 1 hour. Cells were tracked and analyzed with NIS-Element software (Nikon, Melville, NY). Motility Index was calculated by the average frequency of motile cells from three microscopic fields multiplied by their average velocity (μm/S).
Statistical Analysis
Expression data were analyzed within the R statistical environment via Bioconductor packages (http://www.bioconductor.org) using stringent quality control criteria. For CD4 and CD8 subtypes, Limma was used to fit a linear model to normalize expression data for each probe to detect differentially expressed genes between healthy and FL samples. 16 False discovery rates were estimated using the Benjamini-Hochberg method.17 For the IHC data, two independent analyses (categoric and continuous data) were performed to increase the robustness of statistical inferences. Data analysis was performed by using the X-Tile statistical package (Yale University, New Haven, CT),18 enabling cut points to be determined for markers without validated normal ranges. X-Tile divides the cohort into two independent data sets (a test set and a validation set) in a 1:2 ratio, determines optimal cut points for each marker for the test set, and applies this to the validation set.19 Kaplan-Meier curves defined by these cut points were generated, and statistical significance of differences arising from differential expression of each marker were determined in the validation set using the log-rank test. Continuous data analysis assessed the prognostic effect of each of these biomarkers treated as continuous variables using Cox regression analysis. Outcomes measured from date of diagnosis to occurrence of event or date of last follow-up were OS and TT, the time from FL diagnosis to histologically confirmed diffuse, large B-cell lymphoma transformation. Kaplan-Meier plot and the Gehan-Breslow-Wilcoxon methods were used for survival analysis. Biomarkers and clinical risk factors for OS and TT were analyzed using a Cox proportional hazards regression model. For each outcome, models were formulated using forward selection, starting from the most significant clinical prognostic variable in the univariate analysis, adding factors and retaining only significant variables (P < .05) at each step. Backward selection method starting from the full model was used to check that the prognostic factors maintained significance in the final model.20 For all patients, variables included in the backward and forward selection procedures comprised age, stage, grade, and sex, as well as PMCH, NAMPT, and ETV1 number and location. The proportional hazards assumption for each predictor in the final model was assessed by testing correlation between the ranked failure times and the scaled Schoenfeld residuals. The proportional hazards assumption was not rejected in any of the models. The predictive performance of the final model was evaluated using measures of discrimination to distinguish patients with different prognostic factors, quantified using a Harrell's c (concordance) index.20 All statistical analyses were performed in STATA (STATA, College Station, TX).
RESULTS
Abnormal Gene Expression Profile of CD4 and CD8 TILs in FL
To determine the mechanism through which TILs in patients with FL are altered and affect FL outcome, we analyzed gene expression profiles of highly purified CD4 and CD8 TILs from 12 FL patients who were previously treatment-naive and compared them with seveb reactive tonsillar controls (GEO Series accession number GSE27928).21 After purification, mean purity was 98.05% for FL CD4 TILs (range, 95.8% to 99.2%), 96.15% for CD8 TILs (range, 94.1% to 98.5%); 96.95% for tonsillar CD4 cells (range, 91.5% to 99.3%), and for tonsillar CD8 cells 98.03% (range, 96.0% to 99.5). Unsupervised clustering of all samples using all differentially expressed probe sets for both CD4 and CD8 TILs correctly distinguished healthy and FL samples. Using a P value cutoff of .05, log2-fold change cutoff of less than 1 or more than 1, and a false discovery rate of 0.05 for the derived gene lists, 182 genes were upregulated and 85 genes downregulated in CD4 cells; 481 genes were upregulated and 163 genes downregulated in CD8 cells (Fig 1; Data Supplement). Pathway analysis is also shown in the Data Supplement. In both CD4 and CD8 TILs, 109 genes were deregulated in common; 95 were upregulated but only 13 downregulated (Data Supplement).
Fig 1.
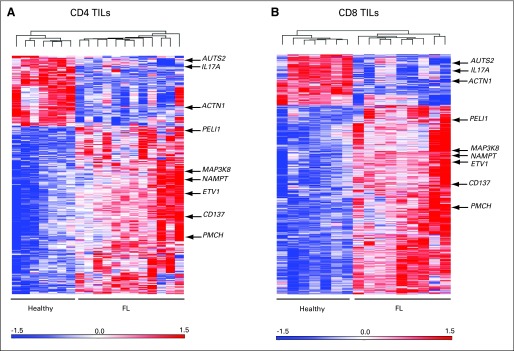
Heat maps showing differentially expressed genes in highly pure tumor-infiltrating lymphocytes (TILs) CD8 and CD4 from patients with follicular lymphoma (FL) at diagnosis compared with healthy donors. (A) Dendrogram of differentially expressed genes by unsupervised analysis (P < .05; false discovery rates < 0.05) showed 182 genes were significantly upregulated (red) and 85 genes were significantly downregulated (blue) in CD4 cells from patients with FL. (B) CD8 cells showed 481 genes were significantly upregulated and 163 genes downregulated in patients with FL.
