Abstract
Arabidopsis trichomes are large branched single cells that protrude from the epidermis. The first morphological indication of trichome development is an increase in nuclear content resulting from an initial cycle of endoreduplication. Our previous study has shown that the C2H2 zinc finger protein GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome initiation in the inflorescence organ and for trichome branching in response to gibberellic acid signaling, although GIS gene does not play a direct role in regulating trichome cell division. Here, we describe a novel role of GIS, controlling trichome cell division indirectly by interacting genetically with a key endoreduplication regulator SIAMESE (SIM). Our molecular and genetic studies have shown that GIS might indireclty control cell division and trichome branching by acting downstream of SIM. A loss of function mutation of SIM signficantly reduced the expression of GIS. Futhermore, the overexpression of GIS rescued the trichome cluster cell phenotypes of sim mutant. The gain or loss of function of GIS had no significant effect on the expression of SIM. These results suggest that GIS may play an indirect role in regulating trichome cell division by genetically interacting with SIM.
Keywords: Arabidopsis thaliana, GLABROUS INFLORESCENCE STEMS (GIS), Endoreduplication, SIAMESE (SIM), Trichome branching, Genetic interaction
1. Introduction
Trichomes are present on most aerial organs of the plant including leaves, stems, and flower in Arabidopsis, and exist as a parrying barrier to protect plants from biotic and abiotic stress (Johnson, 1975; Marks, 1997; Rodney and Mark, 1997). Trichome cells provide an excellent model system to study the interplay of different development processes and cellular functions (Hülskamp et al., 1994). Trichomes are initiated in the epidermis of developing organs, and their distribution is well regulated; in wild type (WT) leaves, the clustering of trichomes is rare (Hülskamp et al., 1994; Larkin et al., 2003). Cells destined to become trichomes stop mitotic division and enter into an endoreduplication cycle. The trichome cells undergo four endoreduplication cycles, resulting in protrusion from the leaf surface, which is followed by two successive branching events (Hülskamp et al., 1994; 1999; Schnittger et al., 2002a; Larkin et al., 2003; Bramsiepe et al., 2010). Ultimately, the mature trichome develops into a three-branched structure with a DNA content of 32C. The process of trichome development is complex and involves many different genes that regulate their spacing, density, and morphology (Hülskamp et al., 1994; Szymanski and Marks, 1998; Hülskamp et al., 1999; Larkin et al., 2003; Bramsiepe et al., 2010). SIAMESE (SIM) is a plant-specific cyclin-dependent kinase (CDK) inhibitor, and acts as a key regulator required for endoreduplication during trichome development (Walker et al., 2000; Churchman et al., 2006). SIM is considered to function as a repressor of cellular division in the endoreduplication cell cycle, because loss-of-function mutations of SIM display multicellular trichomes, in contrast to the unicellular trichome of WT plants (Walker et al., 2000; Churchman et al., 2006). The SIM protein has a motif similar to inhibitor/interactor of cyclin-dependent kinase/Kip-related proteins (ICK/KRP) proteins, and can interact with D-type cyclins (CYCDs) and A-type CDK (CDKA) (Churchman et al., 2006), which are typically considered to function at the G1/synthesis transition (Schnittger et al., 2002a). The overexpression of CYCD in trichomes produces multicellular trichomes and the sim mutant phenotype is rescued when CDK inhibitor ICK1/KRP1 is expressed in the trichomes (Weinl et al., 2005). These results suggest that SIM acts within the CYCD-CDKA complexes to repress mitotic cycles. In addition, overexpression of the B-type cyclin (CYCB), which regulates the G2/M transition (Lilly and Duronio, 2005), results in a phenotype similar to that of the sim mutant (Schnittger et al., 2002b). The detection of the CYCB transcript in the sim mutant, but not WT trichomes, indicates that SIM may inhibit CYCB expression (Schnittger et al., 2002b). Furthermore, SIM has been shown to cooperate with CCS52/Fizzy-Related (FZR) family protein, which functions as an activator of anaphase-promoting complex, to establish endoreduplication in the trichome (Cebolla et al., 1999; Kasili et al., 2010).
