Abstract
Aims
Owing to strong linkage disequilibrium between markers, pinpointing disease associations within genetic regions is difficult in European ancestral populations, most notably the very strong association of the HLA-DRB1*03-DQA1*05:01-DQB1*02:01 haplotype with Type 1 diabetes risk, which is assumed to be because of a combination of HLA-DRB1 and HLA-DQB1. In contrast, populations of African ancestry have greater haplotype diversity, offering the possibility of narrowing down regions and strengthening support for a particular gene in a region being causal. We aimed to study the human leukocyte antigen (HLA) region in African American Type 1 diabetes.
Methods
Two hundred and twenty-seven African American patients with Type 1 diabetes and 471 African American control subjects were tested for association at the HLA class II genes, HLA-DRB1, HLA-DQA1, HLA-DQB1 and 5147 single nucleotide polymorphisms across the major histocompatibility complex region using logistic regression models. Population admixture was accounted for with principal components analysis.
Results
Single nucleotide polymorphism marker associations were explained by the HLA associations, with the major peak over the class II loci. The HLA association overall was extremely strong, as expected for Type 1 diabetes, even in African Americans in whom diabetes diagnosis is heterogeneous. In addition, there were unique features: the HLA-DRB1*03 haplotype was split into HLA-DRB1*03:01, which confers greatest susceptibility in these samples (odds ratio 3.17, 95% CI 1.72–5.83) and HLA-DRB1*03:02, an allele rarely observed in Europeans, which confers the greatest protection in these African American samples (odds ratio 0.22, 95% CI 0.09–0.55).
Conclusions
The unique diversity of the African HLA region we have uncovered supports a specific and major role for HLA-DRB1 in HLA-DRB1*03 haplotype-associated Type 1 diabetes risk.
Introduction
Owing to genome-wide single nucleotide polymorphism (SNP) association studies in Type 1 diabetes, over 50 Type 1 diabetes susceptibility regions have now been reported (http://www.t1dbase.org) 1–4. However, because of the strong linkage disequilibrium between markers, and relatively low haplotype diversity, narrowing down disease-associated regions to aid candidate gene selection is often difficult in European ancestral populations. African ancestral populations have much greater haplotype diversity, offering the possibility of providing support for a smaller genomic region containing the causal variant(s) and, in some instances, a particular causal candidate gene. This approach was first described explicitly in Type 1 diabetes in a human leukocyte antigen (HLA) study of 37 patients (cases) and 79 controls ascertained from Afro-Caribbean communities in England by Todd et al. 5 and later confirmed in 98 African American patients 6. Both studies reported an African-specific HLA-DRB1*07- HLA-DQB1*02 haplotype that predisposed to Type 1 diabetes and carried the 03:01 allele at HLA-DQA1, which is associated with increased risk of Type 1 diabetes in Europeans, suggesting a causal role for HLA-DQA1 5. However, these studies, and subsequent African Type 1 diabetes reports, did not control for the complex admixture that can be extreme in populations such as African Americans 7–9. Furthermore, despite these efforts and genetic analyses of very large European populations, including the application of dense SNP marker maps of the major histocompatibility complex (MHC) region that mapped susceptibility to the HLA-DQB1, HLA-DRB1, HLA-B and HLA-A genes 10,11, the specific HLA genes responsible for the high predisposing effects of HLA-DRB1*03 haplotypes remain uncertain. It is assumed that HLA-DRB1 and HLA-DQB1 are major determinants.
What's new?
This is the largest study in African Americans with Type 1 diabetes to date and the only study to account for ancestry.
This is the only study to date that fine maps the HLA region in African American samples using a dense map of 5147 single nucleotide polymorphisms and the class II genes.
We find HLA Type 1 diabetes associations that agree with known associations in Europeans, and novel associations of African ancestry specific alleles, e.g. HLA-DRB1*03:02, which confers protection from Type 1 diabetes.
Here, we typed African American samples with the dense immune disease SNP chip, ImmunoChip, with over 5000 SNPs in the HLA region 12 (http://www.immunobase.org), performed classical HLA class II typing, and adjusted for ancestry using principal component analysis. We obtained evidence for a specific role for the HLA-DRB1 gene on HLA-DRB1*03 haplotypes.