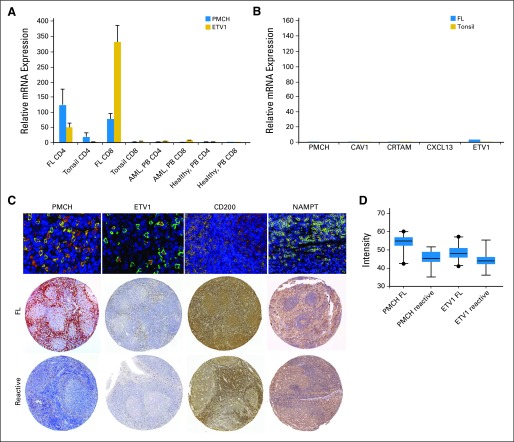
Altered mRNA expression was validated using quantitative real-time polymerase chain reaction, and representative results for PMCH and ETV1 are shown in Figure 2. Genes altered on FL TILs were barely detectable on FL (n = 12) or tonsillar B cells (n = 10; Fig 2B) and are detected at much lower levels in tonsillar T cells or peripheral blood (PB) CD4 or CD8 cells in healthy individuals (Fig 2). We demonstrated altered protein expression of PMCH, ETV1, CD200, and NAMPT by dual-staining immunohistochemistry (Fig 2C), using TMAs from a well-characterized independent cohort of 172 treatment-naive FL patients. Median follow-up time for these patients was 11.48 years (range, 3.67 to 35.77 years), and 41 of these patients' disease subsequently progressed (Data Supplement). This approach requires monoclonal antibodies (mAbs) that stain reliably in paraffin-embedded tissue. We focused attention on PMCH, NAMPT, CD200, and ETV1 (Data Supplement). Although there is considerable interest in TNFRSF9 in FL, we could not overcome technical difficulties in the TMA, even after using three different mAbs. Mean intensity (MI) expression in TMA for PMCH, NAMPT, and ETV1 was significantly higher in FL patients (n = 172) compared with those of reactive LN (n = 12; P < .001; Figs 2C and 2D).
Fig 2.
Validation of gene expression observed by microarray. The results of quantitative real time polymerase chain reaction are concordant with data seen on microarray. The figure shows results of (A) PMCH and ETV1 as representative data for five investigated genes in CD4 and CD8 tumor-infiltrating lymphocytes from previously untreated follicular lymphoma patients (FL; n = 12) compared with those from tonsil (n = 10), peripheral blood (PB) of acute myeloid leukemia patients (AML; n = 10), and healthy age-matched donors (n = 10). (B) The examined genes showed low expression in the CD19+ cells of tonsils (n = 7) and FL cells (n = 12). (C) The merged fluorescence double staining of CD3 (green) and PMCH, ETV1, CD200, or NAMPT (red) in lymph nodes (LN) of a patient with FL that represents the double staining in 10 different FL patients. (D) The gene array results were also concordant with the results of tissue microarrays (TMAs) stained for PMCH, ETV1, CD200, and NAMPT (Visfatin-1) comparing FL patients (top raw) and reactive LN (down raw). Figure is representative of 172 FL patients and 12 reactive LN analyzed. Comparison of mean intensity expression of PMCH and ETV1in FL patients (n = 172) with reactive LN (n = 12).