Mutations that affect the number of endoreduplication cycles also lead to trichomes with supernumerary or reduced numbers of branches (Hülskamp et al., 1994). Generally, DNA content correlates well with trichome branching, such that mutants with a reduced DNA content have fewer branches, and mutants with increased ploidy levels have more branches (Hülskamp et al., 1994; Folkers et al., 1997; Bramsiepe et al., 2010). GLABRA3 (GL3) and TRIPTYCHON (TRY) regulate not only trichome initiation, but also endoreduplication cycles (Hülskamp et al., 1994; Szymanski and Marks, 1998; Payne et al., 2000). GL3, which encodes a helix-loop-helix transcription factor, is thought to facilitate the rapid growth of the trichome cell; mutation of the gl3 locus results in an average DNA content of 16C, reduced trichome branching, and reduced trichome size (Hülskamp et al., 1994). Further evidence indicates that GL3 acts upstream of FURCA4 (FRC4), which positively regulates the trichome branching (Luo and Oppenheimer, 1999; Plett et al., 2009). TRY, which encodes a small single R3-MYB-repeat protein (Szymanski and Marks, 1998), regulates lateral suppression during the process of trichome fate determination. Trichomes of the try mutant have increased branching and an average DNA content of 64C, which suggests an additional endoreduplication cycle has occurred (Szymanski and Marks, 1998; Rao and Venkatachalam, 1999; Schellmann et al., 2002).
Genes which influence the branch number without affecting endoreduplication have also been identified. The STICHEL (STI) gene, which encodes a novel protein containing a domain similar to eubacterial DNA-polymerase III γ subunits (Bertram et al., 1998; Ilgenfritz et al., 2003), is considered to be one of the most important contributors for trichome branching. All trichomes are unbranched in the sti mutant, but endoreduplication levels were not affected in both sti mutants and STI overexpressors (Ilgenfritz et al., 2003). BRANCHLESS TRICHOMES (BLT) gene, which encodes a protein of hitherto unknown function, was also reported to play a key role in trichome branch cell endoreduplication (Kasili et al., 2010). blt mutant was found to enhance the multicellular trichome phenotype of mutants in the SIAMESE (SIM) gene and BLT interacts both genetically and physically with STI to regulate trichome branch, although blt mutants show normal trichome DNA content (Kasili et al., 2010).
Previously, we reported the discovery and identification of four Arabidopsis C2H2 zinc finger protein genes, GIS, GIS2, ZFP5, and ZFP8, which through hormone signalling, stimulate trichome initiation in inflorescence organs and cauline leaves (Gan et al., 2006; 2007). As stated above, in general, a correlation exists between the trichome branch number and the DNA content (Hülskamp et al., 1994; Folkers et al., 1997; Bramsiepe et al., 2010); however, GIS functions to suppress trichome branching, but it not required for endoreduplication (An et al., 2012). Thus, the aim of this study is to investigate further the role of GIS in the regulation of trichome cell division, by exploring the genetic interaction between GIS and SIM.
2. Materials and methods
2.1. Plant materials and growth condition
Arabidopsis thaliana ecotype Columbia Col-0 was used for all experiments. Seeds were surface-sterilized and sown on Murashige and Skoog (MS) medium containing 4.4 g/L MS powder (Sigma), 1% sucrose (Sigma), and 0.5% agar. Seeds were imbibed for 2 d at 4 °C, before being transferred to long day conditions (16 h light/8 h dark) at 22 °C. Seedlings of a similar size were transferred to soil 7–8 d after germination on MS media (Gan et al., 2006). Trichome branching phenotypes were analyzed at 35 d after sowing, when the plants had bolted. The numbers of trichome branches on the adaxial surface of the 3rd, 4th, and 8th rosette leaves were recorded. At least 16 plants per genotype were used for the trichome branch counting analysis. All experiments were duplicated. The sim mutant was a gift from Prof. Martin HÜLSKAMP (University of Cologne, Germany).
2.2. Construction of double mutant
The gis sim double mutant was constructed by crossing gis and sim homozygous mutants. F2 populations were screened for homozygous double mutants on selective MS medium containing sulfadiazine (50 mg/L) for gis mutant, and then verified by genomic polymerase chain reaction (PCR). The homozygous double mutant was also self-crossed to ensure that the homozygous double mutant gis sim did not exhibit a trichome segregation phenotype.
