Abstract
Calotropis procera, commonly known as “milkweed”, possesses long seed trichomes for seed dispersal and has the ability to survive under harsh conditions such as drought and salinity. Aquaporins are water channel proteins expressed in all land plants, divided into five subfamilies plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like proteins (NIPs), small basic intrinsic proteins (SIPs), and the unfamiliar X intrinsic proteins (XIPs). PIPs constitute the largest group of water channel proteins that are involved in different developmental and regulatory mechanisms including water permeability, cell elongation, and stomata opening. Aquaporins are also involved in abiotic stress tolerance and cell expansion mechanisms, but their role in seed trichomes (fiber cells) has never been investigated. A large number of clones isolated from C. procera fiber cDNA library showed sequence homology to PIPs. Both expressed sequence tags (ESTs) and real-time polymerase chain reaction (PCR) studies revealed that the transcript abundance of this gene family in fiber cells of C. procera is greater than that of cotton. Full-length cDNAs of CpPIP1 and CpPIP2 were isolated from C. procera fiber cDNA library and used for constructing plant expression vectors under constitutive (2×35S) and trichome-specific (GhLTP3) promoters. Transgenic tobacco plants were developed via Agrobacterium-mediated transformation. The phenotypic characteristics of the plants were observed after confirming the integration of transgene in plants. It was observed that CpPIP2 expression cassette under 2×35S and GhLTP3 promoter enhanced the numbers of stem and leave trichomes. However, 2×35S::CpPIP2 has a more amplified effect on trichome density and length than GhLTP3::CpPIP2 and other PIP constructs. These findings imply the role of C. procera PIP aquaporins in fiber cell elongation. The PIPs-derived cell expansion mechanism may be exploited through transgenic approaches for improvement of fiber staple length in cotton and boosting of defense against sucking insects by enhancing plant pubescence.
Keywords: Seed trichome, Plasma membrane intrinsic protein (PIP), Fiber quality, Cell elongation, Tobacco, Agrobacterium
1. Introduction
Calotropis procera is a wild and perennial shrub ranging from 2.5 to 10 m in height. It belongs to the family Asclepiadaceae (milkweed family). C. procera is mainly known for its medicinal value. This plant species can grow in dry as well as humid areas (Parrotta, 2001). The plant is highly drought and salt-tolerant. It produces long and fine fibers on its seeds for wind dispersal. C. procera fibers can reach up to 45 mm in length (Cheema et al., 2010).
One of the mechanisms reported for plant cell elongation is the turgor pressure-driven cell expansion by aquaporins. Aquaporins belong to the major intrinsic proteins (MIPs), which comprise a superfamily of integral membrane proteins (Preston et al., 1992), and have been discovered in mammals (Agre et al., 1993), plants (Maurel, 2007), insects (Beuron et al., 1995), yeast (Carbrey et al., 2001), bacteria (Calamita et al., 1995), protozoa (Mitra et al., 2000), and archaea (Kozono et al., 2003). Aquaporins are water channel proteins with an average size of 28–30 kDa that form channels/pores in biological membranes and specifically regulate osmotic pressure-based movement of H2O molecules and other small solutes across living cells (Agre et al., 1993; Maurel, 2007). They play a vital role in transporting bulk volume of water and some solutes through diffusion in biological membranes (Tornroth-Horsefield et al., 2006).
Aquaporins contribute to root hydraulic conductivity (Siefritz et al., 2002), leaf hydraulic conductivity and transpiration (Aharon et al., 2003; Sade et al., 2010), cell elongation (Hukin et al., 2002; Liu et al., 2008; Choat et al., 2009), plant cell osmoregulation (Wallace et al., 2006) and photosynthesis (Sade et al., 2010). These proteins are also involved in turgor pressure development and, in turn, cell volume expansion to respond to a number of abiotic stresses like H2O deficit, salinity and frost (Li et al., 2009). The presence of aquaporins in plants also induces morphological changes. They increase the root and shoot mass by cell volume expansion, differentiation, and shoot axis lengthening so that they can absorb water and nutrients from greater depths and wider ground area.
The higher plant aquaporins can be subdivided into five subfamilies: (1) plasma membrane intrinsic proteins (PIPs), (2) tonoplast intrinsic proteins (TIPs), (3) NOD26-like proteins (NIPs), (4) small basic intrinsic proteins (SIPs), and (5) unfamiliar X intrinsic proteins (XIPs) and glycerol facilitator-like proteins (GLPs) (Chaumont et al., 2001; Gustavsson et al., 2005; Danielson and Johanson, 2008). These aquaporins are mostly tissue-specific depending upon their role in cell metabolism and physiological processes (Park et al., 2010). Amongst the subfamilies, PIPs constitute the largest group and the majority of this type of aquaporin is localized in the plasma membranes (Schaffner, 1998). PIPs are found near the vascular bundles in almost all plant parts, with the highest expression in the roots (Siefritz et al., 2002).
Based on phylogenetic analysis, the subfamily of PIPs can be subdivided into two distinct groups named PIP1 and PIP2 (Zardoya, 2005). The two groups differ in the lengths of N- and C-termini. The members of PIP1 subgroup have extended N-terminus and shorter C-terminus as compared to PIP2 aquaporins. They also exhibit differential water permeability characteristics. Members of the PIP2 subgroup exhibit high water channel activity in different heterologous expression systems (Suga and Maeshima, 2004; Bots et al., 2005). PIP1 is more efficiently involved in the transportation of uncharged solutes like glycerol and urea, and gases like CO2 and NH3 as compared to H2O conductivity (Fetter et al., 2004). These differences in transportation potential may be due to the different molecular structures of PIP2 and PIP1 isoforms. In addition, PIP2 isoforms also possess a stretch of 4–10 amino acids in the first extra cytosolic loop. The members of PIP2 aquaporins have been reported from different plant species. The in vitro Xenopus laevis oocyte expression analysis of PIP2 aquaporins indicated 5–20-fold increased water permeability in response to increased turgor pressure (Weig et al., 1997; Moshelion et al., 2002).