Subjects and methods
Subjects
In total, 227 subjects with clinically defined Type 1 diabetes were recruited from among 13 615 African Americans discharged from 31 New Jersey hospitals between 1982 and 1996 13. All patients had acute onset of disease, for which they were admitted to hospital. A full description of the sample collection can be found in Roy 13. Diagnosis was confirmed through a documented elevated random or postprandial venous plasma glucose level > 11.1 mmol/l and an elevated fasting glucose > 7.8 mmol/l. Over 12 000 samples were excluded from the collection because of the subjects having Type 2 diabetes, or having no initial need of insulin therapy, or because they were over 30 years of age at diagnosis. All subjects were taking insulin, had very low or no detectable C-peptide, and were under 30 years of age at diagnosis (median 17 years). The 497 African American control subjects were sex-matched to case subjects, were of all ages, and were recruited from local community organizations, blood bank attendees and medical clinic attendees in New Jersey, had no diabetes and no family history of diabetes.
All DNA samples were collected after approval from the University of Medicine and Dentistry of New Jersey Institutional Review Board research ethics committee and written informed consent was obtained from the participants.
Genotyping
Samples were genotyped using an Illumina 200K Infinium high-density array, Immunochip (http://www.immunobase.org) 12 according to the manufacturer's protocol at the Wellcome Trust Sanger Institute, Hinxton, UK. The SNP chip comprised 195 806 SNPs and 718 insertions/deletions covering 186 confirmed autoimmune disease-associated regions. SNPs with call rate below 0.95 or out of Hardy–Weinberg equilibrium with P < 1 × 10–6 were excluded. Owing to the small sample size compared with studies of Europeans, only SNPs with a minor allele frequency > 0.1 outside of the MHC region and 0.05 within the MHC region were included. Checks for sample heterozygosity, duplicate samples, related samples, inconsistent sex information and sample call rate < 0.9 were used to exclude samples. Samples with excessive European or Asian proportions from principal component analysis were removed (see also Supporting Information, Figs S1, S2 and S3). After all quality control measures, 227 case samples and 471 control samples were retained.
The HLA class II genes were genotyped using the LABType® SSO which applies Luminex® technology to the reverse sequence-specific oligonucleotide (SSO) DNA typing method.
Statistical methods
SNP chip quality control was performed using the snpStats library (http://www.bioconductor.org) in R (http://www.r-project.org).
SNPs and genes were modelled (and odds ratios calculated) using logistic regression with disease status as outcome variable and allele(s), assuming multiplicative allelic effects, as independent variable(s). The first three principal components were included as confounders and a likelihood ratio test employed to test for association of the allele(s) with Type 1 diabetes. Association analyses were performed with STATA version 10 (http://www.stata.com). Power calculations were performed using QUANTO (http://hydra.usc.edu/GxE/). Haplotypes were reconstructed under the alternative (separately in cases and controls) using an expectation–maximization (EM) algorithm as implemented in the R library, haplo.stats (http://www.r-project.org).
Results
Our study had 80% power to detect an odds ratio of 3.6 at genome-wide significance, α = 5 × 10–8, and an odds ratio of 2.5 at α = 0.001, assuming a minor allele frequency of 0.05 and a multiplicative allelic effects model. With the sample sizes currently available, analysis of small subgroups creates non-replicable false positive results owing to low statistical power. Therefore, although the HLA class II genes were genotyped to four-digit resolution, to maximize statistical power, we mostly analysed and report two-digit alleles at HLA-DRB1 and HLA-DQB1, unless four-digit alleles were sufficiently common. There are fewer HLA-DQA1 four-digit alleles, hence, this locus was analysed at four-digit resolution.
Single-locus association
The most convincing evidence of association in the MHC region was in the HLA class II region (Fig. 1). All three HLA class II genes tested were associated with Type 1 diabetes in these African American samples. HLA-DRB1 was the most associated gene (P = 7.79 × 10–26) and rs9273363C>A the most associated SNP (P = 9.72 × 10–20; situated ∼1 kb 3′ of HLA-DQB1). This SNP had less evidence of association than HLA-DQB1 (P = 1.62 × 10–23) or HLA-DQA1 (P = 2.13 × 10–23), particularly given that the HLA gene association tests are on more degrees of freedom, which reduces power.
FIGURE 1.