2.3. RNA extraction and real-time reverse transcription PCR (RT-PCR)
RNA was extracted using the TRIzol™ reagent (Invitrogen) according to the manufacturer’s instructions. All RNA was isolated from pooled samples of at least eight 21-d old plants. The primers used for quantitative RT-PCR analysis were as follows: GIS (GISQF1: 5′-TTCATGAACGTCGAATCCTTCTC-3′; GISQR1: 5′-ACGAATGGGTTTAGGGTTCTTATCT-3′), SIM (SIMQF: 5′-CCATCTTGAATTTCCCACCAGCCATC-3′; SIMQR: 5′-GCAGCCGCCGCCGTCATCATCTC-3′), UBQ10 (UBQ10F1: 5′-GGTTCGTACCTTTGTCCAAGCA-3′; UBQ10R1: 5′-CCTTCGTTAAACCAAGCTCAGTATC-3′). cDNA synthesis and RT-PCR were conducted as described by Gan et al. (2010). Optimization experiments were performed to establish the optimal concentration of primers. Melt curve analysis and gel electrophoresis of the PCR products were used to confirm the absence of non-specific amplification products. Transcripts from an ubiquitin gene (UBQ10) were detected and used as an endogenous control. Relative expression levels were calculated by subtracting the C t (threshold cycle) values for UBQ10 from those of the target gene (to give ∆C t) and then calculating 2−∆Ct (Gan et al., 2005).
2.4. Molecular cloning
The cloning of the 35S:GIS construct has been previously reported (Gan et al., 2006). In brief, the 35S:GIS construct was introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium-mediated transformation of all Arabidopsis genotypes was performed using the floral dip method (Clough and Bent, 1998), and transgenic seeds were selected using hygromycin (Gan et al., 2006).
2.5. Statistical analyses
All presented data were tested for significance by means of analysis of variance using Statistix program Version 3.5 (Analytical Software, USA). A Student’s t-test was calculated at the probability of either 5% (P<0.05) or 1% (P<0.01) as previously described (Bao et al., 2011; Gan et al., 2011).
3. Results
3.1. Knockout of SIM affects the expression of GIS
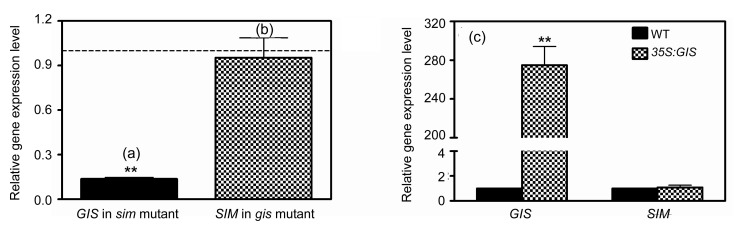
The expression of GIS in the sim mutant was determined, which was significantly reduced in sim mutant plants in comparison to that in WT plants (Fig. 1a). However, the expression of SIM in GIS loss of function mutants was not significantly different from the WT control (Fig. 1b). The expression of SIM in 35S:GIS transgenic plants was also measured and results show that the SIM transcript was not significantly affected (Fig. 1c). These results suggest that GIS acts downstream of SIM to regulate trichome branching.
Fig. 1.
