Abstract
The CHRNA5-CHRNA3-CHRNB4 gene cluster on chromosome 15q25.1 encoding the cholinergic nicotinic receptor subunits is robustly associated with smoking behavior and nicotine dependence. Only a few studies to date have examined the locus with alcohol related traits and found evidence of association with alcohol abuse and dependence. Our main goal was to examine the role of three intensively studied single nucleotide polymorphisms, rs16969968, rs578776 and rs588765, tagging three distinct loci, in alcohol use. Our sample was drawn from two independent Finnish population-based surveys, the National FINRISK Study and the Health 2000 (Health Examination) Survey. The combined sample included a total of 32,592 adult Finns (54% women) of whom 8,356 were assessed for cigarettes per day (CPD). Data on alcohol use were available for 31,812 individuals. We detected a novel association between rs588765 and alcohol use defined as abstainers and low-frequency drinkers versus drinkers (OR=1.15, p=0.00007). Additionally, we provide precise estimates of strength of the association between the three loci and smoking quantity in a very large population based sample. As a conclusion, our results provide further evidence for the nicotine-specific role of rs16969968 (locus 1). Further, our data suggest that the effect of rs588765 (locus 3) may be specific to alcohol use as the effect is seen also in never smokers.
Keywords: Nicotinic acetylcholine receptors, 15q25.1, alcohol use, smoking behavior, public health, population-based sample, genetic association
Introduction
Smoking, alcohol use and abuse are serious public health issues [1,2]. Both genetic and environmental factors clearly are of importance in accounting for inter-individual differences in these common behaviors. Family and twin studies show that alcohol use, abuse, and dependence as well as smoking and nicotine dependence are transmitted within families [3,4]. Smoking and alcohol use often co-occur, suggesting that they may have a shared genetic predisposition [5]. However, the molecular genetic basis for that shared predisposition is still largely unknown.
Within the past few years, genetic findings for nicotine dependence and other smoking related traits and diseases have pinpointed a region on chromosome 15q25.1. This region harbors the CHRNA5-CHRNA3-CHRNB4 gene cluster coding for a5, a3, and b4 nicotinic acetylcholine receptor (nAChR) subunits. Evidence of association with smoking behavior and nicotine dependence has been reported in a genome-wide association study (GWAS) [6] and confirmed in large meta-analyses conducted by independent consortia [7-10]. The most well-established locus within this region is tagged by the single nucleotide polymorphism (SNP) rs16969968 (locus 1) [11]. Rs16969968 is a non-synonymous (D398N) SNP in exon 5 of CHRNA5, located in the cytoplasmic domain preceding the M4 transmembrane domain [12]. Asparagine (N) in the 398 position with its polar uncharged side chain lowers Ca2+ permeability and increases short-term desensitization in (α4β2)α5 nAChRs [12]. When the function of the subunit is decreased, α5-containing nAChRs fail to trigger a normal inhibitory motivational signal that is supposed to limit nicotine intake [13].
Three distinct loci tagged by rs16969968 (locus 1), rs578776 (locus 2), and rs588765 (locus 3) provided evidence of association with smoking amount defined by cigarettes per day (CPD) in a meta-analysis of 38,617 smokers from 34 data-sets [7]. Three other meta-analyses detected association with locus 1 and locus 2 but no independent effect was detected for locus 3 [8-10], which emphasizes the need for further studies. Both locus 1 and locus 2 have been associated also with nicotine dependence defined by the Fagerström Test for Nicotine Dependence (FTND [14]) [15], and by the Diagnostic and Statistical Manual of Mental Disorders 4th edition (DSM-IV [16]) [17,18], with the minor allele of rs16969968 (locus 1) increasing the risk, and the minor allele of rs578776 (locus 2) exhibiting a protective effect.
A few studies have reported association of this region with alcohol related traits, with somewhat discordant results. Chen and colleagues [19] found evidence of association between locus 1 (rs16969968) and symptoms of alcohol abuse or dependence. Wang and colleagues [20] observed association between DSM-IV alcohol dependence and a narrow region that included locus 3 (rs588765); however, no association was detected with locus 1 (rs16969968) or locus 2 (rs578776). Sherva and colleagues [17] detected association between DSM-IV alcohol dependence and locus 2 (rs578776) and locus 3 (rs615470), when using an African-American sample, while their European-American sample provided no evidence of association. On top of the studies focusing on alcohol dependence, one study has reported association between alcohol initiation and two CHRNB4 SNPs (rs1948 and rs11634351) in young Americans of European descent [21]. The incongruity may in part be due to heterogeneity and varying linkage disequilibrium (LD) structures between the study samples. To date there have been no published studies of alcohol use with respect to this gene cluster; thus, further studies are needed to clarify whether genetic variation in the nAChR genes contributes to inter-individual differences in alcohol use.