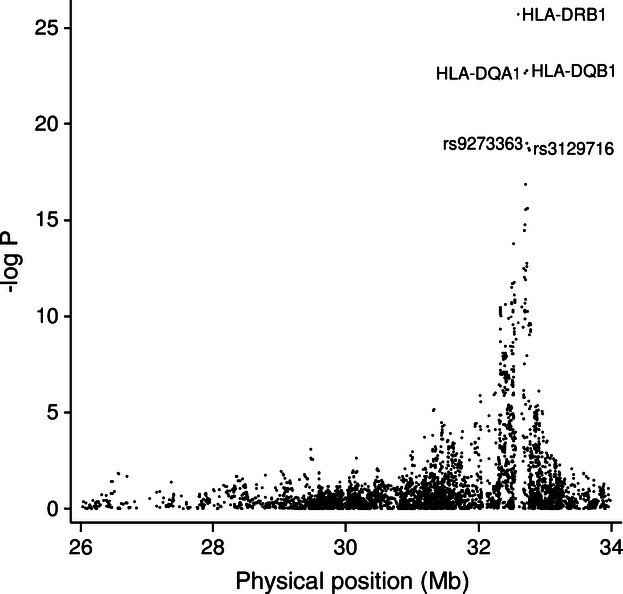
Association of 5147 single nucleotide polymorphisms (SNPs) and three genes across the extended major histocompatibility complex (MHC) region in up to 227 African American type 1 diabetes and 471 controls. All analyses were adjusted for population ancestry using the first three principal components, and signal clouds were manually checked for SNPs discussed in the manuscript with positive associations. Note, the minor A allele at rs9273363, the most associated SNP, was in linkage disequilibrium with the highly susceptible HLA-DRB1*03:01-HLA-DQA1*05:01-HLA-DQB1*02:01 (r2 = 0.43, D' = 1.00) and HLA-DRB1*04.HLA-DQA1*03:01-HLA-DQB1*03:02 haplotypes (r2 = 0.23, D' = 1.00).
Specific HLA class II alleles
While the general association pattern across the region is consistent with that reported in European samples 10,11, the nature of the HLA class II gene associations in African American samples differed to those in European samples. Most informatively, the highly susceptible haplotype in Europeans, signified by HLA-DRB1*03 14, is split in African Americans, into two haplotypes, one with a HLA-DRB1 allele that confers the greatest disease susceptibility in our study, HLA-DRB1*03:01 (odds ratio 3.17, 95% CI 1.72–5.83; Table 1) and the other with the allele that confers greatest protection, HLA-DRB1*03:02 (odds ratio 0.22, 95% CI 0.09–0.55). The risk allele, HLA-DRB1*03:01 has a frequency of approximately 0.14 in British control subjects (see also Supporting Information, Table S1; 15) and 0.05 in African American control subjects. In contrast, the HLA-DRB1*03:02 allele, which is rarely detected in Europeans, has a frequency of 0.07 in African Americans. This allele is in linkage disequilibrium with rs6927077 and rs6940690 (r2 = 0.79, D' = 0.91 in control subjects), which were themselves associated with Type 1 diabetes in African Americans (P < 5 × 10–5, odds ratio 0.26, 95% CI 0.12–0.55) but, remarkably, are monomorphic in the European ancestry CEPH samples and British control subjects (http://www.t1dbase.org/page/Overview/display/marker_id/6927077; http://www.t1dbase.org/page/Overview/display/marker_id/6940690), yet had a minor allele frequency of 0.07 in the African ancestry Yoruban (YRI) population.
Table 1.