All aquaporins include a hydrophobic pore with two passing filters; (1) conserved asparagine-proline-alanine (NPA) motif that acts as molecule-specific and size-exclusion filter and (2) an aromatic region comprising a conserved arginine residue (Arg195) that forms the narrowest part of the pore (de Groot et al., 2003). These hydrophobic motifs permit water molecules in the form of a single-file hydrogen-bonded chain and as the result of dipolar forces the water file is broken into single molecules at the centre of the pore (Maurel, 2007).
Regulation of this mechanism of aquaporin activity can enhance the cell elongation in fibers and in the root growing zone. The cell expansion of individual cells is the resulting function of turgor pressure, cell wall properties, and cell hydraulic conductivity (Cosgrove, 1993). A water deficit condition causes a lower osmotic pressure inside cells, which results in the opening of aquaporins and allows the movement of water into the cell permitting turgor-driven expansion of cell volume (Cosgrove, 1986; Boyer, 2001). Therefore, cell elongation can be regulated by the cell capability to take up water from the surrounding environment. For this purpose, aquaporins can play a better role in cell enlargement than other cell expansion proteins. The role of PIP2 isoforms of aquaporins in cell expansion mechanisms is well studied in various plant parts but not in fiber cells. Previous studies showed their involvement in cell expansion of rose petals (Ma et al., 2008), barley leaf tissues (Volkov et al., 2007), and grape berries (Choat et al., 2009).
To enhance the fiber staple length in cotton cultivars, we must have insight about cell elongation mechanisms. Turgor-driven cell expansion by PIP aquaporins is one of the mechanisms explored in C. procera for elongating fibers. The present study was focused to characterize the aquaporins isolated from C. procera fiber cDNA library. PIP aquaporins were greatly expressed in C. procera fiber cells showing their ability to improve fiber properties. This mechanism can be manipulated for cotton fiber improvement through transgenic technology. The two isoforms of PIP aquaporins screened from C. procera fiber cDNA library were used for constructing plant expression vectors that were further used in the development of tobacco transgenic plants for expression analysis.
2. Materials and methods
C. procera fiber cDNA library and expressed sequence tags (ESTs) were previously established in the Gene Isolation Lab at the National Institute for Biotechnology and Genetic Engineering (NIBGE). The EST sequence data were analyzed using various bioinformatics tools available at ExPASY: SIB Bioinformatics Resource Portal (http://www.expasy.org). The homology search tool basic local alignment search tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed the resemblance of numerous ESTs with plant aquaporins. The BLAST screened aquaporins categorized into two classes, CpPIP1 and CpPIP2. Full-length clones of CpPIP1 and CpPIP2 were selected for further characterization. The deduced amino acid sequences of CpPIP1 and CpPIP2 were determined by a translation tool available at JustBio (http://www.justbio.com). Amino acid sequence identity between CpPIP1 and CpPIP2 was calculated by the following formula: Identity=(number of identical residues in a pairwise alignment)/(length of the shortest sequence)×100%.
2.1. Phylogenetic analysis
GenBank database at the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) was screened for protein sequences of plant PIPs using the BLAST X search tool. CpPIP1 and CpPIP2 protein sequences were used as query sequences. BLAST search enabled us to pick 71 plant aquaporin protein sequences from the GenBank. The alignment data on the reported and C. procera query protein sequences were used to construct a topology-based phylogenetic tree using the CLCbio 6.0 (Denmark) combined workbench.
2.2. Analysis of CpPIP1 and CpPIP2 aquaporins for membrane association
Transmembrane prediction analysis was performed using TMHMM software (http://www.cbs.dtu.dk/services/TMHMM-2.0). The subcellular localizations of CpPIP1 and CpPIP2 were determined by WoLF PSORT analysis software available at http://www.expasy.ch. The dataset generated by this tool was analyzed to check the localization of query proteins in the cell organelles.
2.3. Cloning the full-length CpPIP1 and CpPIP2 in derivatives of pJIT166 under 2×35S and LTP promoters
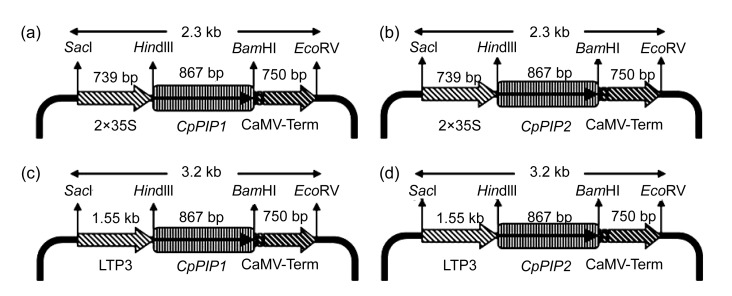
Forward and reverse primers were designed on the PIP1 and PIP2 gene sequences. The HindIII site was introduced in the forward primer upstream to the start codon, while the BamHI site was added at the 3′ end downstream to the stop codon of the genes. The polymerase chain reaction (PCR) amplifications from the respective cDNA clones were performed using Pfu DNA polymerase (Fermentas, UK). The amplified products were double digested with BamHI and HindIII for direct cloning in the respective sites generated in pGR1 that had been modified from pJIT166 in the Lab (intronless GUS was replaced with a GUS gene with introns) and pGR5 (2×35S promoter replaced with GhLTP3 promoter in pGR1). The gene:promoter:terminator cassettes were picked from pGR1 and pGR5 by using SacI and EcoRV restriction enzymes and cloned in the binary vector (pGA482) using the restriction sites SacI and HpaI. The four types of pGA482 constructs (2×35S::CpPIP1, 2×35S::CpPIP2, GhLTP3::CpPIP1, and GhLTP3::CpPIP2) were used for the Agrobacterium-mediated transformation of tobacco plants.