Coculturing Healthy Allogeneic T Cells With FL Cells
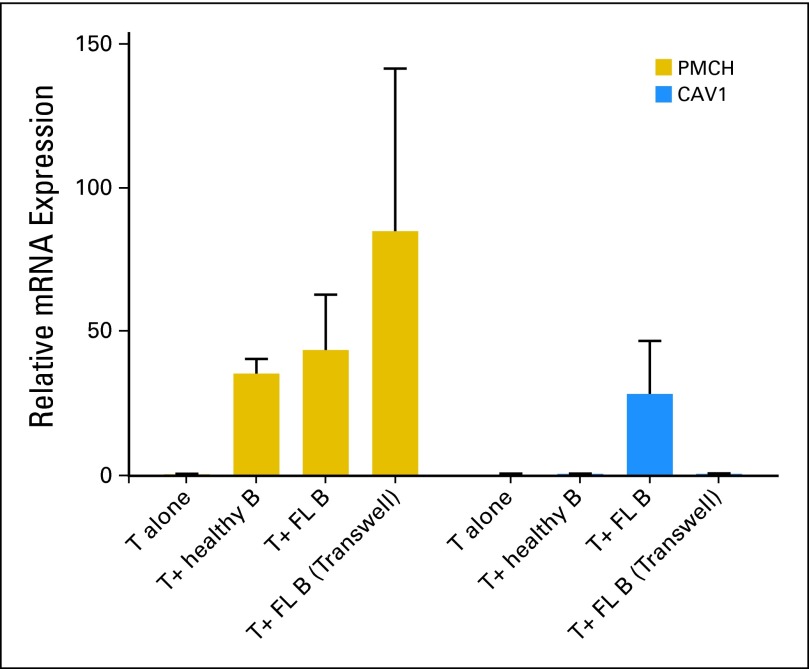
To determine whether the FL cells directly alter TIL protein expression, healthy PB allogeneic T cells were cocultured alone, with purified FL cells (n = 7), or with tonsillar B cells (n = 4) in cell-cell contact or in transwell cultures for 48 hours. PMCH, ETV1, CAV1, CRTAM, and CXCL13 mRNA levels were quantified by quantitative real-time polymerase chain reaction from purified T cells. PMCH expression was induced highly by coculture with FL cells (Fig 3). Optimal induction of both PMCH and ETV1 expression occurred in transwell culture (data not shown), suggesting induction by soluble factors, whereas CAV1 expression on T cells was induced by direct culture with FL cells and not with healthy B cells (Fig 3). Expression of CRTAM on T cells was not altered by coculture with FL cells but was suppressed by coculture with healthy B cells. Expression of CXCL13 was not altered under any of coculture conditions studied (data not shown).
Fig 3.
Relative fold change in mRNA expression results (by qualitative reverse-transcriptase–polymerase chain reaction) for PMCH and CAV1 genes in allogeneic healthy T cells cultured alone (n = 4) or cocultured with follicular lymphoma (FL) cells (n = 7) and healthy B cells (n = 4) for 48 hours in cell-cell contact or in transwell experiment (n = 4).
Expression of Melanin-Concentrating Hormone Receptor 2 by Tissue Macrophages
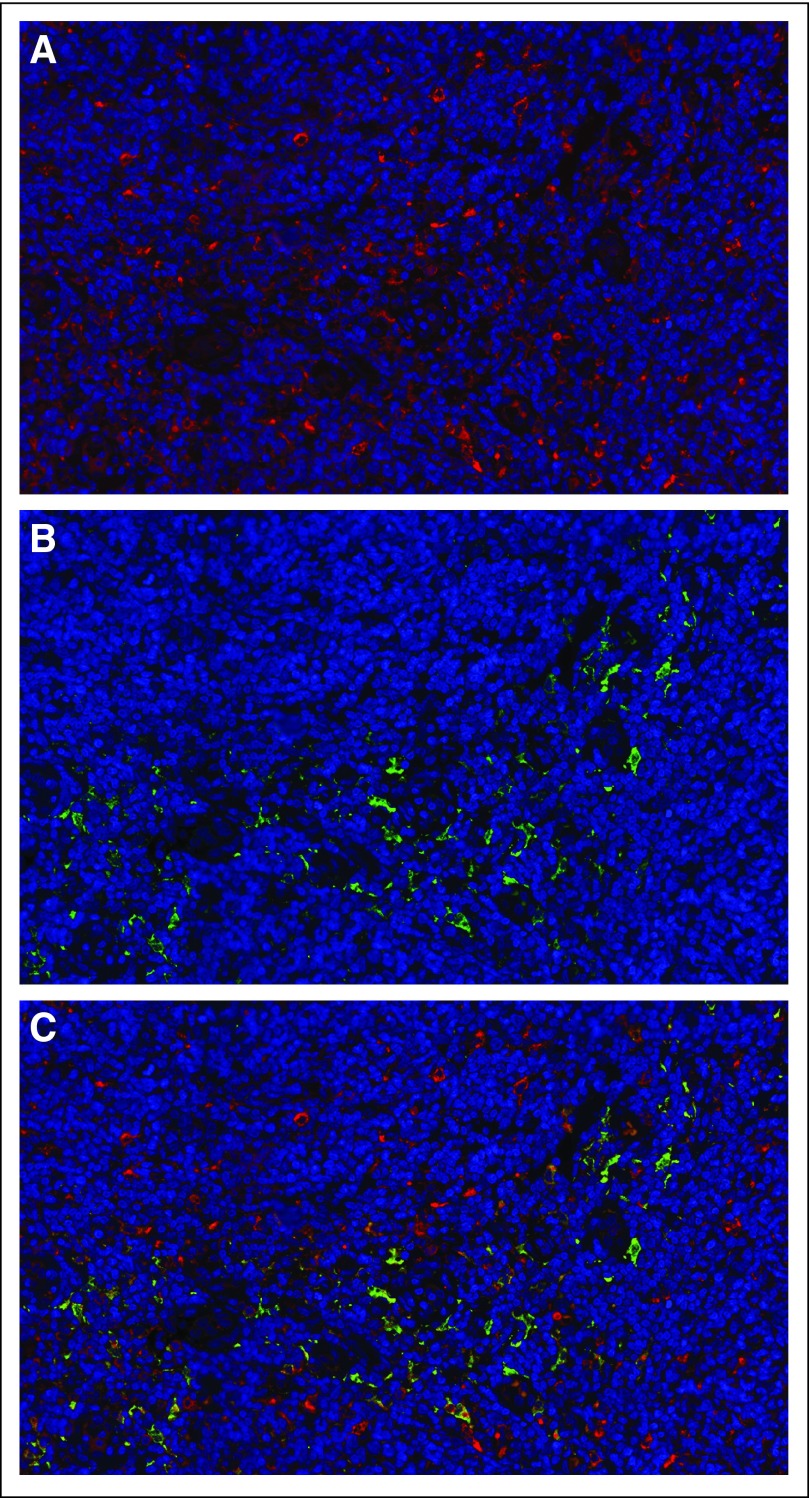
PMCH was the most highly differentially expressed gene in both CD4 and CD8 FL TILs. Melanin-concentrating hormone (MCH) serum level did not differ in patients with FL (n = 40) compared with healthy donors (n = 10; P = .196; data not shown). Although no MCH receptor expression was observed on the FL cells or TILs, dual staining demonstrated expression of MCH receptor 2 (MCHR2), but not MCHR1, on a subset of tumor-infiltrating macrophages (Fig 4) with higher level of expression in the interfollicular versus intrafollicular areas (data not shown). MCHR2 was also expressed on a subset of tonsillar macrophages.