Allele frequencies and association results for HLA-DRB1 in 227 African American cases of Type 1 diabetes and 471 controls
| Allele frequency, n (%) | |||||
|---|---|---|---|---|---|
| HLA-DRB1 allele | Cases | Controls | Odds ratio [95% CI]‡ | Odds ratio [95% CI]‡ | PT1D |
| 03:01 | 85 (18.7) | 51 (5.4) | 3.17 [1.72–5.83] | 4.19 [2.56–6.88] | |
| 04* | 74 (16.3) | 58 (6.2) | 2.37 [1.27–4.42] | 3.13 [1.87–5.23] | |
| 09:01 | 26 (5.7) | 21 (2.2) | 2.35 [1.09–5.09] | 3.11 [1.55–6.27] | |
| 07:01 | 59 (13.0) | 81 (8.6) | 1.63 [0.89–2.99] | 2.15 [1.32–3.52] | |
| 01 | 26 (5.7) | 59 (6.3) | 1.00 (reference) | 1.32 [0.74–2.37] | |
| 08 | 29 (6.4) | 67 (7.1) | 0.86 [0.43–1.74] | 1.14 [0.66–1.99] | |
| 13 | 60 (13.2) | 163 (17.3) | 0.76 [0.42–1.36] | 1.00 (reference) | |
| 11 | 35 (7.7) | 135 (14.3) | 0.51 [0.28–0.96] | 0.68 [0.41–1.12] | |
| 15:03 | 20 (4.4) | 109 (11.6) | 0.39 [0.19–0.80] | 0.52 [0.29–0.94] | |
| 15:01 | 5 (1.1) | 23 (2.4) | 0.36 [0.11–1.11] | 0.47 [0.16–1.36] | |
| 12 | 7 (1.5) | 43 (4.6) | 0.36 [0.13–0.94] | 0.47 [0.19–1.14] | |
| 03:02 | 8 (1.8) | 66 (7.0) | 0.22 [0.09–0.55] | 0.29 [0.13–0.67] | |
| Rares† | 20 (4.4) | 66 (7.0) | 0.68 [0.32–1.42] | 0.90 [0.48–1.69] | |
| 16 | 5 (1.1) | 14 (1.5) | |||
| 10:01 | 10 (2.2) | 29 (3.1) | |||
| 14 | 3 (0.7) | 20 (2.1) | |||
| 03:03 | 1 (0.1) | ||||
| 15:02 | 2 (0.4) | 2 (0.2) | |||
| 7.79 × 10−26 | |||||
The gene was in Hardy–Weinberg equilibrium in controls, P = 0.38.
Frequencies of the four-digit subtypes are given in the Supporting Information (Table S3).
The rare alleles, 16,10:01, 14, 03:03, 15:02, were grouped together.
Odds ratios were calculated with respect to a single reference allele using logistic regression (see Statistical methods).
In Europeans, HLA-DRB1*15 confers greatest protection from Type 1 diabetes associated with the alleles HLA-DRB1*15:01 and HLA-DQB1*06:02 (see also Supporting Information, Table S1 and S2). This was not the case in our African American samples (Table 1 and 2). The HLA-DRB1*15 allele, which, despite a frequency of 0.14 in African American control subjects that is consistent with Europeans (see also Supporting Information, Table S1), was more common than expected in our African American case subjects: 0.06 compared with ≤ 0.01 in European subjects with Type 1 diabetes. However, this was not associated with the common European HLA-DRB1*15 allele, HLA-DRB1*15:01, but HLA-DRB1*15:03, which is rarely observed in Europeans (see also Supporting Information, Table S1), and had a frequency of 0.04 and 0.12 in our African American case and control subjects, respectively. Both HLA-DRB1*15:01 and HLA-DRB1*15:03 were less protective for Type 1 diabetes in our African American samples [odds ratio 0.36 (95% CI 0.11–1.11), odds ratio 0.39, 95% CI 0.19–0.80)] than HLA-DRB1*15:01 in Europeans (odds ratio 0.05, 95% CI 0.04–0.07; Supporting Information, Table S1).
Table 2.
Allele frequencies and association results for HLA-DQB1 in 225 African American cases of Type 1 diabetes and 461 controls
| Allele frequency, n (%) | |||||
|---|---|---|---|---|---|
| Allele | Cases | Controls | OR [95% CI]* | OR [95% CI]* | PT1D |
| 02:01 | 86 (19.1) | 48 (5.2) | 4.25 [2.47–7.32] | 1.94 [1.19–3.18] | |
| 03:02 | 63 (14.0) | 39 (4.2) | 3.46 [1.90–6.31] | 1.58 [0.93–2.71] | |
| 02:02 | 92 (20.4) | 115 (12.5) | 2.19 [1.30–3.67] | 1.00 (reference) | |
| 03 | 10 (2.2) | 28 (3.0) | 1.24 [0.51–3.04] | 0.57 [0.25–1.29] | |
| 06 | 33 (7.3) | 92 (10.0) | 1.00 (reference) | 0.46 [0.27–0.77] | |
| 05 | 64 (14.2) | 191 (20.7) | 0.96 [0.58–1.58] | 0.44 [0.29–0.67] | |
| 03:19 | 25 (5.6) | 82 (8.9) | 0.94 [0.50–1.74] | 0.43 [0.24–0.76] | |
| 03:01 | 25 (5.6) | 82 (8.9) | 0.82 [0.44–1.52] | 0.37 [0.21–0.66] | |
| 04:02 | 18 (4.0) | 66 (7.2) | 0.79 [0.40–1.56] | 0.36 [0.19–0.69] | |
| 06:02 | 34 (7.6) | 175 (19.0) | 0.54 [0.31–0.94] | 0.25 [0.15–0.40] | |
| 02:03 | 4 (0.4) | ||||
| 1.62 × 10−23 | |||||
The gene was in Hardy–Weinberg equilibrium in controls, P = 0.37.