2.4. Agrobacterium-mediated tobacco transformation
Sterilized (treated with 15% bleach for 15 min) tobacco (Nicotiana tabaccum L.) seeds were germinated in vitro on Petri dishes containing MS medium in a growth chamber under constant light at 25 °C. Each Petri dish was planted with 10 seeds. Stem portions with one node and two leaves taken from grown plantlets were subcultured every 2–3 weeks onto fresh MS medium (Murashige and Skoog, 1962).
The plasmids from four clones carrying CpPIP expression cassettes were isolated using a miniprep kit (Promega, USA) from overnight grown cultures of Escherichia coli and transformed into electrocompetant cells of Agrobacterium tumefaciens (LBA4404). The four transformations were spread on Lauria Bertani (LB) agar plates containing rifampicin (25 μg/ml) and kanamycin (50 μg/ml) and incubated at 28 °C for 48 h, and single colonies were screened for transformed vectors by restriction and PCR analysis. Simultaneously, leaf disks (2–3 cm in diameter) were obtained from in vitro grown, four-week-old tobacco plantlets and placed on Petri dishes containing MS medium. The samples were incubated at 25 °C in a diurnal plant growth chamber (Farma Scientific Inc.) for about two days to allow cell regeneration from disk termini. After two days, 10–20 ml Agrobacterium inoculums of each of the four clones were used to treat 30–40 leaf disks. The infected disks were allowed to stay at room temperature for half an hour to permit the physical attachment of Agrobacterium to the plant tissues and then transferred to Whatman filter papers and thoroughly blotted. After co-cultivation, 5–7 leaf disks were placed on co-cultivation media in a Petri dish and incubated at 25 °C for further two days to allow for the induction of virulence and physical transfer of genetic material by Agrobacterium.
The co-cultivated leaf disks were collected, washed with MS liquid medium containing cefotaxime (250 μg/ml), dried, and placed on the shooting medium for induction of callus formation and selection. Callus-induced leaf disks were then shifted on shooting media at (25±0.5) °C under constant light for two weeks. It took 7–10 d for plantlets emergence. The non-transformed cells ultimately died due to kanamycin stress. The plantlets with 3–5 leaves were shifted to jars containing selection media and incubated in a plant growth room at (25±1) °C.
After the shooting and selection, the putative transgenic plants with 7–10 leaves were transferred to rooting media containing the selection antibiotic kanamycin. After 5–7 d, plants started producing primary roots from the base of shoots and with the appearance of secondary roots they were transferred to earthen pots with a soil:sand mixture (1:1, w/w).
2.5. Analysis of putative transgenic plants
Young leaves from putative transgenic plants and control tobacco plants (negative controls) were selected for DNA isolation by the modified cetyl trimethyl ammonium bromide (CTAB) method (Azmat et al., 2012). The isolated DNA was resuspended in 50 μl of ultrapure sterile H2O and stored at −20 °C. The primers for transgene analysis were designed at specific sites inside the promoter and gene for amplification of junction regions near promoter(s) and gene(s) specific to each construct (Table 1). These primers were used for the PCR amplification of the transgene from genomic DNA of putative transgenics along with positive and negative controls. The plasmids containing transgenes were used as a positive control for all the transgenes. The plants confirmed for the presence of the transgene were further analyzed for their morphological characteristics.
Table 1.