Our first aim was to replicate in a large population-based sample the results of the meta-analysis of Saccone and colleagues [7] reporting an association between three distinct loci at 15q25.1 and smoking quantity, thus providing precise estimates of the strength of the association in the population at large. The second aim, our main goal, was to study the role of genetic variants on 15q25.1 in alcohol use and to explore whether this role is independent of smoking.
Materials and methods
Study sample
Our sample was drawn from two independent Finnish population-based surveys, the National FINRISK Study [25] and the Health 2000 (Health Examination) Survey [26].
The National FINRISK Study has been initiated in 1972 and carried out since then every five years using independent, random, and representative samples from four to six different parts of Finland depending on the year of the survey [25]. We used data from cohorts 1992, 1997, 2002, and 2007 (participation rates 76%, 72%, 69% and 65%, respectively), comprising of 26,800 genotyped subjects (53% women) with self-reported information about their health-related habits, including tobacco and alcohol use. The mean age was 48 years.
The Health 2000 population based survey was carried out during 2000-2001 in Finland (participation rate 76%), as previously described [26]. This random and representative sample included 5,792 genotyped subjects (55% women) with self-reported information about their health-related habits, including tobacco and alcohol use. The mean age was 54 years.
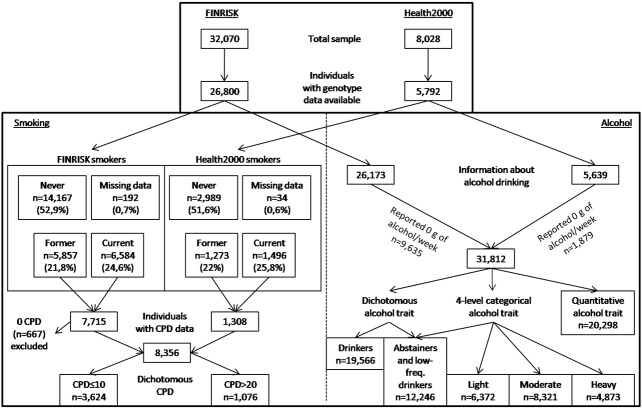
As the marker frequencies did not significantly vary between study regions (i.e., no population stratification was detected) and time of the cross-sectional survey, these factors were not included in the final analyses. Thus, we combined the FINRISK and Health 2000 samples, resulting in a total study sample of 32,592 adult Finns (54% women) aged 25 to 98 years. The mean age was 49 years. A flow chart of the sample is presented in Figure 1.
Figure 1.
Study sample. A flow chart of the sample and variables included in the study.
Smoking quantity traits
The smoking quantity traits were derived from quantitative self-reported measure of cigarettes smoked per day (CPD), administered to current (including daily and occasional) and former smokers. In both FINRISK and Health 2000 samples, the participants were asked: “How many cigarettes (either manufactured or self-rolled cigarettes, or equivalent amounts of cigars or pipe tobacco) do you smoke daily or smoked before quitting?”. The answers were summed as one quantitative measure of CPD which was analyzed as a quantitative trait and as a dichotomous trait with light smoking “controls” (CPD≤10; N=3,624), and heavy smoking “cases” (CPD>20; N=1,076), as used by Saccone and colleagues [7]. The case/control designation is considered as a proxy for nicotine dependence, as nicotine dependence was not otherwise assessed in the entire data. In the combined data set of 32,592 subjects there were 667 (2%) individuals who reported zero CPD. Among these individuals there were 197 former smokers and 470 current smokers (this category included daily and occasional smokers). We excluded these 667 individuals from our analyses due to the ambiguity in their questionnaire answers.
