Abstract
Background
Phenanthroindolizidine alkaloids are a family of plant-derived compounds with significant antineoplastic activity. The specific biomolecular targets of these alkaloids have not yet been clearly identified. (+)-(13aS)-deoxytylophorinine is a new phenanthroindolizidine alkaloid originally extracted from the roots of Tylophora atrofolliculata and Tylophora ovata in our institute. (+)-(13aS)-deoxytylophorinine exerts both in vitro and in vivoanticancer activities.
Methods
The in vivo anticancer effects and toxicity of this compound were investigated in mice, and interactions between this compound and double-helical DNA sequences were studied in detail with circular dichroic spectroscopy and fluorescence spectroscopy. Viscosity measurements were applied to check the interactive mode between this compound and DNA.
Results
Potent anticancer effects were observed in vivo. Also, concentration-dependent interactions were observed and this compound seemed to interact in a sequence-specific manner with AT-repeated sequences of double-helical DNA. Such interactions were proved to be intercalating by viscosity measurements.
Conclusions
Anticancer alkaloid (+)-(13aS)-deoxytylophorinine can have sequence-specific interactions with DNA in an intercalating manner.
Key Words: Phenanthroindolizidine, (+)-(13aS)-deoxytylophorinine, Anticancer alkaloid, Chemotherapy, DNA-ligand interaction, DNA intercalation
Introduction
Phenanthroindolizidine alkaloids are a family of plant-derived alkaloids isolated from Asclepiadaceae and the Moraceae plant family [1]. Many of these alkaloids possess significant anticancer activity (both in vitro and in vivo) as well as anti-inflammatory, antiameobicidal, and antiviral activities [2]. The significant antineoplastic activities of many phenanthroindolizidines have attracted the interest of quite a number of medicinal chemists and pharmacologists since 1935 when the first phenanthroindolizidine alkaloid, tylophorine, was isolated from Tylophora indica [3]. However, the specific cellular target of the phenanthroindolizidines is still unclear, though a significant amount of cellular activity of these compounds is known. Suppressions of the syntheses of protein and nucleic acid have been reported previously and the 40S ribosomal subunit has been postulated to host the binding site of these compounds [4]. Other targets, such as thymidylate synthase [5], dihydrofolate reductase [6], and NF-κB [7], were also reported. Tylocrebrine, a phenanthroindolizidine alkaloid, inhibits protein, RNA, and DNA synthesis [8]. Therefore, we highly suspect that nucleic acids themselves may be the primary target of this kind of compound, although other unclear mechanism may also work. Interactions of such compounds with DNA may directly interfere with the process of duplication and transcription. Thus, the direct interactions of such compounds with DNA were determined in our laboratory and interesting results were found.
(+)-(13aS)-deoxytylophorinine (CAT; fig. 1a) is a new phenanthroindolizidine alkaloid, originally extracted in our institute from the roots of Tylophora atrofolliculata and Tylophora ovata, which has a long history of being used as medicine to treat rheumatism and wounds in China [9]. Eleven phenanthroindolizidine alkaloids were isolated and screened. Of all of these alkaloids, CAT was found to have potent anticancer activities both in vitro and in vivo.
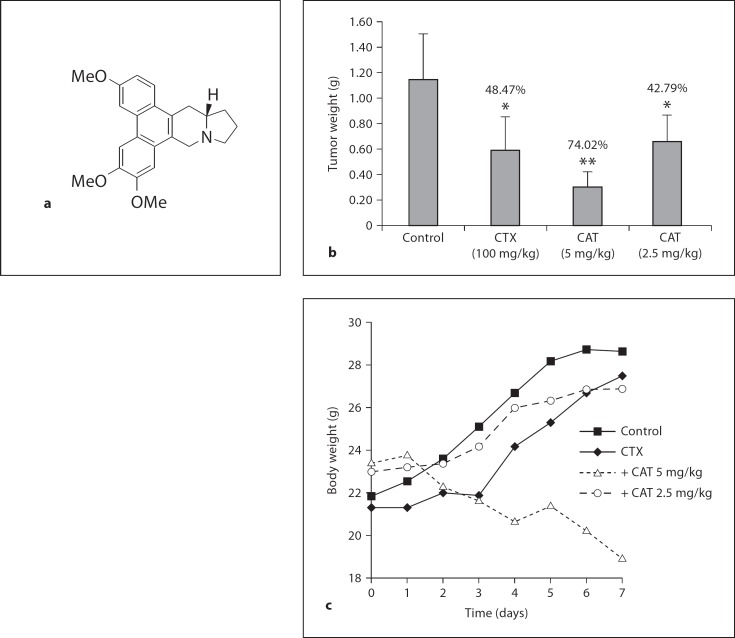
Fig. 1.