Primers used for putative transgenic analysis
| Sr. No. | Primer name | Primer sequence |
| 1 | GJLTPIP1F | 5′ TACCCTCAAGCCCTAACG 3′ |
| 2 | GJLTPIP1R | 5′ TACAAGAACAAGAGAGTAGCC 3′ |
| 3 | GJ35PIP1F | 5′ GCTATCTGTCACTTCATC 3′ |
| 4 | GJ35PIP1R | 5′ GGTGGTTCTTTGTAATCC 3′ |
| 5 | GJLTPIP2F | 5′ AAACCCTCCTACCCTCAAG 3′ |
| 6 | GJLTPIP2R | 5′ CCAATAACAGTCAAGACAGTG 3′ |
| 7 | GJ35PIP2F | 5′ GCTATCTGTCACTTCATCG 3′ |
| 8 | GJ35PIP2R | 5′ TGGTAATCCTTGGCTGAG 3′ |
3. Results and discussion
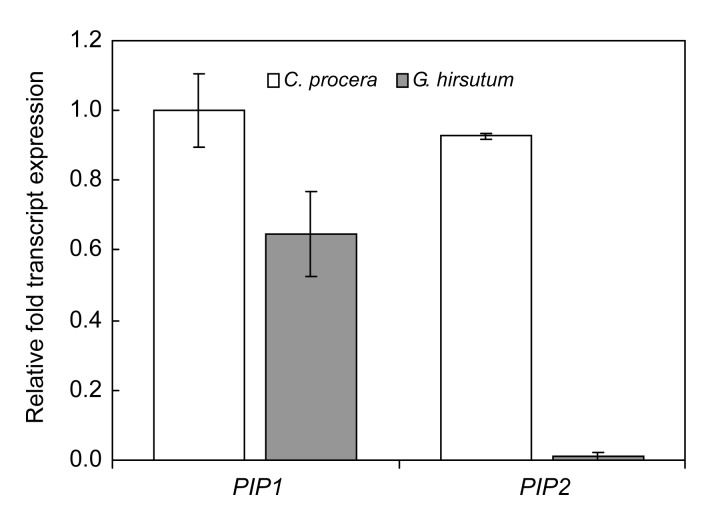
The C. procera seed fibers are epidermal appendages of micropylar regions usually called seed trichomes. Two variants of PIPs (CpPIP1 and CpPIP2) were identified to be the most abundant transcripts represented in the C. procera fiber cDNA library. Real-time RT-PCR studies indicated that PIP1 and PIP2 are expressed in both C. procera and cotton fibers, but the expression of these genes is significantly higher in C. procera fibers than in cotton fibers (Fig. 1). This information supports the reported functionality of these proteins in cellular development processes (Hukin et al., 2002; Liu et al., 2008) regulating the buildup of turgor pressure within the cell which is, in turn, attributed to cell enlargement. This direct relationship between fiber cell size and the abundance of CpPIP1 and CpPIP2 transcripts is an important clue for the pivotal role this gene family may play in defining the fiber cell volume. Plasma membranes are the most important gateways for the permeation of molecules into and out of the cell, and they house different intrinsic proteins. Cell expansion is based on the rapid uptake of water and solutes into the cell either through plasma membrane or plasmodesmata, thus increasing the turgor pressure that drives cell enlargement (Hukin et al., 2002). Further studies, therefore, focused to characterize the role of identified genes on plant development, specifically, the morphological traits in the model tobacco plants. Therefore, transgenic tobacco plants were obtained, expressing the CpPIP1 and CpPIP2 genes under constitutive (2×35S) and fiber specific (GhLTP3) promoters. The four expression vectors are named accordingly as 2×35S::CpPIP1, 2×35S::CpPIP2, GhLTP3::CpPIP1, GhLTP3::CpPIP2 (Fig. 2).
Fig. 1.
Relative expressions of PIP1 and PIP2 in Calotropis procera and Gossypium hirsutum (cotton) fibers through real-time PCR
The data show that PIP1 and PIP2 have significantly high expression in the C. procera fibers as compared to cotton fiber
Fig. 2.
Details of vectors constructed for stable transformation
(a) 2×35S::CpPIP1; (b) 2×35S::CpPIP2; (c) GhLTP3::CpPIP1; (d) GhLTP3::CpPIP2
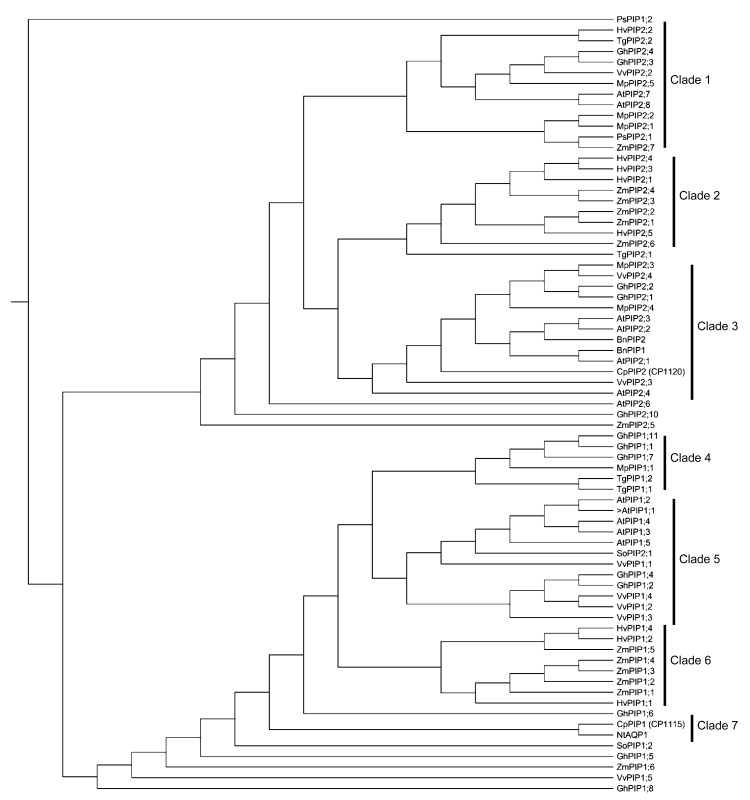
Phylogenetic analysis of plant PIPs classified all members into two distinct subgroups: PIP1, bottom 4 clades; and PIP2, upper 3 clades (Fig. 3). The clade distribution of PIP1 aquaporins showed that CpPIP1 is closely related to NtAQP1, and together they constitute a distinct clade, clade 7. This clade is rooted to all other members and clades in the PIP1 subgroup, except outliers. These results suggest that the two members are earlier divergent in evolutionary history. The PIP1 class of plant aquaporins has been known to possess extended N-terminus and short C-terminus. The PIP1 clade of plant aquaporins includes member proteins that have low water regulation activity in different expression assays as observed in PIP isoforms of Samanea saman (Moshelion et al., 2002). These proteins are generally involved in the conductivity of solutes across plasma membranes (Fetter et al., 2004). The phylogenetic analysis of CpPIP2 shows its grouping in clade 3 of PIP2 plant aquaporins (Fig. 3), which are characterized as having shorter N-terminus and longer C-terminus. The members of this clade are reported to be involved in the influx of water, thus building up the cellular turgor pressure (Suga and Maeshima, 2004).