Alcohol use traits
In the analyses of alcohol consumption, we examined weekly alcohol consumption as quantitative, categorical, and dichotomized traits. In the quantitative analyses we used self-reported alcohol consumption defined as grams (g) per week with logarithmic transformation in order to approach normal distribution. Abstainers (N=11,514) were excluded from the analyses of quantitative alcohol consumption. In the FINRISK sample, the measure was derived from a question: “How many glasses (restaurant measures) or bottles of alcohol beverages did you drink during last week?”. Alcohol consumption (g/week) was then calculated for each individual based on the knowledge of how many grams of ethanol different alcohol beverages contain. In the Health 2000 sample, respective measure was derived from a question: “How often and how much did you drink alcohol beverages during the past 12 months?”. An average consumption of alcohol per week (g/week) was calculated for each individual based on the knowledge of how many grams of ethanol various alcohol beverages contain. For both categorical and dichotomous alcohol use analyses the quantitative alcohol consumption was first coded as standard drinks per week; one alcohol drink was defined as containing 12 g of pure ethanol. In the analysis of categorical alcohol trait, individuals were categorized as abstainers and low-frequency drinkers (less than 1 drink per week), light drinkers (1-2 drinks per week), moderate drinkers (3-7 drinks per week for women, and 3-14 drinks per week for men), and heavy drinkers (over 7 drinks per week for women, and over 14 drinks per week for men). For dichotomous alcohol use, we contrasted those drinking one or more drinks per week (drinkers) versus those either using no alcohol at all or drinking less than one drink per week (i.e. less than 12 g of alcohol per week) (abstainers and low-frequency drinkers).
Genotyping
Our SNPs of interest, rs16969968 (CHRNA5; locus 1), rs578776 (CHRNA3; locus 2), and rs588765 (CHRNA5; locus 3), were selected based on the meta-analysis of Saccone and colleagues [7]. DNA was derived from whole blood samples frozen immediately at the clinical study sites. Frozen samples were transferred to the laboratory of molecular genetics at the National Institute of Health and Welfare (earlier National Public Health Institute), where the DNA was extracted. Genotyping of the DNA samples was carried out using standard protocols of iPLEX Gold technology on the MassARRAY System (Sequenom, San Diego, CA, USA). The success rate was >0.99 for all the SNPs and they were in Hardy Weinberg Equilibrium (P>0.05). Minor allele frequencies were 0.32, 0.32, and 0.38 for rs16969968, rs578776, and rs588765, respectively.
Statistical analyses
The associations between the quantitative CPD and the three loci were modeled using negative binomial regression. This model provided the best fit for our trait distribution (data not shown; available upon request). The associations between dichotomous CPD (light versus heavy smokers) and the three loci were examined using logistic regression. All analyses were adjusted for age, sex, and log transformed alcohol use (g/week). In order to quantify the separate and joint effects of the SNPs, we analyzed two series of models: each SNP separately in the single-SNP model, and joint models for locus 1 and each of the other two loci. All allele main effects were modeled assuming an additive effect.
In the analyses of alcohol use traits, associations with the three loci were modeled using linear regression (for log transformed quantitative alcohol consumption (g/week)), multinomial logistic regression (for categorical alcohol use), and logistic regression (for dichotomous alcohol use, i.e. abstainers and low-frequency drinkers versus drinkers). The analyses for all alcohol use traits were adjusted for age, sex, and CPD.
We explored the best-fitting model for locus-specific dominance model (recessive, dominant, and additive versus co-dominant) using goodness of fit tests based on likelihood ratios of the two models. Additive model provided the best-fitting model for most traits and loci with the exception of dichotomous alcohol use and locus 3 (rs588765) for which the co-dominant model provided the best fit (data not shown).
In order to minimize the effect of possible former alcoholics (most of who typically also have smoked) within the abstainers group and to inhibit the confounding effect of smoking, we performed the analysis of dichotomous alcohol use also among never smokers.
All statistical analyses were performed using Stata 11.1 [27]. Throughout the manuscript we report nominal p-values.
Replication samples
In an attempt to replicate the detected novel association between locus 3 and alcohol use, we utilized two independent samples: (1) an Australian twin family sample (NAG-OZALC, N=5743), sample demographics previously described elsewhere [28], and (2) the Young Finns Study (YFS, N=1982) [29].