Tumor weight and body weight after administration of CAT. a Chemical structure of CAT. b Tumor weights for the different groups. * p < 0.05; ** p < 0.01 versus the control group. c Change in body weight after the treatments. There were 6 mice in each group. CTX = Cyclophosphamide.
Detailed studies of the interactions between this compound and DNA were carried out. Concentration-dependent interactions between this compound and DNA were observed in DNA solution by circular dichroism (CD) [10] and fluorescence emission spectra [11,12]. What is of great significance is that this compound seemed to interact in a sequence-specific manner with an AT-repeated sequence of double-helical DNA and this is, to the best of our knowledge, the first time a phenanthroindolizidine alkaloid was found to have sequence-specific interactions with duplex DNA. CD was used to determine such interactions. Actinomycin D, a classic DNA-intercalating anticancer drug, was used as the positive control to verify the method. The interactions between DNA and small molecules can be intercalating or groove binding, and the interactions between CAT and DNA were later proved to be intercalating rather than groove binding.
In short, CAT is a new phenanthroindolizidine alkaloid with significant anticancer effects. We detected the interactions between this compound and many sorts of DNAs, and interesting results were found and are reported in this article.
Materials and Methods
Animal Study
Male KM mice were obtained from the Laboratory Animal Center of the Academy of Military Medical Science and acclimated to laboratory conditions 1 week before tumor implantation. The mice were maintained in accordance with Institutional Animal Care and Use Committee procedures and guidelines. H22 murine hepatoma (purchased from the Cell Culture Center of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences) xenografts were established by subcutaneously injecting 1 × 106 H22 cells into the axillary fossa. Treatment was initiated the day after implantation. CAT was dissolved in 0.5% aqueous carboxymethyl cellulose sodium. The solvent alone served as the control. Cyclophosphamide was dissolved in normal saline. CAT was administered p.o. once a day continuously for 6 days and cyclophosphamide was administered by i.p. injection once the day after innoculation. There were 6 mice in each group. The body weights of the mice were monitored daily for toxicity. At the end of the treatment, the mice were sacrificed and the tumors were peeled off and weighed. To evaluate the anticancer activity of CAT, the percentage of inhibition was calculated as follows:
CD Spectroscopy
CD spectra were measured using a Jasco J-815 CD spectrometer (displaying the CD in millidegrees of ellipticity) using 0.5 cm cells. All of the CD experiments were carried out at 25°C in BPES buffer (6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA, and 185 mM NaCl; pH 7.0) [10]. Calf thymus DNA (CT-DNA) (catalog No. D1501; Sigma) was dissolved in BPES buffer as stock solution and diluted when used. Polynucleotides were purchased from Shanghai Sangon Biological Engineering Technology and Services Co. Ltd. with mass spectrometry characterization. They were first synthesized linearly and then annealed to form the hairpin and duplex structures before use. Annealing of strands to form duplexes was accomplished by heating of the solution at 95°C for 5 min and then slow cooling to room temperature, followed by refrigeration for 30 min to ensure the formation of hairpins or duplexes [13]. The concentration of CT-DNA was calculated by the amount of base pairs [M(bp)] [10], and that of hairpin polynucleotides was calculated by the amount of base pairs [M(bp)] of their duplex parts. Actinomycin D (catalog No. BIA-A1185, purity >99% by HPLC; Bioaustralis) and CAT were dissolved in dimethyl sulfoxide (DMSO) for a stock solution of 6 mM. DMSO was previously proved not to interfere with CD spectra in the wavelength range of 232–450 nm up to a concentration of 5% in BPES buffer (online suppl. fig. 1; for all suppl. material, see www.karger.com?doi=10.1159/000329506). Stock solutions of DNA and drugs were diluted with BPES buffer with the designated concentrations and ratios to make a 1-ml volume system and stirred constantly before being scanned from 350 to 220 nm. The samples were scanned every 0.5 nm 4 times and average values were used for the spectra automatically. The baseline of solvent CD signals was deducted from each spectrum. Induced CD (ICD) spectra were obtained by subtracting the spectra of the single compound and the spectra of the single DNA from the spectra of their mixtures of corresponding concentrations.