Odds ratios were calculated with respect to a single reference allele using logistic regression (see Statistical methods).
In contrast to Europeans, in whom the HLA-DRB1*09:01 allele is neutral for Type 1 diabetes risk (Supporting Information, Table S1), in our African American samples, it confers susceptibility (Table 1; odds ratio 2.35, 95% CI 1.09–5.09). Lastly, at HLA-DQB1, HLA-DQB1*04:02 is more common than observed in Europeans (Supporting Information, Table S2).
HLA-DQA1 alleles were also very strongly associated with Type 1 diabetes in African Americans, reflecting the strong linkage disequilibrium with highly disease-associated HLA-DRB1 and HLA-DQB1 alleles (Table 3). However, in the haplotype analysis results section below, we do present evidence of an independent effect of HLA-DQA1 on a specific haplotype background.
Table 3.
Allele frequencies and association results for HLA-DQA1 in 225 African American cases of Type 1 diabetes and 461 controls
| Allele frequency, n (%) | |||||
|---|---|---|---|---|---|
| Allele | Cases | Controls | Odds ratio [95% CI]‡ | Odds ratio [95% CI]‡ | PT1D |
| 03:02 | 50 (11.1) | 18 (2.0) | 3.07 [1.65–5.71] | 5.09 [2.97–8.74] | |
| 05:01 | 88 (19.6) | 57 (6.2) | 2.67 [1.54–4.62] | 4.43 [2.81–6.97] | |
| 03:01 | 81 (18.0) | 84 (9.1) | 2.47 [1.44–4.25] | 4.09 [2.60–6.45] | |
| 02:01 | 38 (8.4) | 84 (9.1) | 1.00 (reference) | 1.66 [1.02–2.70] | |
| 01:01 | 43 (9.6) | 139 (15.1) | 0.72 [0.42–1.25] | 1.20 [0.76–1.90] | |
| Rares† | 15 (3.3) | 46 (5.0) | 0.69 [0.33–1.43] | 1.14 [0.59–2.21] | |
| 05:05 | 29 (6.4) | 112 (12.1) | 0.67 [0.37–1.22] | 1.11 [0.67–1.83] | |
| 04:01 | 31 (6.9) | 104 (11.3) | 0.67 [0.37–1.21] | 1.10 [0.67–1.82] | |
| 01:02 | 75 (16.7) | 278 (30.2) | 0.60 [0.37–0.98] | 1.00 (reference) | |
| 01:03 | 13 (2.9) | 41 (4.4) | |||
| 05:03 | 1 (0.1) | ||||
| 06:01 | 2 (0.4) | 4 (0.4) | |||
| 2.13 × 10−23 | |||||
The gene was in Hardy–Weinberg equilibrium in controls, P = 0.55.
Rare alleles comprise *01:03, *05:03, *06:01.
Odds ratios were calculated with respect to a single reference allele using logistic regression (see Statistical methods).
Conditional analyses
Tests for additional independent signals of association in the HLA region in our African American samples showed neither rs9273363, HLA-DQB1 nor HLA-DQA1 added to a model with HLA-DRB1 (P = 0.059, 0.089 and 0.055, respectively). All SNP associations conditional on HLA-DRB1 were unconvincing (P > 1 × 10–4; Supporting Information, Fig. S4). The most evidence of association was at rs1383264 (P = 2.00 × 10–4) and rs3129716 (P = 1.50 × 10–4), both of which are intergenic in the class II region and so their marginal associations are probably because of linkage disequilibrium with HLA-DQB1 and HLA-DQA1. We would expect the HLA-DQ genes to add to HLA-DRB1 if we had a larger sample set, and lack of additional association signals is probably attributable to insufficient power.