Fig 4.
Expression of MCHR2 by interfollicular tissue macrophages. Staining of lymph node (LN) paraffin-embedded section using (A) anti-CD68 TexRed conjugated secondary Ab and (B) anti-MCHR2 and FITC-conjugated secondary Ab. (C) A combination of (A) and (B), which shows the specific expression of MCHR2 by interfollicular tissue macrophages. This is representative of 10 follicular lymphoma patients and five reactive LNs examined.
Motility of CD4 and CD8 TILs From FL Patients Are Drastically Impaired
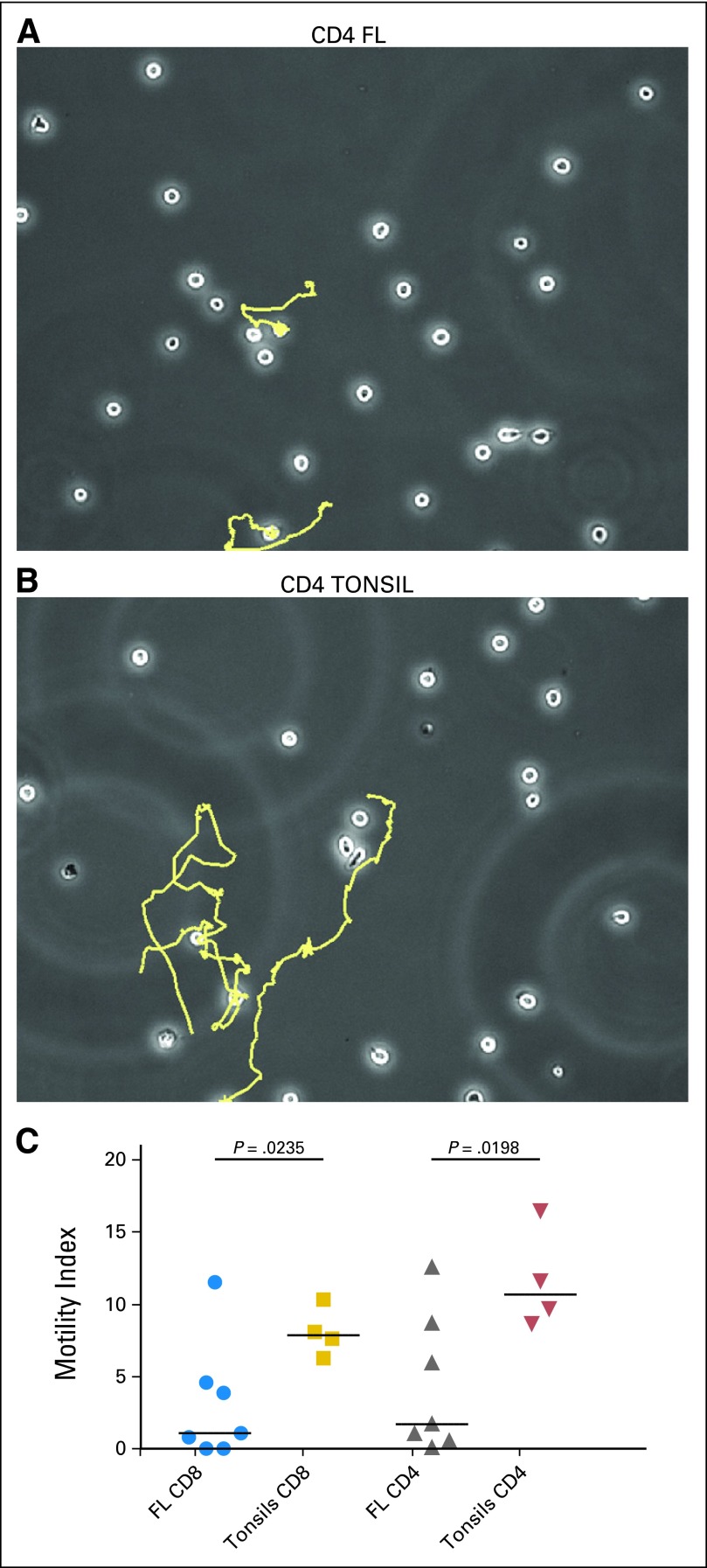
ACTN1 was among the most down-regulated genes in FL TILs and analysis highlighted disruption in actin-based motility and cytoskeleton signaling pathways, similar to that seen in our previous T cell studies in FL,14 AML22 and CLL.23 We performed migration tracking on sorted CD4 and CD8 TILs from treatment naive FL patients (n = 7) compared with tonsillar controls (n = 4) to assess the functional effects of tumor cell induced actin-based motility disruption. Compared to tonsillar T cells, both CD4 and CD8 FL TILs had significantly impaired motility index scores (P < .025) (Fig 5). Movies of representative T cells from FL and tonsil are presented in supplemental data. A significant reduction in motility index of T cells (P = .0002) was seen after coculture of healthy allogeneic T cells with FL cells compared with tonsillar B cells (data not shown).
Fig 5.
Tracking T cell motility on the intercellular adhesion molecule (ICAM-1) coated slides. Shown are representative paths of the movements of CD4 tumor-infiltrating lymphocytes (TILs) from (A) a patient with FL and (B) a healthy donor. (C) Severely impaired Motility Index, calculated via time-lapse imaging of CD4 and CD8 TILs from patients with follicular lymphoma (FL; n = 7) compared with those from reactive tonsils (n = 4).