Gene expression patterns of GIS and SIM
(a) GIS relative expression level in the sim mutant in comparison to that of the wild type; (b) SIM relative expression level in gis mutant in comparison to that of the wild type; (c) Expression levels of GIS and SIM in 35S:GIS transgenic plants in comparison to those of the wild type. The relative gene expression value (mean±standard deviation (SD), n=3) was calculated by using UBQ10 as the housekeeping gene against the wild type. The t-test was calculated at either 5% (* P<0.05) or at 1% (** P<0.01) probability
3.2. GIS functions downstream of SIM to regulate trichome branching
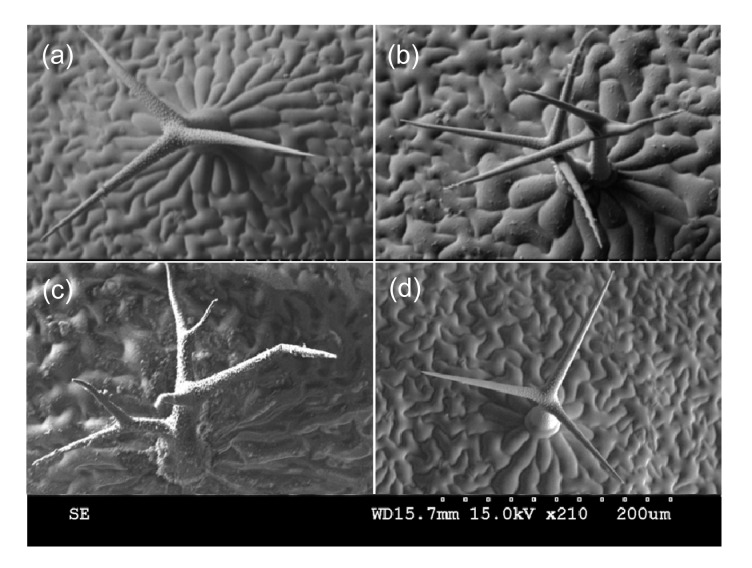
In the WT plants, most of the trichomes on rosette leaves are three-branched (Fig. 2a; Tables 1–4). However, in the gis mutant background, five-branched trichomes were observed on the 8th rosette leaves (Fig. 2c; Tables 1 and 2), which confirms that GIS acts to suppress trichome branching (An et al., 2012). The sim mutant exhibited abundant trichome clustering on the rosette leaves (Fig. 2b; Tables 1 and 3). These clusters are considered to be an abnormal trichome phenotype (Hülskamp et al., 1994; Larkin et al., 2003). To assess the genetic interaction between GIS and SIM further, GIS was overexpressed in a sim background. The abnormal branching phenotype of the sim mutant on the rosette leaves was rescued by this GIS overexpression (Fig. 2d; Tables 1 and 3). In 35S:GIS-sim transgenic plants, we did not observe any trichome clusters on the 3rd, 4th, and 8th rosette leaves (Tables 1–3). Furthermore, the results show that the percentages of one- and two-branched trichomes in 35S:GIS-sim plants were significantly increased while those of the three-branched trichomes on the 3rd and 4th rosette leaves in 35S:GIS-sim plants were significantly decreased in comparison to those of the WT and sim mutant (Table 4). In the case of the 8th rosette leaves, the percentage of one-branched trichomes was also significantly increased, while the percentage of four-branched trichomes was significantly decreased (Table 2). These results are consistent with our previous findings that the overexpression of GIS suppresses trichome branching (An et al., 2012). These results indicate a genetic interaction between GIS and SIM to control the trichome branching.
Fig. 2.
Frozen scanning electron microscope images of genetic interactions between GIS and SIM
Trichome branching phenotypes were observed on the 8th rosette leaves of different genotypes. (a) In wild type, most of the trichomes are three-branched; (b) sim mutants frequently produce cluster trichomes; (c) Trichomes possess five-branched in gis background; (d) In trichome branching phenotype of 35S:GIS-sim transgenic plants, no cluster trichomes could be found
Table 1.
Frequency of trichome branching phenotypes observed at the 8th rosette leaves of different genotypes
| Line | Number of branchesa
|
|||||
| 1 | 2 | 3 | 4 | 5 | Cluster | |
| Wild type (Col) | 0±0 | 4.0±1.3 | 22.2±6.2 | 1.5±0.8 | 0±0 | 0±0 |
| gis | 0±0 | 2.4±1.5* | 20.7±4.0 | 6.5±1.5** | 0.2±0.4** | 0±0 |
| sim | 1.0±0.5** | 2.7±1.1** | 26.4±5.7* | 1.5±0.5 | 0±0 | 1.7±1.9** |
| 35S:GIS-sim | 1.1±1.3** | 4.8±1.8 | 32.4±7.5** | 0±0 | 0±0 | 0±0 |
| gis sim | 0±0 | 2.4±1.5* | 22.2±5.7 | 5.4±1.3** | 0±0 | 0±0 |
Trichomes were counted on the adaxial surface of the 8th rosette leaves of 16 plants. Values indicate mean±standard deviation (SD) (n=16). All significant test analyses were done using the corresponding wild type data as the control. The experiment was repeated at least once with similar results
t-test was calculated at the probability of 5% (P<0.05)
t-test was calculated at the probability of 1% (P<0.01)
Table 4.