Haplotype analyses
The African-specific HLA-DRB1*03:02 allele, which is associated with protection for Type 1 diabetes, occurs on the HLA-DQA1*04:01-HLA-DQB1*04:02 haplotype and on no other HLA haplotype in our samples (Table 4). Exchange of HLA-DRB1*03:02 for HLA-DRB1*08 removes the protective effect of the HLA-DQA1*04:01-HLA-DQB1*04:02 haplotype (Table 4) despite both HLA-DQ alleles conferring protection in a single locus analysis, indicating HLA-DRB1 is likely to be causal. HLA-DRB1*08 is more common in our African American samples (0.07) than in Europeans (0.02), which is attributable to HLA-DRB1*08 occurring on the HLA-DQA1*04:01-HLA-DQB1*03:19 haplotype, a haplotype rarely observed in Europeans. HLA-DRB1*07:01 is protective in European populations, but not in our African American samples (odds ratio 1.63, 95% CI 0.89–2.99; Table 1). However, as reported by others 5,6, we observed that the effect of the HLA-DRB1*07:01 haplotype on Type 1 diabetes risk is dependent on the HLA-DQA1 allele present. The HLA-DRB1*07:01-HLA-DQA1*02:01-HLA-DQB1*02:02 haplotype is neutral for risk on Type 1 diabetes (frequency 7.3% in cases, 6.4% in controls). In contrast, HLA-DRB1*07:01-HLA-DQA1*03:01-HLA-DQB1*02:02, confers susceptibility to Type 1 diabetes (frequency 3.1% in cases, 0.8% in controls; odds ratio 5.28, 95% CI 1.77–15.81; Table 4). Similarly, HLA-DRB1*07-HLA-DQA1*03:02-HLA-DQB1*02:02 had a frequency of 2.0% in cases and 0.4% in controls. We also note that although HLA-DRB1*15:03 is more common than HLA-DRB1*15:01 allele, they both occur on the same haplotype background, namely, HLA-DQA1*01:02-HLA-DQB1*06:02.
Table 4.
HLA-DRB1. HLA-DQA1. HLA-DQB1 African American haplotypes, in 225 African American cases and 461 African American controls with complete genotypes at all three genes
| Haplotype | Frequency/% | Odds ratio [95% CI] | ||
|---|---|---|---|---|
| DRB1.DQA1.DQB1 | Cases | Controls | ||
| 0701.0301.0202 | 3.1 | 0.8 | 1.93 [0.68–5.48] | 5.28 [1.77–15.81] |
| 0701.0302.0202 | 2.0 | 0.4 | ||
| 04.0302.0302 | 4.2 | 0.8 | 1.37 [0.47–4.06] | 3.75 [1.25–11.31] |
| 0301.0501.0201 | 18.9 | 5.1 | 1.00 (reference) | 2.73 [1.52–4.91] |
| 08.0401.0402 | 2.2 | 0.8 | ||
| 04.0301.0302 | 9.3 | 3.3 | 0.72 [0.37–1.37] | 1.96 [0.97–3.92] |
| 0901.0302.0202 | 2.7 | 0.3 | ||
| 0901.0301.0202 | 2.9 | 1.8 | 0.53 [0.24–1.18] | 1.44 [0.60–3.47] |
| 0701.0201.0202 | 7.3 | 6.4 | 0.37 [0.20–0.66] | 1.00 (reference) |
| 13.0102.05 | 2.4 | 2.7 | 0.32 [0.13–0.78] | 0.87 [0.35–2.17] |
| 01.0101.05 | 5.6 | 6.2 | 0.29 [0.16–0.52] | 0.78 [0.40–1.54] |
| 13.0102.06 | 4.2 | 4.7 | 0.26 [0.14–0.50] | 0.72 [0.35–1.47] |
| 13.0505.0301 | 1.5 | 1.7 | 0.28 [0.09–0.87] | 0.77 [0.23–2.53] |
| 04.0301.0301 | 0.9 | 1.3 | ||
| 1001.0101.05 | 2.2 | 3.1 | 0.20 [0.09–0.47] | 0.56 [0.23–1.33] |
| 11.0505.0319 | 2.4 | 3.9 | 0.22 [0.10–0.45] | 0.59 [0.26–1.32] |
| 08.0401.0319 | 2.9 | 4.0 | 0.21 [0.09–0.47] | 0.57 [0.24–1.34] |
| 13.0103.06 | 2.7 | 3.9 | 0.18 [0.07–0.41] | 0.48 [0.19–1.18] |
| 13.0201.0202 | 0.9 | 1.8 | ||
| 13.0505.0319 | 0.3 | 1.0 | ||
| 14.0101.05 | 0.7 | 1.7 | ||
| 11.0102.05 | 1.1 | 1.4 | 0.14 [0.04–0.43) | 0.38 [0.12–1.18] |
| 11.0102.0602 | 2.0 | 4.5 | 0.13 [0.05–0.30) | 0.35 [0.14–0.86] |
| 16.0102.05 | 0.7 | 1.5 | ||
| 1501.0102.0602 | 1.1 | 1.8 | 0.13 [0.03–0.45] | 0.34 [0.09–1.27] |
| 1503.0102.0602 | 4.0 | 11.1 | 0.11 [0.05–0.22] | 0.30 [0.14–0.62] |
| 0302.0401.0402 | 1.8 | 6.4 | 0.09 [0.04–0.22] | 0.25 [0.10–0.64] |