Results
Associations with smoking quantity
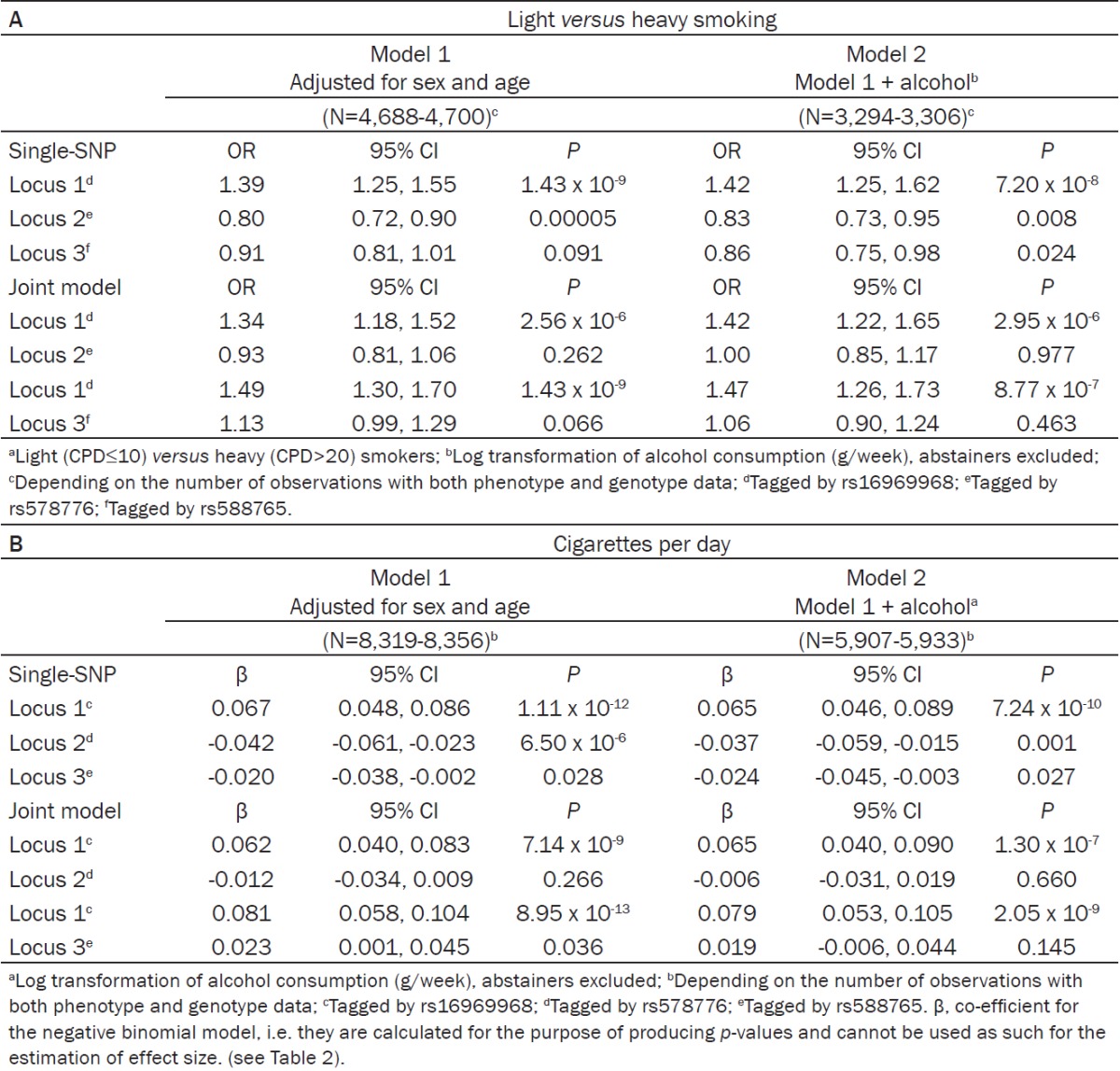
We replicated the established association between dichotomous CPD (light versus heavy smokers) and locus 1 (rs16969968, prevalence odds ratio (OR)=1.39, p=1.43 x 10-9) and locus 2 (rs578776, OR=0.80, p=0.00005). Locus 3 (rs588765) did not yield statistical significance. When locus 1 and locus 3 were analyzed together in the joint model, the effect size of locus 1 increased (OR=1.49, p=1.43 x 10-9), and locus 3 approached statistical significance (OR=1.13, p=0.066). Adjustment for alcohol consumption had no effect on the results. The association results for dichotomous CPD from the additive model are presented in Table 1A.