Fig. 3.
Phylogenetic analyses of CpPIP1 and CpPIP2 genes with plasma membrane aquaporins of some other plant species
Deduced amino acid sequences were used to construct the topology-based phylogenetic tree using the following sequences. Arabidopsis thaliana: AtPIP1;1: P61837, AtPIP1;2: Q06611, AtPIP1;3: Q08733, AtPIP1;4: Q39196, AtPIP1;5: NP194071, AtPIP2;1: P43286, AtPIP2;2: P43287, AtPIP2;3: P30302, AtPIP2;4: NP200874, AtPIP2;5: Q9SV31, AtPIP2;6: Q9ZV07, AtPIP2;7: P93004, AtPIP2;8: Q9ZVX8; Brassica napus: BnPIP1: AAD39373, BnPIP2: AAD39374; Calotropis procera: CpPIP1, CpPIP2; Hordeum vulgare: HvPIP1;1: BAF41978, HvPIP1;2: BAF33067, HvPIP1;4: BAF33068, HvPIP2;1: BAE02729, HvPIP2;2: BAG06230, HvPIP2;3: BAF33069, HvPIP2;4: BAE06148, HvPIP2;5: BAG06231; Mimosa pudica: MpPIP1;1: BAD90696, MpPIP2;1: BAD90697, MpPIP2;2: BAD90698, MpPIP2;3: BAD90699, MpPIP2;4: BAD90700, MpPIP2;5: BAD90701; Nicotiana tabaccum: NtAQP1 (PIP1): AAB81601; Pisum sativum: PsPIP1;2: CAD68986, PsPIP2;1: CAB45651; Spinacea oleracea: SoPIP1;2: AAR23268, SoPIP2;1 (PM28B): CAB56217; Tulipa gesneriana: TgPIP1;1: BAG68659, TgPIP1;2: BAG68660, TgPIP2;1: BAG68661, TgPIP2;2: BAG68662; Vitis vinifera: VvPIP1;1: ABN14347, VvPIP1;2: ABN14348, VvPIP1;3: ABN14349, VvPIP1;4: ABN14350, VvPIP1;5: ABN14355, VvPIP2;2: ABN14351, VvPIP2;3: ABN14352, VvPIP2;4: ABN14353; Zea mays: ZmPIP1;1: Q41870, ZmPIP1;2: Q9XF59, ZmPIP1;3: NP001105022, ZmPIP1;4: AAK26755, ZmPIP1;5: AAK26756, ZmPIP1;6: NP001105023, ZmPIP2;1: NP_001105024, ZmPIP2;2: NP001105638, ZmPIP2;3: AAK26760, ZmPIP2;4: AAK26761, ZmPIP2;5: Q9XF58, ZmPIP2;6: NP001105027, ZmPIP2;7: NP001105639; Gossypium hirsutum: GhPIP1;1: EF079900.1, GhPIP1;2: EF470293.1, GhPIP1;4: BK007045.1, GhPIP1;5: BK007046.1, GhPIP1;6: BK007047.1, GhPIP1;7: BK007048.1, GhPIP1;8: BK007049.1, GhPIP1;11: GU998828.1, GhPIP2;1: EF079901.2, GhPIP2;2: EF079902.1, GhPIP2;3: EU402412.1, GhPIP2;4: EU402413.1, GhPIP2;10: BK007052.1
The association of two aquaporins within different clades indicates that both are orthologous and evolutionarily divergent, which is supported by the 68.4% identity observed between their amino acid sequences. The hydropathy profiles and the determination of transmembrane regions for CpPIP1 and CpPIP2 indicated six membrane embedding domains (Table 2). This analysis showed that although CpPIP1 and CpPIP2 belong to the same gene family (Fig. 2) and have the same structural ontology (Table 2), they are functionally distinct.
Table 2.