Fluorescence Spectroscopy
Fluorescence emission spectra were recorded for CAT in the presence and absence of CT-DNA in the range of 340–450 nm upon excitation at 260 nm at 23°C using a slit width of 2 nm. Fluorescence emission spectra were measured using a fluorescence spectrophotometer (Spectra Max Gemini XS, USA). Samples of DNA and CAT were treated the same way as in CD spectroscopy. Designated concentrations and ratios of DNA and CAT were added and mixed before being scanned.
Viscosity Measurements
All viscosity measurements were carried out in a Ubbelohde viscometer (inner capillary diameter 0.5 mm) submerged in a water bath that was maintained at 25 ± 0.5°C. Flow times were measured in triplicate with a stopwatch and the average time of the different replicates was used. Viscosity studies on CT-DNA and plasmid pEGFP-N1 (catalog No. 6085-1; Clontech) were conducted in BPES buffer. Aliquots of CAT solutions in concentrated stock solutions were titrated directly into the viscometer containing DNA solutions (15 ml). The plasmid pEGFP-N1 used here was propagated in DH5a with kanamycin (30 μg·ml−1) as a selectable marker and extracted using a Plasmid Maxprep Kit (Vigorous Biotechnology) according to the kit's manual.
Statistical Tests
Data were analyzed by 2-tailed t tests using Prism 4 (GraftPad Software). Data are presented as means ± SD.
Results
In vivo Anticancer Activity of CAT
To assess the therapeutic effects of CAT in vivo, animal studies were carried out using KM mice with H22 mouse hepatoma xenografts. CAT (5 and 2.5 mg/kg) was administered p.o. once a day continuously for 6 days from the second day of inoculation. Body weight was recorded once a day on each of the following days. Cyclophosphamide (100 mg/kg) was used as the positive control and administered i.p. once a day after inoculation. On the seventh day, the mice were sacrificed, the solid tumors in the axillary fossa were removed and weighed, and inhibition rates were calculated (fig. 1b). The inhibition rate of CAT 5 mg/kg was 74.47% compared to the control group, and that of CAT 2.5 mg/kg was 42.79%. However, the average body weight of the CAT 5 mg/kg group decreased rapidly to 19 g on the last day (fig. 1c). The decreased body weight was an obvious indication of severe toxicity.
Concentration-Dependent Interactions between CAT and DNA Determined by CD
CD is a powerful tool for the study of the secondary structures and conformations adopted by nucleic acids. CD arises from the asymmetric backbone sugars and the helical structures often adopted by nucleic acids [14]. CD spectroscopy is extremely sensitive to nucleic acid conformations, especially within the 180- to 320-nm wavelength range [15]. The CD spectrum of DNA is sensitive to changes in conformation resulting from ligand binding [10,14,16]. CD is particularly powerful for monitoring structural changes resulting from changes in environmental conditions [14,17,18]. CT-DNA is often conveniently used for DNA-ligand studies. A representative spectrum of standard right-handed double-helical DNA was obtained by using 3 × 10−5M(bp) CT-DNA (Sigma) in BPES buffer with a CD maximum near 280 nm, a CD minimum near 245 nm, and a crossover point at ∼260 nm (online suppl. fig. 2).