Table 1.
A: Association between dichotomous CPDa and CHRNA5-CHRNA3-CHRNB4 variants in the Finnish population sample, prevalence odds ratios (ORs) and 95% confidence intervals (95% CIs) from logistic regression (additive model), (B) association between quantitative CPD and CHRNA5-CHRNA3-CHRNB4 variants in the Finnish population sample, regression coefficients (β) and 95% confidence intervals (CIs) from negative binomial regression (additive model)
|
The analysis of quantitative CPD yielded similar results when compared to the analysis of dichotomous CPD. As an exception, locus 3 achieved statistical significance (β=0.023, p=0.036) when analyzed in the joint model with locus 1. The association results for quantitative CPD from the additive model are presented in Table 1B. Distributions of CPD per genotype are presented in Table 2.
Table 2.
Estimated amount of cigarettes per daya and 95% confidence intervals (CIs) for each loci in the Finnish population sample
| Cigarettes per day | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Major allele homozygotes | Heterozygotes | Minor allele homozygotes | ||||
|
|
||||||
| Single-SNP | marginal mean of CPD | 95% CI | marginal mean of CPD | 95% CI | marginal mean of CPD | 95% CI |
| Locus 1b | 13.9 | 13.6, 14.2 | 14.8 | 14.5, 15.1 | 16.0 | 15.4, 16.6 |
| Locus 2c | 14.9 | 14.7, 15.2 | 14.2 | 13.9, 14.5 | 13.9 | 13.9, 14.5 |
| Locus 3d | 14.8 | 14.5, 15.1 | 14.3 | 14.0, 14.6 | 14.4 | 13.9, 14.9 |
Estimates from marginal mean values based on the negative binomial regression from Model 1 in Table 1B;
Tagged by rs16969968;
Tagged by rs578776;
Tagged by rs588765.
Associations with alcohol use
We detected no evidence of association when alcohol consumption was analyzed as a quantitative (standard drinks per day) or as a categorical (abstainers and low-frequency drinkers, light, moderate or heavy drinkers) phenotype. Further adjustment for quantitative CPD had no effect on the results (data not shown; available upon request).
However, when alcohol use was analyzed as a dichotomous phenotype, with abstainers and low-frequency drinkers compared to those drinking at least one drink per week (altogether N=31,812), significant association with locus 3 was detected (rs588765, OR=1.15, p=0.00007). Table 3 shows the results from the co-dominant model. When the analysis was repeated among never smokers (N=16,786) the effect size of locus 3 remained significant (OR=1.18, p=0.001) (Table 3).
Table 3.
Association between dichotomous alcohol usea and CHRNA5-CHRNA3-CHRNB4 variants in the Finnish population sample, prevalence odds ratios (ORs) and 95% confidence intervals (95% CIs) from logistic regression (co-dominant model)
| Abstainers and low-frequency drinkers versus drinkers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| Model 1 Adjusted for sex and age | Model 2 Model 1 among current smokers | Model 3 Model 1 among never smokers | Model 4 Model 1 among smokers adjusted for CPDb | |||||||||
|
|
||||||||||||
| (N=31,775-31,812)c | (N=7,907-7,919)c | (N=16,786-16,803)c | (N=8,911-8,927)c | |||||||||
|
|
||||||||||||
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| Locus 1d | ||||||||||||
| GG | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| GA | 0.93 | 0.89, 0.98 | 0.007 | 0.88 | 0.79, 0.97 | 0.014 | 0.96 | 0.90, 1.03 | 0.230 | 0.88 | 0.80, 0.98 | 0.016 |
| AA | 0.97 | 0.89, 1.05 | 0.430 | 1.02 | 0.86, 1.21 | 0.818 | 0.95 | 0.85, 1.06 | 0.368 | 1.02 | 0.87, 1.20 | 0.795 |
| Locus 2e | ||||||||||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| CT | 0.98 | 0.94, 1.03 | 0.532 | 1.09 | 0.98, 1.21 | 0.110 | 0.96 | 0.90, 1.03 | 0.271 | 1.08 | 0.98, 1.19 | 0.125 |
| TT | 0.98 | 0.90, 1.06 | 0.569 | 1.07 | 0.90, 1.27 | 0.441 | 0.93 | 0.84, 1.04 | 0.217 | 1.07 | 0.91, 1.25 | 0.428 |
| Locus 3f | ||||||||||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||
| CT | 1.00 | 0.95, 1.05 | 0.945 | 0.88 | 0.79, 0.98 | 0.018 | 1.04 | 0.97, 1.12 | 0.231 | 0.90 | 0.81, 0.99 | 0.039 |
| TT | 1.15 | 1.07, 1.24 | 0.00007 | 1.09 | 0.93, 1.28 | 0.259 | 1.18 | 1.07, 1.31 | 0.001 | 1.07 | 0.93, 1.24 | 0.358 |
Abstainers and low-frequency drinkers (<1 drink/week) versus drinkers (≥1 drink/week); 1 drink = 12 g of alcohol;
Quantitative CPD (cigarettes smoked per day);
Depending on the number of observations with both phenotype and genotype data;
Tagged by rs16969968;
Tagged by rs578776;
Tagged by rs588765.