Identification of transmembrane regions by TMHMM software#
| Protein name | Helice | Length of transmembrane regions of deduced amino acid sequence*
|
|||||
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| CpPIP1 | Inside to outside | 59–76 | 86–105 | 133–154 | 178–194 | 210–235 | 259–279 |
| Outside to inside | 54–74 | 88–105 | 133–151 | 178–194 | 208–226 | 259–277 | |
| CpPIP2 | Inside to outside | 43–65 | 82–99 | 128–149 | 170–189 | 205–277 | 252–269 |
| Outside to inside | 43–63 | 80–98 | 128–146 | 170–186 | 203–221 | 252–269 | |
Available at http://www.cbs.dtu.dk/services/TMHMM-2.0/
The length of various helices of transmembrane regions is indicated by the number of the amino acids in the deduced polypeptides of CpPIP1 and CpPIP2 by taking starting methionine as 1
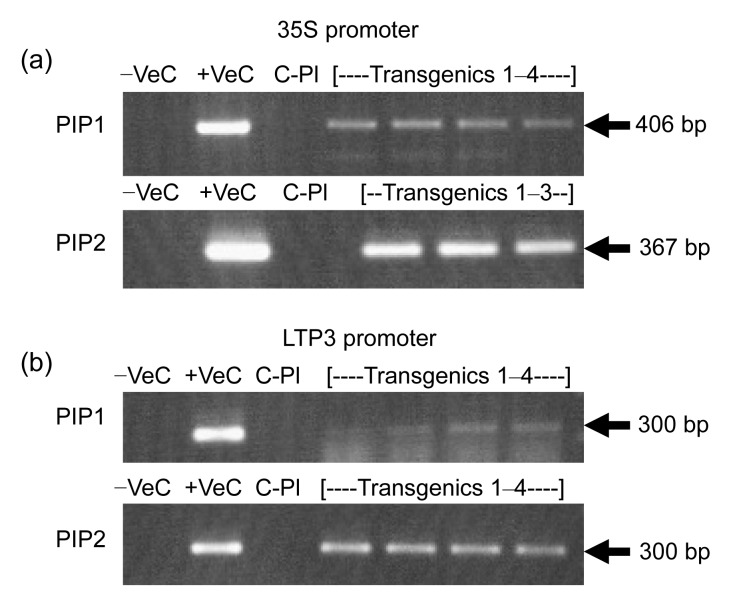
Transgenic tobacco plants developed by using the four expression vectors (2×35S::CpPIP1, 2×35S::CpPIP2, GhLTP3::CpPIP1, GhLTP3::CpPIP2) indicated good transformation efficiencies of 53.33%, 30%, 46.67%, and 60%, respectively. The low rate of morphological observations may be due to variation in transgene integration events in the tobacco genome (Table 3). The tobacco transgenics were confirmed for transgene status by conventional PCR analysis (Fig. 4). The morphological changes in transgenics were studied in comparison to the control plants. The key focus was on leaf and stem trichomes as CpPIP1 and CpPIP2 were isolated from C. procera seed fibers. Transgenic tobacco plants containing CpPIP1 indicated no phenotypic changes during the plant development process (Figs. 5b, 5d, 5g and 5h). The possible reason for this could be the fact that aquaporins are expressed in all land plants, and tobacco has been reported to have high expression of PIP1 isoforms (Siefritz et al., 2004). The stable transformation of tobacco with CpPIP1 might have intensified the expression of this gene family, which is reported to be involved in solute conductivity and has less impact on turgor pressure driven-cell elongation than other aquaporin types (Fetter et al., 2004). Transgenic plants having CpPIP2 were observed to have dense hair population on stem and also on the dorsal and ventral sides of leaves as compared to control plants (Figs. 5c, 5e, 5i and 5j). It is possible that an increase in the number of water channels in the transgenic plants might be involved in increasing the water transport activities (Suga and Maeshima, 2004) thus raising the cell turgor pressure, which in turns lead to the trichome cell expansion (Cosgrove, 1986). In addition, transgenic plants having 2×35S::CpPIP2 showed more pubescence on leaves and stem portions (Figs. 5c and 5i) as compared to those having GhLTP3::CpPIP2 (Figs. 5e and 5j), which indicated that GhLTP3 might have a tighter control for single-celled fiber expression as compared to multicellular trichomes. Another explanation for the difference of hairiness between CpPIP2 and CpPIP1 transgenic plants might be the heteromerization of CpPIP2 with NtPIP1 which regulates the targeting of later one to the plasma membrane. This assumption coincides with the report by Fetter et al. (2004), which demonstrated that co-expression of ZmPIP1 and ZmPIP2 had improved water regulation activity.
Table 3.
Transformation efficiencies of the CpPIP constructs and morphological observations in tobacco transgenic plants
| Sr. No. | Name of construct | No. of regenerated plants | No. of transgenes confirmed | Transformation efficiency (%) | Phenotypic observation |
| 1 | CpPIP1 under 2×35S promoter | 30 | 16 | 53.33 | 7 |
| 2 | CpPIP2 under 2×35S promoter | 30 | 14 | 46.67 | 5 |
| 3 | CpPIP1 under LTP3 promoter | 30 | 9 | 30 | 4 |
| 4 | CpPIP2 under LTP3 promoter | 30 | 18 | 60 | 9 |
Fig. 4.
Transgene analysis of putative transgenic tobacco plants through PCR
(a) PCR analysis for plants having 2×35S::PIP1 and 2×35S::PIP2 constructs; (b) PCR analysis for plants having GhLTP3::CpPIP1 and GhLTP3::CpPIP2 constructs. −Ve: negative control of PCR master mix; +Ve: positive control of PCR master mix using plasmid DNA as a template; C-Pl: control plant of PCR using its DNA as a template; Transgenics 1–4: PCR analysis of 4 randomly selected putative transgenic plants using Gene-Junction primers, showing expected amplification products of 406, 367, 300, and 300 base pairs (bp), respectively
Fig. 5.
Morphological appearance of leaf and stem of tobacco plants
(a) Leaf morphology of control tobacco plant; (b) Leaf morphology of transgenic plant for CpPIP1 under 35S promoter; (c) Leaf morphology of transgenic plant for CpPIP2 under 35S promoter with more pubescence; (d) Leaf morphology of transgenic plant for CpPIP1 under LTP3 promoter; (e) Leaf morphology of transgenic tobacco plant for CpPIP2 under LTP3; (f) Stem appearance of control tobacco plant; (g) Stem morphology of transgenic plant for CpPIP1 under 35S promoter; (h) Stem morphology of transgenic tobacco plant for CpPIP1 under LTP3 promoter; (i) Stem morphology of transgenic plant for CpPIP2 under 35S promoter with enhanced pubescence; (j) Stem morphology of transgenic tobacco plant for CpPIP2 under LTP3
It was also observed that transgenics for PIP2 constructs had thicker and darker leaves than those of control tobacco plants. Darkness of leaves revealed that there might be a relatively high CO2 conductivity associated with the CpPIP2 transgenics. The possible reason for this could be the putative role of CpPIP2 aquaporins in CO2 conductance in the leaves and stem parts as observed in the rice transgenic plants expressing barley (Hordeum vulgare L.) HvPIP2 (Hanba et al., 2004; Katsuhara and Hanba, 2008). This study depicts the potential role of CpPIP2 in cell elongation. It has a strong capability to increase trichome density and length, which can be utilized as a defense system in agricultural crops to avoid or reduce sucking insect attacks. On the other hand, its expression in single-celled fibers may provide a significant genetic resource in improving fiber length.