Many DNA-binding ligands are achiral and optically inactive. Upon interaction with DNA, a ligand can acquire an ICD signal through coupling of the electric transition moments of the ligand and the DNA bases. The observation of an ICD signal within the absorption bands of the achiral ligand is immediately indicative of a ligand-DNA interaction [10,14,16,18]. However, CAT, with a chiral carbon atom in its structure, exhibits an intrinsic CD spectrum that is expected to change the CD spectrum of polynucleotide when mixed even if there are no interactions between CAT and DNA. Spectra of CT-DNA [3 × 10−5M(bp)] were obtained by adding a series concentration of CAT, with a CAT/DNA [M/M(bp)] ratio increasing from 0.02 to 10 (online suppl. fig. 3). The DNA CD spectrum is significantly altered in the presence of ligand. As CAT is intrinsically chiral, its own CD spectrum (online suppl. fig. 4) should be considered and subtracted. The ICD (fig. 2a) is the CD spectra with the DNA spectra and CAT spectra subtracted; the CAT/DNA [M/M(bp)] ratio ranges from 0.02 to 10, and the CT-DNA concentration is 3 × 10−5M(bp). Minimum ICD signals of 255 nm were evident in the ICD spectra. The CD spectrum of DNA is sensitive to changes in conformation resulting from ligand binding. A constructed curve of the minimum ICD, at around 255 nm, versus the logarithm of the CAT/DNA [M/M(bp)] was generated from the ICD raw data (fig. 2a). A concentration-dependent relationship in this figure can be an indication of dose-independent interactions between CAT and DNA.
Fig. 2.
Concentration-dependent interactions between CAT and DNA. a ICD of increasing concentrations of CAT in the presence of CT-DNA [3 × 10−5M(bp)], that is the original CD spectra with the CT-DNA CD spectrum and the CAT CD spectra subtracted. The curve of the minimum ICD, at around 255 nm, versus the logarithm of CAT/DNA [M/M(bp)] generated from the ICD data shows the concentration dependence. b Fluorescence emission spectra of CAT with a series of DNA/CAT ratios [M/M(bp)] obtained by an excitation wavelength of 260 nm and emission wavelengths ranging from 340 to 450 nm at 23°C. The CAT concentration is fixed at 3 × 10−5M. The reduced intensity at the wavelength of 380 nm is plotted against the logarithm concentration of the DNA concentration [M(bp)]. c Reduced emissions of different CAT concentrations with an increasing CAT/DNA ratio [M/M(bp)]. The reduced emission at 380 nm is plotted against the logarithm of the CAT/DNA ratio [M/M(bp)] and the maximum emission reduction apparently occurs at a CAT/DNA ratio of 1:1. The CT-DNA concentration is fixed at 3 × 10−5M(bp).
Concentration-Dependent Interactions between CAT and DNA Determined by Fluorescence Emission Spectra
Fluorescence emission spectra are sensitive to changes in the fluorophore's environment such as polarity and the presence of quencher [12]. Fluorescence emission spectra were recorded for CAT in the presence and absence of DNA with a 260-nm excitation. We obtained fluorescence emission spectra in the absence and with increasing concentrations of CT-DNA (fig. 2b). With increasing CT-DNA concentrations, the fluorescence intensity decreased gradually without any perceptible shift in the fluorescence maximum of around 380–382 nm. The fluorescence intensity of CAT tends to be constant at a high concentration of CT-DNA, which suggests the reaching of binding saturation. The reduced intensity compared with the CAT fluorescence intensity in the wavelength of 380 nm was plotted against the logarithm concentration of the DNA concentration [M(bp)], and a concentration-independent curve was formed (fig. 2b).
In order to further study the binding stoichiometry of the CAT-DNA complex, a series of concentrations of CAT ranging from 0.02 to 10 were added to a fixed concentration of CT-DNA [3 × 10−5M(bp)]. Fluorescence emission spectra of DNA with increasing concentrations of CAT were obtained with the same parameters. The reduced emissions compared to the original CAT emissions of different CAT concentrations were plotted against wavelengths. It was found that when the CAT/DNA ratio [M/M(bp)] increased from 0.02 to 1, the magnitude of the reduced emission increased in the wavelength range of 350–450 nm (online suppl. fig. 5). However, when the CAT/DNA ratio [M/M(bp)] increased over 1 (from 1 to 10), the magnitude of the reduced emissions started to decrease in a concentration-dependent manner (online suppl. fig. 6). The reduced emission at 380 nm is plotted against the logarithm of the CAT/DNA ratio, and the maximum emission reduction apparently occurs at a CAT/DNA ratio of 1:1 (fig. 2c).