Impact of PMCH, ETV1, and NAMPT Expression and Clinical Outcome
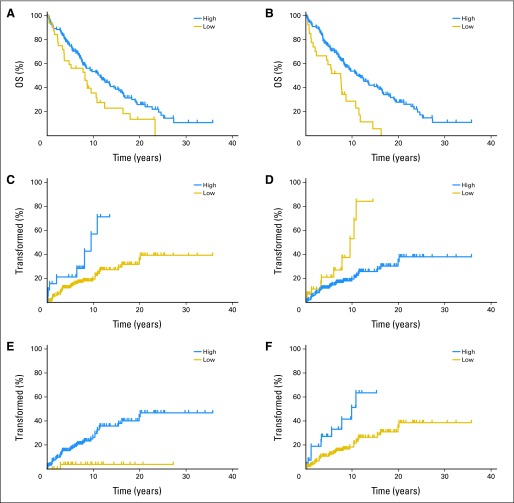
We examined the clinical significance of altered expression of PMCH, ETV1, and NAMPT on FL CD4 and CD8 TILs using IHC on the FL TMA. The value of this approach is that it allows examination of not only the number of TILs expressing the altered protein but also their location. We examined the number of T cells expressing each protein of interest within the malignant follicle (intrafollicular), in the interfollicular area, and overall. The results demonstrate the complexity of interactions between FL and TILs. Higher number of TILs expressing PMCH in the intrafollicular (P = .03; Fig 6A) or interfollicular (P = .0003; Fig 6B) areas was associated with improved OS and disease-specific survival (data not shown). The same was true when TILs were analyzed for percentage expression of cells or MI for PMCH (data not shown). This difference was maintained independently of previous rituximab treatment (data not shown). Higher number of NAMPT-expressing TILs was associated with improved OS in both the intrafollicular (P = .02) and interfollicular (P = .0087) areas. The results for ETV1 suggest a more complex, location-dependent interaction. Patients with a high number of intrafollicular ETV1-expressing TILs had poor OS (P = .045), whereas patients who had a high number of interfollicular ETV1-expressing TILs had improved OS (P = .03). In multivariate analysis, none of the proteins examined alone retained independent significance for OS (Data Supplement). However, we were able to build a model based on a combination of these biomarkers, with the best model being the number of PMCH- and NAMPT-expressing cells in the interfollicular area plus the ratio of ETV1 cells in the interfollicular/intrafollicular area, with a high combined score identifying patients with improved OS (hazard ratio, 0.32; 95% CI, 0.1 to 0.61; P = .007).