4. Conclusions
This study describes the novel findings about PIP aquaporins of wild C. procera in cell elongation. It may be assumed that the fiber cell elongation in C. procera is turgor pressure driven by aquaporin activity in cooperation with other fiber-related gene families like expansins, lipid transfer proteins, tubulins, arabino-galactan proteins, and actins (Cheema et al., 2010; Indrais et al., 2011).
These results will contribute to the understanding of fiber elongation mechanisms in plants. This study concluded that CpPIP2 is a potential gene that may be used in adjusting turgor pressure-driven elongation of developing fibers in cotton through transgenic technologies. However, being hosted in C. procera (a wild plant spp.), the role of PIPs in abiotic stresses (drought and frost) is still to be investigated.
Acknowledgments
Ministry of Food and Agriculture (MinFA) and Higher Education Commission (HEC) of Pakistan.
Footnotes
Compliance with ethics guidelines: Usman ASLAM, Asia KHATOON, Hafiza Masooma Naseer CHEEMA, and Aftab BASHIR declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Agre P, Sasaki S, Chrispeels M. Aquaporins: a family of water channel proteins. Am J Physiol. 1993;261:F461. doi: 10.1152/ajprenal.1993.265.3.F461. [DOI] [PubMed] [Google Scholar]
- 2.Aharon R, Shahak Y, Wininger S, Bendov R, Kapulnik Y, Galili G. Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell. 2003;15(2):439–447. doi: 10.1105/tpc.009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azmat MA, Khan IA, Cheema HMN, Rajwana IA, Khan AS, Khan AA. Extraction of DNA suitable for PCR applications from mature leaves of Mangifera indica L. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2012;13(4):239–243. doi: 10.1631/jzus.B1100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuron F, Caherec FL, Guillam MT, Cavalier A, Garret A, Tassan JP, Delamarche C, Schultz P, Mallouh V, Rolland JP, et al. Structural analysis of a MIP family protein from the digestive tract of Cicadella viridis . J Biol Chem. 1995;270(29):17414–17422. doi: 10.1074/jbc.270.29.17414. [DOI] [PubMed] [Google Scholar]
- 5.Bots M, Vergeldt F, Wolters-Arts M, Weterings K, van As H, Mariani C. Aquaporins of the PIP2 class are required for efficient anther dehiscence in tobacco. Plant Physiol. 2005;137(3):1049–1056. doi: 10.1104/pp.104.056408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer JS. Growth-induced water potentials originate from wall yielding during growth. J Exp Bot. 2001;52(360):1483–1488. doi: 10.1093/jexbot/52.360.1483. [DOI] [PubMed] [Google Scholar]
- 7.Calamita G, Bishai WR, Preston GM, Guggino WB, Agre P. Molecular cloning and characterization of AqpZ, a water channel from Escherichia coli . J Biol Chem. 1995;270(49):29063–29066. doi: 10.1074/jbc.270.49.29063. [DOI] [PubMed] [Google Scholar]
- 8.Carbrey JM, Bonhivers M, Boeke JD, Agre P. Aquaporins in Saccharomyces: characterization of a second functional water channel protein. PNAS. 2001;98(3):1000–1005. doi: 10.1073/pnas.98.3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125(3):1206–1215. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheema HMN, Bashir A, Khatoon A, Iqbal N, Zafar Y, Malilk KA. Molecular characterization and transcriptome profiling of expansin genes isolated from Calotropis procera fibers. Electron J Biotechnol. 2010;13(6):1–14. doi: 10.2225/vol13-issue6-fulltext-7. [DOI] [Google Scholar]
- 11.Choat B, Gambetta GA, Shackel KA, Matthews MA. Vascular function in grape berries across development and its relevance to apparent hydraulic isolation. Plant Physiol. 2009;151(3):1677–1687. doi: 10.1104/pp.109.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosgrove DJ. Biophysical control of plant cell growth. Annu Rev Plant Physiol. 1986;37(1):377–405. doi: 10.1146/annurev.pp.37.060186.002113. [DOI] [PubMed] [Google Scholar]
- 13.Cosgrove DJ. Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol. 1993;124(1):1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- 14.Danielson JA, Johanson U. Unexpected complexity of the aquaporins gene family in moss Physcomitrella patens . BMC Plant Biol. 2008;8(1):45. doi: 10.1186/1471-2229-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Groot BL, Frigato T, Helms V, Grubmuller H. The mechanism of proton exclusion in the aquaporin-1 water channel. J Mol Biol. 2003;333(2):279–293. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Fetter K, van Wilder V, Moshelion M, Chaumont F. Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell. 2004;16(1):215–228. doi: 10.1105/tpc.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustavsson S, Lebrun AS, Norden K, Chaumont F, Johanson U. A novel plant major intrinsic protein in Physcomitrella patens most similar to bacterial glycerol channels. Plant Physiol. 2005;139(1):287–295. doi: 10.1104/pp.105.063198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanba YT, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K, Terashima I, Katsuhara M. Over expression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiol. 2004;45(5):521–529. doi: 10.1093/pcp/pch070. [DOI] [PubMed] [Google Scholar]