Percentage of each trichome branching phenotype on the 3rd and 4th rosette leaves of different genotypes
| Line | Percentage of trichome branchesa
|
|||||
| 1 | 2 | 3 | 4 | 5 | Cluster | |
| Wild type (Col) | 2.9±1.3 | 18.7±7.9 | 78.4±7.3 | 0±0 | 0±0 | 0±0 |
| gis | 1.3±0.8* | 10.3±3.7** | 78.4±3.8 | 10.0±3.0** | 0±0 | 0±0 |
| sim | 2.6±1.2 | 9.7±1.8** | 71.9±5.1* | 2.7±1.4** | 0±0 | 13.1±3.5** |
| 35S:GIS-sim | 8.8±2.0** | 34.4±5.6** | 56.8±5.2** | 0±0 | 0±0 | 0±0 |
| gis sim | 1.2±2.7* | 11.3±4.3* | 81.2±3.7 | 6.3±3.8** | 0±0 | 0±0 |
Trichomes were counted on the adaxial surface of the 8th rosette leaves of 16 plants. Values indicate mean±standard deviation (SD) (n=16). All significant test analyses were done using the corresponding wild type data as the control. The experiment was repeated at least once with similar results
t-test was calculated at the probability of 5% (P<0.05)
t-test was calculated at the probability of 1% (P<0.01)
Table 2.
Percentage of each trichome branching phenotype on the 8th rosette leaves of different genotypes
| Line | Percentage of trichome branchesa
|
|||||
| 1 | 2 | 3 | 4 | 5 | Cluster | |
| Wild type (Col) | 0±0 | 14.8±3.3 | 81.6±7.2 | 3.6±1.8 | 0±0 | 0±0 |
| gis | 0±0 | 8.2±2.4** | 69.2±3.0** | 21.9±2.5** | 0.7±0.4** | 0±0 |
| sim | 2.9±0.5** | 8.0±1.1** | 79.4±6.7 | 4.5±2.3 | 0±0 | 5.2±2.0** |
| 35S:GIS-sim | 2.9±1.8** | 12.5±2.8 | 84.6±5.1 | 0±0** | 0±0 | 0±0 |
| gis sim | 0±0 | 7.9±5.5** | 74.2±7.4* | 17.9±5.9** | 0±0 | 0±0 |
Trichomes were counted on the adaxial surface of the 8th rosette leaves of 16 plants. Values indicate mean±standard deviation (SD) (n=16). All significant test analyses were done using the corresponding wild type data as the control. The experiment was repeated at least once with similar results
t-test was calculated at the probability of 5% (P<0.05)
t-test was calculated at the probability of 1% (P<0.01)
Table 3.
Frequency of trichome branching phenotypes observed at the 3rd and 4th rosette leaves of different genotypes
| Line | Number of branchesa
|
|||||
| 1 | 2 | 3 | 4 | 5 | Cluster | |
| Wild type (Col) | 2.2±1.2 | 9.2±3.4 | 59.4±11.7 | 0±0 | 0±0 | 0±0 |
| gis | 1.0+1.1 | 8.3±2.7 | 63.1±9.9 | 8.1±3.3** | 0±0 | 0±0 |
| sim | 1.9±1.6 | 7.1±3.4 | 52.4±7.9* | 2.0±1.8** | 0±0 | 9.6±3.3** |
| 35S:GIS-sim | 14.6±5.2** | 57.1±17.7** | 94.3±26.7** | 0±0 | 0±0 | 0±0 |
| gis sim | 0.9±1.5 | 8.4±2.5 | 60.1±9.1 | 4.7±1.9** | 0±0 | 0±0 |
Trichomes were counted on the adaxial surface of the 8th rosette leaves of 16 plants. Values indicate mean±standard deviation (SD) (n=16). All significant test analyses were done using the corresponding wild type data as the control. The experiment was repeated at least once with similar results
t-test was calculated at the probability of 5% (P<0.05)
t-test was calculated at the probability of 1% (P<0.01)
To further study the genetic relationship between GIS and SIM, the clustering and branching of trichomes in the sim, gis, and gis sim double mutants were analyzed. As shown in Tables 1 and 3, the clustering of trichomes was not found in the gis sim double mutant on either of 3rd, 4th, and 8th rosette leaves. The trichome branching pattern of gis sim plants most closely resembled that of gis single mutant phenotype (Tables 2 and 4). Taken together with results from gene expression data and genetic interaction analyses, we conclude that GIS acts downstream of SIM to control trichome branching and cell division in an indirect manner.