Sequence-Specific Interactions between CAT and DNA
CD spectroscopy was used to further determine whether CAT can interact in a sequence-specific manner with certain sequences of DNA as some compounds do. Similar studies with plant alkaloid were reported previously [19,20]. The extent of the changes in ICD may be indicative of different binding abilities between the compound and various DNA sequences. Four specific short hairpin DNA sequences were chosen for this study, with A-repeated (T-repeated for the complementary strand), AT-repeated, G-repeated (C-repeated for the complementary strand), and GC-repeated double-helical sequences (fig. 3a). It should be noted that it is difficult to synthesize over 5 continuous G in the polynucleotide sequences chemically. Therefore, the G-repeated sequence used here has an A inserted between 4 continuous G and, in compensation, the other 3 sequences all have an extra nucleotide end added at both 5′ and 3′ ends, respectively. All of the 4 hairpins were originally chemically synthesized as single strands and afterwards annealed to form the hairpin structures [13,19]. Unlike CT-DNA or any other standard double-helical DNA, the CD spectra of such short sequences (online suppl. fig. 7) showed their unique shapes which are not identical to that of CT-DNA. The exact positions and intensities of peaks are sensitive to base composition and sequence.
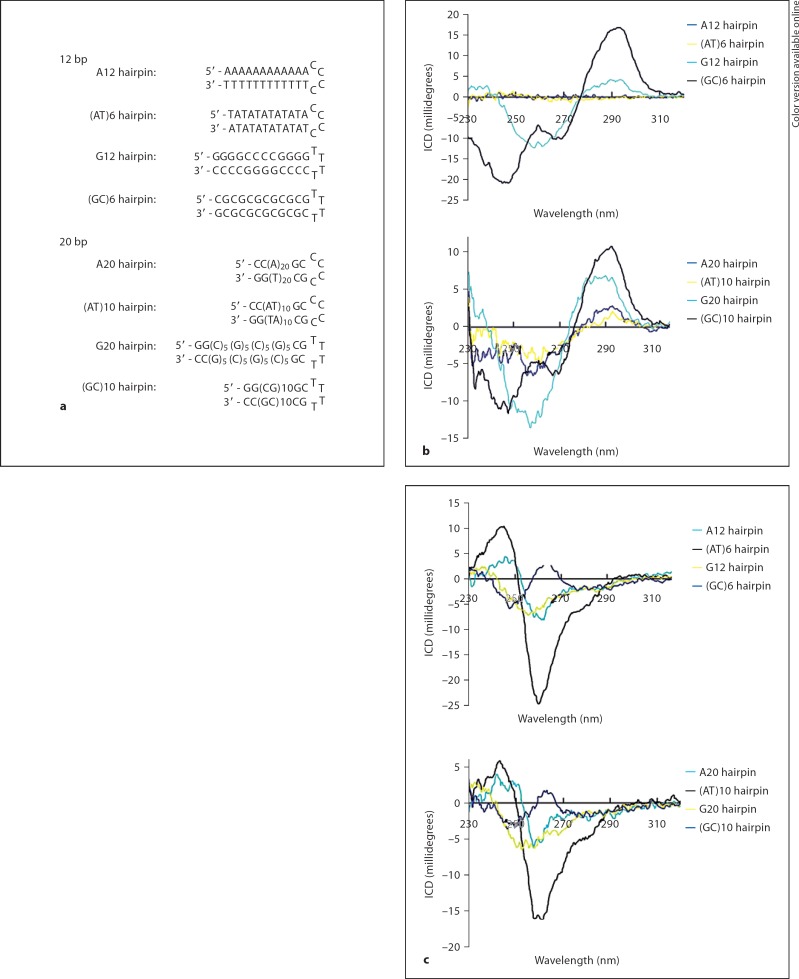
Fig. 3.
Sequence-specific interactions of actinomycin D and CAT with 8-bp short DNA sequences. a Short hairpin DNA sequences (8 bp) used in the experiments. b ICD of actinomycin D (6 × 10−5M) in the presence of four 8-bp DNA sequences [3 × 10−5M(bp)], respectively. The ICD of the (GC)4 hairpin is significantly different from that of the other 3 sequences. c ICD of CAT (CAT/DNA ratios 1:1, 2:1, and 4:1) in the presence of four 8-bp DNA sequences [3 × 10−5M(bp)], respectively. The ICD of the (AT)4 hairpin is more intensive than that of the other 3 sequences compared with the same ratio group.