The association between locus 3 and (dichotomized) alcohol use did not replicate in the NAG-OZALC or YFS sample (data not shown; available upon request).
Discussion
Chromosome 15q25.1 harbors the well-established smoking locus within the CHRNA5-CHRNA3-CHRNB4 gene cluster. It is robustly associated with smoking behavior and nicotine dependence [7-10]. Only a few studies to date have examined the locus with alcohol related traits and found evidence of association with alcohol dependence [17,20] and alcohol abuse/dependence [19]. We aimed to replicate and quantify in the population at large the associations between the three distinct loci on chromosome 15q25.1 and smoking quantity (defined as CPD) reported in the meta-analysis of Saccone and colleagues [7]. However, our main aim was to test whether the 15q25.1 region shows an association with regular alcohol use while carefully accounting for smoking behavior.
First, we provide precise estimates of the strength of the association of locus 1 (rs16969968), locus 2 (rs578776), and locus 3 (rs588765) with smoking quantity in this large Finnish population based sample. We were unable to achieve statistical significance for locus 3 (rs588765) with dichotomous CPD, but detected the association with quantitative CPD. This is likely due to differences in power; quantitative CPD phenotype included 8,356 current smokers, while the analysis of the dichotomous CPD included 3,624 “cases” (smoked>20 CPD) and 1,076 “controls” (smoked≤10 CPD). The effect of rs588765 reached statistical significance only after the locus was adjusted for rs16969968, as previously reported by Saccone and colleagues [7]. Interestingly, three other large consortia have been unable to detect strong evidence of association for rs588765 even when analyzed jointly with rs16969968 [8-10]. These three meta-analyses used linear regression to test for association with either quantitative or categorical CPD while Saccone and colleagues [7] used logistic regression to test the association with dichotomous CPD. The methodological differences between logistic and linear regressions have been suggested as a reason for this discordance in the results. The current study demonstrates that association between CPD and locus 3 (rs588765) can be detected using other models as well, such as negative binomial regression. This is the largest single study replicating the association between the three loci at 15q25.1 and smoking amount, providing estimates of the strength of the association comparable to those reported previously, i.e. 1 CPD for each minor allele (Table 2).
As alcohol use frequently co-occurs with tobacco use and the traits are highly correlated [22], we adjusted CPD analyses for alcohol consumption. However, adjustment for alcohol use had no notable effect on the association. The lack of difference between the analyses with and without adjustment for alcohol use suggests that the association signal detected in locus 1 is specific to smoking. This is in line with the finding of Wang and colleagues [20], who reported no evidence of association between locus 1 (rs16969968) and alcohol dependence, suggesting that the effect of rs16969968 is specific to smoking.