| 11.0505.0301 | 0.9 | 3.6 | 0.08 [0.03–0.23] | 0.22 [0.07–0.64] |
| 12.0101.05 | 0.9 | 3.4 | 0.07 [0.02–0.19] | 0.19 [0.06–0.55] |
| Rares | 0.26 [0.16–0.41] | 0.70 [0.41–1.20] | ||
The study is underpowered to detect rare effects, therefore only haplotypes with a combined frequency > 1% are listed and only those with a frequency > 1.5% are used to calculate odds ratios and 95% confidence intervals.
Discussion
Our study of samples from donors with African ancestry is not only the largest of its kind to date in terms of number of Type 1 diabetes cases analysed for association, marker resolution and coverage of the extended MHC region, it also controls for bias from population substructure. The alleles that confer the greatest susceptibility and protection for Type 1 diabetes at the HLA-DRB1 locus, 03:01 and 03:02, respectively, differ at four exon 2-encoded amino acids, positions 26, 28, 47 and 86 with HLA-DRB1*03:02 encoding phenylalanine, glutamic acid, tyrosine and glycine, respectively, while HLA-DRB1*03:01 encodes tyrosine, aspartic acid, phenylalanine and valine, respectively. These differences could well alter the peptide-binding specificity and capacity of these class II allotypes and their interactions with T-cell receptors, in particular the glycine to valine difference at position 86, a residue that forms part of peptide binding pocket 1 and has been associated with disease susceptibility previously and peptide binding 16,17.
The specific association of HLA-DQA1 on the HLA-DRB1*07:01 haplotype was also confirmed in our samples (Table 4), which had more power than previous studies 5,6. In addition, owing to improved resolution of the HLA genotyping system used, and in contrast to a previous study 6, we were able to distinguish HLA-DQB1*02 subtypes and found that the HLA-DQB1*02:02 allele rather than the HLA-DQB1*02:01 allele was present on this HLA-DRB1*07:01 haplotype.
Finally, we note the unexpectedly high frequency of the protective HLA-DRB1*15 haplotypes in our African American cases compared with Europeans, does not indicate inclusion of non-autoimmune Type 1 diabetes or cases with Type 2 diabetes. The excess was attributable to the HLA-DRB1*15:03 allele, which is rarely observed in Europeans, and is less protective for Type 1 diabetes than HLA-DRB1*15:01 in Europeans (Table 1 and Supporting Information, Table S1). Although HLA-DRB1*15:03 is on the same haplotype, with HLA-DQB1*06:02, this African-specific haplotype could well have different sequences affecting class II gene expression or structure, that increase its predisposition to Type 1 diabetes. For example, the African American HLA-DRB1*15:03 allele's exon 2 sequence differs from 15:01 at amino acid, position 29 (tyrosine to histidine).
In conclusion, given the differences in risk of Type 1 diabetes associated with HLA-DRB1*03 and HLA-DRB1*08 haplotypes, our results are consistent with the assumption that HLA-DRB1 is causal on the HLA-DRB1*03 haplotype. Furthermore, they illustrate the utility of African samples in the analysis of common disease and a need to collect and analyse much larger sample sizes. Our study, in combination with larger studies in the future, will be important for evaluating HLA and non-HLA mediated risk of Type 1 diabetes by ethnic group, which could prove informative for identifying candidates for future causal genes and the pathways involved in disease initiation and progression.