4. Discussion and conclusions
The development of eukaryotes requires coordinated cell division and growth to produce specific organs and tissues (Churchman et al., 2006). The cell cycle is essential in the establishment of developmental procedures, because it determines the cell type and function, as well as the types and functions of the organs and tissues (Lilly and Duronio, 2005). In Arabidopsis, no differentiation of the epidermal cells of aerial organs is apparent until cells switch from mitotic division to endoreduplication, which initiates the differentiation into trichome cells, where endoreduplication levels also influence the degree of trichome branching (Hülskamp et al., 1994; Ishida et al., 2008; Bramsiepe et al., 2010; Tominaga-Wada et al., 2011). Reduced endoreduplication rates result in smaller trichomes with fewer branches, while an increased level of endoreduplication leads to larger trichomes with more branches (Hülskamp et al., 1994; Folkers et al., 1997; Bramsiepe et al., 2010). These results indicate that the endoreduplication rate is regulated precisely and functions to regulate trichome development.
A role for the C2H2 zinc finger protein GIS in controlling trichome initiation and branching through gibberellic acid signaling has been established previously (Gan et al., 2006; An et al., 2012). This current study demonstrates that GIS acts downstream of the key endoreduplication regulator, SIM, to regulate trichome branching. Furthermore, GIS may regulate the endoreduplication pathway indirectly by genetically interacting with SIM (Fig. 2; Tables 1 and 2). Further research is needed to explore the mechanism and components involved in this genetic interaction.
Acknowledgments
We sincerely thank Prof. Martin HÜLSKAMP (University of Cologne, Germany) for providing the sim mutant for this study.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30970167 and 31228002), the Zhejiang Qianjiang Talent Program (No. 2010R10084), the Zhejiang Provincial Natural Science Foundation of China (No. Z31100041), and the Zhejiang Province Foundation for Returned Scholars (No. 20100129), China
Compliance with ethics guidelines: Li-li SUN, Zhong-jing ZHOU, Li-jun AN, Yan AN, Yong-qin ZHAO, Xiao-fang MENG, Clare STEELE-KING, and Yin-bo GAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.An L, Zhou Z, Su S, Yan A, Gan Y. GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis . Plant Cell Physiol. 2012;53(2):457–469. doi: 10.1093/pcp/pcr192. [DOI] [PubMed] [Google Scholar]
- 2.Bao SJ, An LJ, Su S, Zhou ZJ, Gan YB. Expression patterns of nitrate, phosphate, and sulfate transporters in Arabidopsis roots exposed to different nutritional regimes. Botany-Botanique. 2011;89(9):647–653. doi: 10.1139/B11-053. [DOI] [Google Scholar]
- 3.Bertram JG, Bloom LB, Turner J, O′donnell M, Beechem JM, Goodman MF. Pre-steady state analysis of the assembly of wild type and mutant circular clamps of Escherichia coli DNA polymerase III onto DNA. J Biol Chem. 1998;273(38):24564–24574. doi: 10.1074/jbc.273.38.24564. [DOI] [PubMed] [Google Scholar]
- 4.Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hulskamp M, Schnittger A. Endoreplication controls cell fate maintenance. PLoS Genet. 2010;6(6):e1000996. doi: 10.1371/journal.pgen.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18(16):4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchman ML, Brown ML, Kato N, Kirik V, Hulskamp M, Inze D, de Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana . Plant Cell. 2006;18(11):3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 8.Folkers U, Berger J, Hulskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124(19):3779–3786. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- 9.Gan YB, Filleur S, Rahman A, Gotensparre S, Forde BG. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana . Planta. 2005;222(4):730–742. doi: 10.1007/s00425-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 10.Gan YB, Kumimoto R, Liu C, Ratcliffe O, Yu H, Broun P. GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis . Plant Cell. 2006;18(6):1383–1395. doi: 10.1105/tpc.106.041533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan YB, Liu C, Yu H, Broun P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 2007;134(11):2073–2081. doi: 10.1242/dev.005017. [DOI] [PubMed] [Google Scholar]
- 12.Gan YB, Zhou ZJ, An LJ, Bao SJ, Liu Q, Srinivasan M, Goddard P. The effects of fluctuations in the nutrient supply on the expression of ANR1 and 11 other MADS box genes in shoots and roots of Arabidopsis thaliana . Botany-Botanique. 2010;88(12):1023–1031. doi: 10.1139/B10-075. [DOI] [Google Scholar]