Second, we examined the association between quantitative alcohol consumption and the three loci on 15q25.1 and detected no significant association. Similarly, when using multiple categories of alcohol use, no association was seen. In contrast, the dichotomous alcohol use, defined as abstainers and low-frequency drinkers (N=12,246) versus drinkers (N=19,566), showed an association with locus 3 (rs588765). Being a drinker was more frequent among individuals homozygous for the minor allele, while the same genotype exhibited a protective effect against heavy smoking. However, in the joint model with locus 1, locus 3 appeared as a risk factor for heavy smoking. Here, it should be noted that abstainers and low-frequency drinkers are a heterogeneous group of subjects as this category may contain also former alcoholics who are abstaining at the time of the data collection. To address this as a possible confounder, we repeated the analysis among never-smokers. Due to the high co-occurrence of heavy smoking and alcoholism it is unlikely that the never smoker group contains many former alcoholics. Moreover, by including only never smokers we excluded the potential confounding association signal emerging from smoking and CPD. Limiting the analysis to never smokers (N=16,786) did not have a notable impact on the results, suggesting that locus 3 (rs588765) is associated specifically with regular alcohol use. Although the actual effect size of locus 3 (OR=1.18) is modest, this finding provides new direction for the research of CHRNA5-CHRNA3-CHRNB4 locus 3 and suggests that the effects of alcohol may partially be mediated through cholinergic receptors and their downstream transmitter molecules. The association did not replicate in the NAG-OZALC or YFS sample, possibly due to the significantly smaller sample sizes when compared to the study sample.
Locus 3 has previously been associated with DSM-IV alcohol dependence in African Americans [17] and in the Collaborative Study on the Genetics of Alcoholism (COGA) sample [20]. To the best of our knowledge, the current study is the first population based study to examine association between regular alcohol use and the 15q25.1 region, and to report an association with locus 3. Although our sample is adequately sized, unascertained, and genetically homogenous, we acknowledge the likely underrepresentation of severely alcohol and nicotine addicted individuals who are less likely to participate in health-related surveys. Their absence is unlikely to explain the association with the drinker status. Further studies are needed to elucidate and verify the associations between the 15q25.1 region and alcohol related traits, and the specific role of locus 3.
Within the past few years, several independent studies, large-scale meta-analyses, and replication studies have reported association between the CHRNA5-CHRNA3-CHRNB4 gene cluster on chromosome 15q25.1 and smoking-related traits and diseases, such as nicotine dependence, CPD, chronic obstructive pulmonary disease, and lung cancer [7-10,23,24]. Two distinct loci have been consistently highlighted; locus 1 tagged by rs16969968 or rs1051730 and locus 2 tagged by rs578776. Several association studies, including the current study, suggest that locus 1 is a consistent risk factor for smoking behavior with the minor allele predisposing a smoker to smoke more, become dependent on nicotine, and, as a consequence, even develop a smoking related disease. Evidence for a role of locus 1 in alcohol-related traits has been much more controversial. In the current study, the lack of difference in the analyses of CPD with and without adjustment for alcohol use suggests that the effect of rs16969968 is specific to smoking. Locus 2, on the other hand, seems to be protective against smoking. In the current study locus 2 showed an effect size of an OR 0.8, which is in line with previous studies [7,18]. The role of locus 3 (tagged by rs588765) in smoking behavior remains unconfirmed. The current study failed to achieve significant association between rs588765 and dichotomous CPD, but association was detected with quantitative CPD. Locus 3 has previously been associated with alcohol dependence [17,20] and it is also associated with altered levels of CHRNA5 mRNA [20]. The current study provided evidence of a novel association between locus 3 (rs588765) on 15q25.1 and alcohol use defined as abstainers and low-frequency drinkers versus drinkers in a large Finnish population sample. Limiting the analysis to never smokers did not have an impact on the results, suggesting that the effect of rs588765 may be specific to alcohol use.
Acknowledgements
This study was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (grant numbers 213506, 129680 to JK), Helsinki Biomedical Graduate Program, HBGP to JH, the Academy of Finland (grant numbers 139635 and 129494 to VS; 136895 and 263836 to SM, 141054 to JK), the Sigrid Juselius Foundation (to JK), and ENGAGE – European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant agreement number 201413. The Young Finns Study has been financially supported by the Academy of Finland: grants 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9N035 to TL), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Eemil Aaltonen Foundation (to TL). The expert technical assistance in the statistical analyses by Irina Lisinen and Ville Aalto are gratefully acknowledged.
Disclosure of conflict of interest
J. Kaprio has consulted for Pfizer on nicotine dependence in 2011 and 2012, and for ERAB on alcohol drinking in 2011. T. Korhonen has consulted for Pfizer on tobacco dependence treatment in 2011 and 2012.