Acknowledgments
From the JDRF/Wellcome Trust Diabetes and Inflammation Laboratory (DIL) at the University of Cambridge, we thank Gillian Coleman for preparation of DNA samples, Trupti Mistry for performing HLA genotyping, Neil Walker for data management and for generating PLINK files of ImmunoChip data, and Hui Guo for regenerating the final reports for the ImmunoChip data.
Funding sources
The authors are supported by grants from the Juvenile Diabetes Research Foundation (JDRF), the Wellcome Trust, and the National Institute for Health Research Cambridge Biomedical Centre. The Cambridge Institute for Medical Research is in receipt of a Wellcome Trust Strategic Award (079895). MSR is in receipt of a JDRF grant 8-2004-780. We acknowledge support from the National Institute on Alcohol Abuse and Alcoholism.
Competing interests
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Principal component analysis of the African American samples.
Figure S2. Eigenvalues of the principal components plotted against principal component number.
Figure S3. Q-Q plot for association of the ImmunoChip single nucleotide polymorphisms.
Figure S4. Association with Type 1 diabetes in African Americans conditional on HLA-DRB1 alleles.
Table S1. Allele frequencies in 6201 white British subjects with Type 1 diabetes and 5323 control subjects at HLA-DRB1.
Table S2. Allele frequencies in 6198 white British subjects with Type 1 diabetes and 5295 control subjects at HLA-DQB1.
Table S3. Frequencies of HLA-DRB1*04 subtypes in African American subjects with Type 1 diabetes and control subjects.
References
- 1.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howson JM, Rosinger S, Smyth DJ, Boehm BO, ADBW-END study Group, Todd JA. Genetic analysis of adult-onset autoimmune diabetes. Diabetes. 2011;60:2645–2653. doi: 10.2337/db11-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd JA, Mijovic C, Fletcher J, Jenkins D, Bradwell AR, Barnett AH. Identification of susceptibility loci for insulin-dependent diabetes mellitus by trans-racial gene mapping. Nature. 1989;338:587–589. doi: 10.1038/338587a0. [DOI] [PubMed] [Google Scholar]
- 6.Noble JA, Johnson J, Lane JA, Valdes AM. Race-specific type 1 diabetes risk of HLA-DR7 haplotypes. Tissue Antigens. 2011;78:348–351. doi: 10.1111/j.1399-0039.2011.01772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombard Z, Brune AE, Hoal EG, Babb C, Van Helden PD, Epplen JT, et al. HLA class II disease associations in southern Africa. Tissue Antigens. 2006;67:97–110. doi: 10.1111/j.1399-0039.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 8.Pirie FJ, Hammond MG, Motala AA, Omar MA. HLA class II antigens in South African Blacks with type I diabetes. Tissue Antigens. 2001;57:348–352. doi: 10.1034/j.1399-0039.2001.057004348.x. [DOI] [PubMed] [Google Scholar]
- 9.Berka N, Bland GN, Gause DP, Harris PF, Erabhaoui EH, Bonar AS, et al. Early age of disease onset in African American type 1 diabetes patients is associated with DQB1*0201 allele. Hum Immunol. 2000;61:816–819. doi: 10.1016/s0198-8859(00)00148-8. [DOI] [PubMed] [Google Scholar]
- 10.Howson JM, Walker NM, Clayton D, Todd JA Type 1 Diabetes Genetics Consortium. Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes Metab. 2009;11:S31–45. doi: 10.1111/j.1463-1326.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy MS. Diabetic retinopathy in African Americans with type 1 diabetes: The New Jersey 725: I. Methodology, population, frequency of retinopathy, and visual impairment. Arch Ophthalmol. 2000;118:97–104. doi: 10.1001/archopht.118.1.97. [DOI] [PubMed] [Google Scholar]
- 14.Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, et al. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the UK. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 15.Howson JM, Stevens H, Smyth DJ, Walker NM, Chandler KA, Bingley PJ, et al. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes. 2011;60:2635–2644. doi: 10.2337/db11-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cucca F, Lampis R, Congia M, Angius E, Nutland S, Bain SC, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet. 2001;10:2025–2037. doi: 10.1093/hmg/10.19.2025. [DOI] [PubMed] [Google Scholar]
- 17.Busch R, Hill CM, Hayball JD, Lamb JR, Rothbard JB. Effect of natural polymorphism at residue-86 of the HLA-DR beta-chain on peptide binding. J Immunol. 1991;147:1292–1298. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