- 19.Hukin D, Doering Saad C, Thomas CR, Pritchard J. Sensitivity of cell hydraulic conductivity to mercury is coincident with symplasmic isolation and expression of plasmalemma aquaporin genes in growing maize roots. Planta. 2002;215(6):1047–1056. doi: 10.1007/s00425-002-0841-2. [DOI] [PubMed] [Google Scholar]
- 20.Indrais E, Cheema HMN, Bashir A. Temporal expression analysis and cloning of cotton (Gossypium hirsutum) fiber genes. Int J Agric Biol. 2011;13(1):89–94. [Google Scholar]
- 21.Katsuhara M, Hanba YT. Barley plasma membrane intrinsic proteins (PIP aquaporins) as water and CO2 transporters. Eur J Physiol. 2008;456(4):687–691. doi: 10.1007/s00424-007-0434-9. [DOI] [PubMed] [Google Scholar]
- 22.Kozono D, Ding XD, Iwasaki I, Meng XY, Kamagata Y, Agre P, Kitagawa Y. Functional expression and characterization of an archaeal aquaporin-AqpM from Methanothermobacter marburgensis . J Biol Chem. 2003;278(12):10649–10656. doi: 10.1074/jbc.M212418200. [DOI] [PubMed] [Google Scholar]
- 23.Li DD, Wu YJ, Ruan XM, Li B, Zhu L, Wang H, Li XB. Expressions of three cotton genes encoding the PIP proteins are regulated in root development and in response to stresses. Plant Cell Rep. 2009;28(2):291–300. doi: 10.1007/s00299-008-0626-6. [DOI] [PubMed] [Google Scholar]
- 24.Liu DQ, Tu LL, Wang L, Li YJ, Zhu LF, Zhang XL. Characterization and expression of plasma and tonoplast membrane aquaporins in elongating cotton fibers. Plant Cell Rep. 2008;27(8):1385–1394. doi: 10.1007/s00299-008-0545-6. [DOI] [PubMed] [Google Scholar]
- 25.Ma M, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol. 2008;148(2):894–907. doi: 10.1104/pp.108.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurel C. Plant aquaporins: novel functions and regulation properties. FEBS Lett. 2007;581(12):2227–2236. doi: 10.1016/j.febslet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Mitra BN, Yoshino R, Morio T, Yokoyama M, Maeda M, Urushihara H, Tanaka Y. Loss of a member of the aquaporin gene family, aqpA affects spore dormancy in Dictyostelium . Gene. 2000;251(2):131–139. doi: 10.1016/S0378-1119(00)00201-8. [DOI] [PubMed] [Google Scholar]
- 28.Moshelion M, Becker D, Biela A, Hehlein N, Hedrich R, Otto B, Levi H, Moran N, Kaldenhoff R. Plasma membrane aquaporins in the motor cells of Samanea saman: diurnal and circadian regulation. Plant Cell. 2002;14(3):727–739. doi: 10.1105/tpc.010351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 30.Park W, Scheffler BE, Bauer FJ, Campbell BT. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2010;10(1):142. doi: 10.1186/1471-2229-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrotta JA. Healing Plants of Peninsular India. UK and New York: CAB International Wallingford Press; 2001. p. 944. [Google Scholar]
- 32.Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256(5055):385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 33.Sade N, Gebretsadik M, Seligmann R, Schwartz A, Wallach R, Moshelion M. The role of tobacco aquaporin1 in improving water use efficiency, hydraulic conductivity, and yield production under salt stress. Plant Physiol. 2010;152(1):245–254. doi: 10.1104/pp.109.145854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaffner AR. Aquaporin function, structure, and expression: are there more surprises to surface in water relations? Planta. 1998;204(2):131–139. doi: 10.1007/s004250050239. [DOI] [PubMed] [Google Scholar]
- 35.Siefritz F, Tyree MT, Lovisolo C, Schubert A, Kaldenhoff R. PIP1 plasma membrane aquaporins in tobacco: from cellular effects to function in plants. Plant Cell. 2002;14(4):869–876. doi: 10.1105/tpc.000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siefritz F, Otto B, Bienert GP, van der Krol A, Kaldenhoff R. The plasma membrane aquaporin NtAQP1 is a key component of the leaf unfolding mechanism in tobacco. Plant J. 2004;37(2):147–155. doi: 10.1046/j.1365-313X.2003.01947.x. [DOI] [PubMed] [Google Scholar]
- 37.Suga S, Maeshima M. Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol. 2004;45(7):823–830. doi: 10.1093/pcp/pch120. [DOI] [PubMed] [Google Scholar]
- 38.Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshi E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439(7077):688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 39.Volkov V, Hachez C, Moshelion M, Draye X, Chaumont F, Fricke W. Water permeability differs between growing and non-growing barley leaf tissues. J Exp Bot. 2007;58(3):377–390. doi: 10.1093/jxb/erl203. [DOI] [PubMed] [Google Scholar]
- 40.Wallace IS, Choi WG, Roberts DM. The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim Biophys Acta. 2006;1758(8):1165–1175. doi: 10.1016/j.bbamem.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Weig A, Deswarte C, Chrispeels MJ. The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. Plant Physiol. 1997;114(4):1347–1357. doi: 10.1104/pp.114.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zardoya R. Phylogeny and evolution of the major intrinsic protein family. Biol Cell. 2005;97(6):397–414. doi: 10.1042/BC20040134. [DOI] [PubMed] [Google Scholar]