- 13.Gan YB, Zhou ZJ, An LJ, Bao SJ, Forde BG. A comparison between northern blotting and quantitative real-time PCR as a means of detecting the nutritional regulation of genes expressed in roots of Arabidopsis thaliana . Agric Sci China. 2011;10(3):335–342. doi: 10.1016/S1671-2927(11)60012-6. [DOI] [Google Scholar]
- 14.Hülskamp M, Misra S, Jurgens G. Genetic dissection of trichome cell development in Arabidopsis . Cell. 1994;76(3):555–566. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]
- 15.Hülskamp M, Schnittger A, Folkers U. Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. Int Rev Cytol. 1999;186:147–178. doi: 10.1016/S0074-7696(08)61053-0. [DOI] [PubMed] [Google Scholar]
- 16.Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jurgens G, Hulskamp M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiol. 2003;131(2):643–655. doi: 10.1104/pp.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 18.Johnson HB. Plant pubescence: an ecological perspective. Bot Rev. 1975;41(3):233–258. doi: 10.1007/BF02860838. [DOI] [Google Scholar]
- 19.Kasili R, Walker JD, Simmons LA, Zhou J, de Veylder L, Larkin JC. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics. 2010;185(1):257–268. doi: 10.1534/genetics.109.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin JC, Brown ML, Schiefelbein J. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis . Annu Rev Plant Biol. 2003;54:403–430. doi: 10.1146/annurev.arplant.54.031902.134823. [DOI] [PubMed] [Google Scholar]
- 21.Lilly MA, Duronio RJ. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24(17):2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- 22.Luo D, Oppenheimer DG. Genetic control of trichome branch number in Arabidopsis: the roles of the FURCA loci. Development. 1999;126(24):5547–5557. doi: 10.1242/dev.126.24.5547. [DOI] [PubMed] [Google Scholar]
- 23.Marks MD. Molecular genetic analysis of trichome development in Arabidopsis . Annu Rev Plant Physiol Plant Mol Biol. 1997;48:137–163. doi: 10.1146/annurev.arplant.48.1.137. [DOI] [PubMed] [Google Scholar]
- 24.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1 . Genetics. 2000;156(3):1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plett JM, Mathur J, Regan S. Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana . J Exp Bot. 2009;60(13):3923–3933. doi: 10.1093/jxb/erp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao KN, Venkatachalam SR. Dihydrofolate reductase and cell growth activity inhibition by the β-carboline-benzoquinolizidine plant alkaloid deoxytubulosine from Alangium lamarckii: its potential as an antimicrobial and anticancer agent. Bioorg Med Chem. 1999;7(6):1105–1110. doi: 10.1016/S0968-0896(98)00262-4. [DOI] [PubMed] [Google Scholar]
- 27.Rodney M, Mark RD. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution. 1997;51:1435–1444. doi: 10.1111/j.1558-5646.1997.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 28.Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hülskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis . EMBO J. 2002;21(19):5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnittger A, Schobinger U, Bouyer D, Weinl C, Stierhof YD, Hülskamp M. Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. PNAS. 2002;99(9):6410–6415. doi: 10.1073/pnas.092657299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnittger A, Schobinger U, Stierhof YD, Hülskamp M. Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr Biol. 2002;12(5):415–420. doi: 10.1016/S0960-9822(02)00693-0. [DOI] [PubMed] [Google Scholar]
- 31.Szymanski DB, Marks MD. GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis . Plant Cell. 1998;10(12):2047–2062. doi: 10.1105/tpc.10.12.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tominaga-Wada R, Ishida T, Wada T. New Insights into the Mechanism of Development of Arabidopsis Root Hairs and Trichomes. In: Jeon KW, editor. Vol. 286. International Review of Cell and Molecular Biology; 2011. pp. 67–106. [DOI] [PubMed] [Google Scholar]
- 33.Walker JD, Oppenheimer DG, Concienne J, Larkin JC. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development. 2000;127(18):3931–3940. doi: 10.1242/dev.127.18.3931. [DOI] [PubMed] [Google Scholar]
- 34.Weinl C, Marquardt S, Kuijt SJ, Nowack MK, Jakoby MJ, Hulskamp M, Schnittger A. Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell. 2005;17(6):1704–1722. doi: 10.1105/tpc.104.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]