Fig 6.
Overall survival (OS) and time to transformation (TT) of patients with follicular lymphoma (FL) according to high versus low expression of examined proteins at time of diagnosis. Number of positive cells for PMCH expression (P = .03) in (A) intrafollicular and (B) interfollicular area (P = .0002). TT of patients with FL according to number of PMCH-expressing cells in (C) intrafollicular (P = .029) and (D) interfollicular area (P = .033). TT for the same patients for (E) number of ETV1-expressing cells in intrafollicular area (P = .02) and (F) mean intensity of ETV1 expression in interfollicular area (P = .0005).
A high number of PMCH-expressing TILs in the intrafollicular area (P = .029) and a low number in the interfollicular area (P = .033) was associated with shorter TT (Figs 6C and 6D). High MI level of NAMPT expression in the intrafollicular area was associated with longer TT (P = .0034; data not shown). A higher number of ETV1-expressing cells in the intrafollicular area (P = .02) and higher MI level in the interfollicular area (P = .0005) were associated with shorter TT (Figs 6E and 6F). In multivariate analysis, the number of PMCH-expressing cells in the interfollicular area (95% CI, 0.1 to 0.71; P = .008) and MI of NAMPT in intrafollicular area (95% CI, 0.15 to 0.86; P = .021) were independently associated with predicted prognosis of TT (Data Supplement). A model combining the ratio of PMCH-expressing cells in the interfollicular/intrafollicular area plus a high level of expression of NAMPT and a low level of expression of ETV1 in the intrafollicular area best identified patients with longer TT (hazard ratio, 0.19; 95% CI, 0.09 to 0.47; P = .003).
DISCUSSION
An important hallmark of tumor cells is their ability to evade immune recognition. Understanding the relationships between neoplastic cells and the immune microenvironment should help facilitate improved therapeutic interventions. To determine the molecular mechanisms through which FL cells alter the function of tumor-infiltrating T cells, we performed gene-expression profiling on highly pure CD4 and CD8 TILs and demonstrated profound changes in gene expression.
The number, percentage, mean intensity of expression, and location of TILs expressing PMCH, ETV1, and NAMPT in an FL TMA had potential prognostic significance that was seen on univariate analysis. But the number, as well as the location of cells, had to be taken into account in multivariate analysis to allow for the complex impact on prognosis dependent on location of T cells expressing these altered genes. The differing patterns of expression of the same protein on T cells having an impact on OS and TT suggests that disease progression and transformation may be differentially regulated by the immune microenvironment. How these cells affect other cells specific to the tumor microenvironment, including T regulatory cells and tumor-associated macrophages, form the basis of ongoing studies.
The high expression of PMCH by TILs in patients and its potential association with survival and transformation suggests it may play an important role in FL. The highest normal PMCH transcript level is observed in the brain,24 where it may play a role in appetite regulation.25–27 PMCH is proteolytically processed by tissue-specific convertases to form several active peptides, including the orexigenic peptide MCH.28 The role of the other PMCH-derived peptides, neuropeptide EI and neuropeptide-glycine-glutamic acid, is not well understood and there are no published immune-related functions. However, MCH has an immunomodulatory effect, it inhibits proliferation of CD3-stimulated and nonstimulated PBMC, and it reduces IL-2 release.29 In vitro–differentiated human Th2 cells highly selectively express PMCH.30 Its main receptor, MCHR1, is expressed by human PB granulocytes, monocytes, T cells, and B cells.31 We observed MCHR2, but not MCHR1expression in FL lymph nodes, and double staining demonstrated specificity for tissue macrophages, especially in the interfollicular areas. We postulate that FL induces T cell production of MCH, which binds to the surrounding MCHR2-expressing tissue macrophages, a novel way that malignant cells shape their immune microenvironment. Increased tissue macrophages are correlated with worse prognosis in FL,6,32 and we are investigating MCH-induced macrophage alteration. The molecular mechanisms through which ETV1 and NAMPT affect FL biology and outcome remain unknown. ETS transcription factors appear critical regulators of oncogenesis by different mechanisms in gastrointestinal stromal tumors,33 prostate cancer,34 melanoma,35 and breast cancer.36 However, ETV1 expression has not previously been described in T cells, and its role in FL TILs requires further investigation.