References
- 1.U.S. Department of Health and Human Services. How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease, A Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2010. (http://www.surgeongeneral.gov/library/tobaccosmoke/report/full_report.pdf) [Google Scholar]
- 2.Global health risks: mortality and burden of disease attributable to selected major risks. Geneva: World Health Organization; 2009. (http://www.who.int/healthinfo/global_burden_disease/GlobalHealthRisks_report_full.pdf) [Google Scholar]
- 3.Dick DM, Bernard M, Aliev F, Viken R, Pulkkinen L, Kaprio J, Rose RJ. The role of socioregional factors in moderating genetic influences on early adolescent behavior problems and alcohol use. Alcohol Clin Exp Res. 2009;33:1739–1748. doi: 10.1111/j.1530-0277.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rose JE, Broms U, Korhonen T, Dick DM, Kaprio J. Genetics of smoking behavior. In: Kim YK, editor. Handbook of behavior genetics. New York: Springer; 2009. pp. 411–432. [Google Scholar]
- 5.Li TK, Volkow ND, Baler RD, Egli M. The biological bases of nicotine and alcohol co-addiction. Biol Psychiatry. 2007;61:1–3. doi: 10.1016/j.biopsych.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsäter A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, Kong X, Landi MT, Ma JZ, Short SE, Stephens SH, Stevens VL, Sun L, Wang Y, Wenzlaff AS, Aggen SH, Breslau N, Broderick P, Chatterjee N, Chen J, Heath AC, Heliövaara M, Hoft NR, Hunter DJ, Jensen MK, Martin NG, Montgomery GW, Niu T, Payne TJ, Peltonen L, Pergadia ML, Rice JP, Sherva R, Spitz MR, Sun J, Wang JC, Weiss RB, Wheeler W, Witt SH, Yang BZ, Caporaso NE, Ehringer MA, Eisen T, Gapstur SM, Gelernter J, Houlston R, Kaprio J, Kendler KS, Kraft P, Leppert MF, Li MD, Madden PA, Nöthen MM, Pillai S, Rietschel M, Rujescu D, Schwartz A, Amos CI, Bierut LJ. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schäfer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Völzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A, Mooser V, Francks C, Marchini J Wellcome Trust Case Control Consortium. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Mägi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tönjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Döring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Järvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K ENGAGE Consortium. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–446. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierut LJ. Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24-25. Trends Pharmacol Sci. 2010;31:46–51. doi: 10.1016/j.tips.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 15.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, Culverhouse RC, Fox L, Hinrichs AL, Steinbach JH, Wu M, Rice JP, Goate AM, Bierut LJ. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4th edition. Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- 17.Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broms U, Wedenoja J, Largeau MR, Korhonen T, Pitkäniemi J, Keskitalo-Vuokko K, Häppölä A, Heikkilä KH, Heikkilä K, Ripatti S, Sarin AP, Salminen O, Paunio T, Pergadia ML, Madden PA, Kaprio J, Loukola A. Analysis of Detailed Phenotype Profiles Reveals CHRNA5-CHRNA3-CHRNB4 Gene Cluster Association with Several Nicotine Dependence Traits. Nicotine Tob Res. 2012;14:720–733. doi: 10.1093/ntr/ntr283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Chen J, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC, Kendler KS. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:926–933. doi: 10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broms U, Wedenoja J, Largeau MR, Korhonen T, Pitkäniemi J, Keskitalo-Vuokko K, Häppölä A, Heikkilä KH, Heikkilä K, Ripatti S, Sarin AP, Salminen O, Paunio T, Pergadia ML, Madden PA, Kaprio J, Loukola A. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dani JA, Harris RA. Nicotine addiction and comorbidity with alcohol abuse and mental illness. Nat Neurosci. 2005;8:1465–1470. doi: 10.1038/nn1580. [DOI] [PubMed] [Google Scholar]
- 23.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcátová I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 25.Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Männistö S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty-five-year trends in cardiovascular risk factors in Finland. Int J Epidemiol. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- 26.Heistaro S, editor. Methodology report Health 2000 Survey. Publications of the National Public Health Institute; B 26/2008. [Google Scholar]
- 27.StataCorp. College Station, TX: StataCorp LP; 2009. Stata Statistical Software: Release 11. [Google Scholar]
- 28.Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: Findings and implications. Biol Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raitakari OT, Juonala M, Rönnemaa T, Keltikangas-Järvinen L, Räsänen L, Pietikäinen M, Hutri-Kähönen N, Taittonen L, Jokinen E, Marniemi J, Jula A, Telama R, Kähönen M, Lehtimäki T, Akerblom HK, Viikari JS. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]