The disruption in actin-based motility and cytoskeleton polarization in TILs is in keeping with our previous studies of FL,14 acute myeloid leukemia,22 and chronic lymphatic leukemia,23 in which we have shown markedly decreased actin polymerization at T cell immunologic synapse with reduced recruitment of phosphotyrosine-signaling molecules.14,22,37 We recently characterized that cell surface expression of regulatory molecules, including CD200, induce defective actin polymerization in hematologic malignancies, including FL and in solid tumors.38 In this article, we show that FL-induced actin changes also impair TIL motility, potentially a novel mechanism to impair effective antitumor immune response by TILs, and that FL cells can induce defects in healthy allogeneic T cells. These findings have implications for autologous and allogeneic immunotherapy treatment approaches, and clinical trials using immunomodulatory drugs that can repair T cell defects14,37 are ongoing in patients with FL.
In conclusion, our findings improve our understanding of the impact of tumor-infiltrating T cells in the microenvironment, identify molecular pathways that are altered, and introduce novel genes that may affect FL biology and outcome. These results contribute to our understanding of the complex interactions of lymphoma cells, TILs, and macrophages in their microenvironment and help us generate hypotheses. But until we have a better understanding of these interactions, it does not yet seem feasible to incorporate IHC analysis of TILs in FL for prognosis. However, because nonmalignant infiltrating immune cells play a crucial role in outcomes in FL, understanding the nature and impact of the abnormalities induced in TILs in these patients is vital before any immunotherapeutic strategies can be implemented to alter the immune microenvironment in FL.
Supplementary Material
Acknowledgment
We thank Farideh Miraki-Moud, PhD, and Andrew Owen for their technical assistance.
Footnotes
See accompanying editorial on page 2641
Supported by Grant No. C1574/A6806 from Cancer Research UK (J.G.G.) and Grant No. P01 CA81538 from the National Cancer Institute.
Presented in part at the American Society of Hematology in Orlando, FL, December 4-7, 2010, and San Diego, CA, December 10-13, 2011, and at 12th International Conference on Malignant Lymphoma, Lugano, Switzerland, June 15-18, 2011.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) and/or an author's immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: John G. Gribben, Roche/Genentech, Celgene Research Funding: None Expert Testimony: None Patents: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Shahryar Kiaii, John G. Gribben
Collection and assembly of data: Shahryar Kiaii, Andrew J. Clear, Derek Davies, John G. Gribben
Data analysis and interpretation: Shahryar Kiaii, Andrew J. Clear, Alan G. Ramsay, Ajanthah Sangaralingam, Abigail Lee, Maria Calaminici, Donna S. Neuberg, John G. Gribben
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Gribben JG. How I treat indolent lymphoma. Blood. 2007;109:4617–4626. doi: 10.1182/blood-2006-10-041863. [DOI] [PubMed] [Google Scholar]
- 2.Montoto S, Fitzgibbon J. Transformation of indolent B-cell lymphomas. J Clin Oncol. 2011;29:1827–1834. doi: 10.1200/JCO.2010.32.7577. [DOI] [PubMed] [Google Scholar]
- 3.Federico M, Bellei M, Marcheselli L, et al. Follicular lymphoma International Prognostic Index 2: A new prognostic index for follicular lymphoma developed by the International Follicular Lymphoma Prognostic Factor project. J Clin Oncol. 2009;27:4555–4562. doi: 10.1200/JCO.2008.21.3991. [DOI] [PubMed] [Google Scholar]
- 4.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 6.Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL) Blood. 2005;106:2169–2174. doi: 10.1182/blood-2005-04-1565. [DOI] [PubMed] [Google Scholar]
- 7.de Jong D. Molecular pathogenesis of follicular lymphoma: A cross talk of genetic and immunologic factors. J Clin Oncol. 2005;23:6358–6363. doi: 10.1200/JCO.2005.26.856. [DOI] [PubMed] [Google Scholar]
- 8.Alvaro T, Lejeune M, Salvadó MT, et al. Immunohistochemical patterns of reactive microenvironment are associated with clinicobiologic behavior in follicular lymphoma patients. J Clin Oncol. 2006;24:5350–5357. doi: 10.1200/JCO.2006.06.4766. [DOI] [PubMed] [Google Scholar]
- 9.Lee AM, Clear AJ, Calaminici M, et al. Number of CD4+ cells and location of forkhead box protein P3-positive cells in diagnostic follicular lymphoma tissue microarrays correlates with outcome. J Clin Oncol. 2006;24:5052–5059. doi: 10.1200/JCO.2006.06.4642. [DOI] [PubMed] [Google Scholar]
- 10.Glas AM, Knoops L, Delahaye L, et al. Gene-expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25:390–398. doi: 10.1200/JCO.2006.06.1648. [DOI] [PubMed] [Google Scholar]
- 11.Curiel TJ. Regulatory T-cell development: Is Foxp3 the decider? Nat Med. 2007;13:250–253. doi: 10.1038/nm0307-250. [DOI] [PubMed] [Google Scholar]
- 12.Hagemann T, Lawrence T, McNeish I, et al. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallam S, Escorcio-Correia M, Soper R, et al. Activated macrophages in the tumour microenvironment-dancing to the tune of TLR and NF-kappaB. J Pathol. 2009;219:143–152. doi: 10.1002/path.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsay AG, Clear AJ, Kelly G, et al. Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: Implications for the tumor microenvironment and immunotherapy. Blood. 2009;114:4713–4720. doi: 10.1182/blood-2009-04-217687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson L, Howarth K, McDowall A, et al. Leukocyte adhesion deficiency-III is caused by mutations in KINDLIN3 affecting integrin activation. Nat Med. 2009;15:306–312. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 18.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 19.Altman DG, Lausen B, Sauerbrei W, et al. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- 20.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.National Center for Biotechnology Information. GEO Accession Viewer: GEO Series accession number GSE27928. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse27928.
- 22.Le Dieu R, Taussig DC, Ramsay AG, et al. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114:3909–3916. doi: 10.1182/blood-2009-02-206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Görgün G, Holderried TA, Zahrieh D, et al. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bittencourt JC, Presse F, Arias C, et al. The melanin-concentrating hormone system of the rat brain: An immuno- and hybridization histochemical characterization. J Comp Neurol. 1992;319:218–245. doi: 10.1002/cne.903190204. [DOI] [PubMed] [Google Scholar]
- 25.Bradley RL, Kokkotou EG, Maratos-Flier E, et al. Melanin-concentrating hormone regulates leptin synthesis and secretion in rat adipocytes. Diabetes. 2000;49:1073–1077. doi: 10.2337/diabetes.49.7.1073. [DOI] [PubMed] [Google Scholar]
- 26.Marsh DJ, Weingarth DT, Novi DE, et al. Melanin-concentrating hormone 1 receptor-deficient mice are lean, hyperactive, and hyperphagic and have altered metabolism. Proc Natl Acad Sci U S A. 2002;99:3240–3245. doi: 10.1073/pnas.052706899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mul JD, Yi CX, van den Berg SA, et al. Pmch expression during early development is critical for normal energy homeostasis. Am J Physiol Endocrinol Metab. 2010;298:E477–E488. doi: 10.1152/ajpendo.00154.2009. [DOI] [PubMed] [Google Scholar]
- 28.Nahon JL, Presse F, Bittencourt JC, et al. The rat melanin-concentrating hormone messenger ribonucleic acid encodes multiple putative neuropeptides coexpressed in the dorsolateral hypothalamus. Endocrinology. 1989;125:2056–2065. doi: 10.1210/endo-125-4-2056. [DOI] [PubMed] [Google Scholar]
- 29.Coumans B, Grisar T, Nahon JL, et al. Effect of ppMCH derived peptides on PBMC proliferation and cytokine expression. Regul Pept. 2007;143:104–108. doi: 10.1016/j.regpep.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Sandig H, McDonald J, Gilmour J, et al. Human Th2 cells selectively express the orexigenic peptide, pro-melanin-concentrating hormone. Proc Natl Acad Sci U S A. 2007;104:12440–12444. doi: 10.1073/pnas.0705457104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verlaet M, Adamantidis A, Coumans B, et al. Human immune cells express ppMCH mRNA and functional MCHR1 receptor. FEBS Lett. 2002;527:205–210. doi: 10.1016/s0014-5793(02)03232-5. [DOI] [PubMed] [Google Scholar]
- 32.Canioni D, Salles G, Mounier N, et al. High numbers of tumor-associated macrophages have an adverse prognostic value that can be circumvented by rituximab in patients with follicular lymphoma enrolled onto the GELA-GOELAMS FL-2000 trial. J Clin Oncol. 2008;26:440–446. doi: 10.1200/JCO.2007.12.8298. [DOI] [PubMed] [Google Scholar]
- 33.Chi P, Chen Y, Zhang L, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 35.Jané-Valbuena J, Widlund HR, Perner S, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70:2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin S, Bosc DG, Ingle JN, et al. Rcl is a novel ETV1/ER81 target gene upregulated in breast tumors. J Cell Biochem. 2008;105:866–874. doi: 10.1002/jcb.21884. [DOI] [PubMed] [Google Scholar]
- 37.Ramsay AG, Johnson AJ, Lee AM, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118:2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsay AG, Clear AJ, Fatah R, et al. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: Establishing a reversible immune evasion mechanism in human cancer. Blood. 2012;120:1412–1421. doi: 10.1182/blood-2012-02-411678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.