In order to check the feasibility of determining sequence-specific DNA-compound interactions by CD spectroscopy we used actinomycin D, which is a widely studied definite DNA intercalator, as a positive control compound. Actinomycin D is a potent antitumor antibiotic that preferentially targets the GC-repeated sequence on double-helical DNA. The binding of actinomycin D to DNA is sequence specific and has been shown to prefer the duplex GpC site [21,22]. Like CAT, actinomycin D itself is chiral and optically active and has its own CD spectrum (online suppl. fig. 8). Addition of the same concentration of actinomycin D into the 4 special 8-bp DNA sequences with the same concentrations greatly altered the original shapes of these DNAs’ CD spectra. It is conspicuous that the ICD (the CD spectra with the DNA spectra and actinomycin D spectra subtracted) of actinomycin D with the (GC)4 hairpin sequence is different from that of the other 3 sequences in a significant way (fig. 3b). The ICD intensity in the case of the (GC)4 hairpin at 292 nm reaches up to 7.8 millidegrees as compared to the other 3 sequences that have CD intensities below 0.5 millidegrees. These discrepancies correspond perfectly with the well-known fact that actinomycin D preferentially targets the GC-repeated sequence on double-helical DNA. Thus, CD spectroscopy, which is sensitive to the change in conformation induced by ligand interactions, is an appropriate method for the study of sequence-specific interactions between double-helical DNA and ligands.
We further studied the sequence-specific interactions between CAT and the 4 listed DNA sequences. The concentrations of DNA were still fixed at 3 × 10−5M(bp), and 3 different molar CAT/DNA ratios (1:1, 2:1, and 4:1) were used. The ICD in the case of the (AT)4 hairpin was significantly different from that of the other 3 sequences (fig. 3c). The minus ICD peaks at a 250- to 260-nm range were more intensive in the case of the (AT)4 hairpin than in the other 3 sequences. This is indicative of stronger interactions between CAT and the (AT)4 hairpin versus the other 3 sequences. In other words, the same concentration of CAT can induce more intensive ICD in the presence of the (AT)4 hairpin than in the presence of the same concentration of the other 3 hairpins in this wavelength range. These results strongly suggest that CAT interacts more strongly with AT-repeated sequences than with CG-repeated, AA(TT)-repeated, and GG(CC)-repeated double-helical DNA sequences. Therefore, we can conclude that CAT preferably interacts with the AT-repeated sequence, although it may also interact with the other 3 sequences to a lesser extent.
We further extended the length of the double-helical DNA sequences to 12 and 20 bp (fig. 4a) and the same experiments were carried out with both actinomycin D and CAT to check the reliability of the results we had just obtained. The results were proved to be basically applicable to the 12- and 20-bp sequences, although some other aspects were found (fig. 4b, c). It should also be noted that due to the difficulty in synthesizing over 5 continuous G in the polynucleotide sequences chemically, the G-repeated sequences (G12 and G20) used here had to alternate between G and C every 4 or 5 nucleotides. Thus, in the G-repeated sequences used here, GC sequences also existed, which is manifested in the relatively strong intensity of 292-nm peaks in the actinomycin D ICD spectra (fig. 4b). Comparisons between the CAT ICD of all of these sequences revealed that the AT-repeated sequence was the most intensive at the 250- to 260-nm minus peaks (fig. 4c).
Fig. 4.
Sequence-specific interactions of actinomycin D and CAT with 12- and 20-bp short DNA sequences. a Short hairpin DNA sequences (12 and 20 bp) used in the experiments. b ICD spectra of actinomycin D (3 × 10−5M) in the presence of the four 12- and 20-bp DNA sequences [3 × 10−5M(bp)] in BPES buffer. c ICD spectra of CAT (3 × 10−5M) in the presence of the four 12- and 20-bp DNA sequences [3 × 10−5M(bp)] in BPES buffer.
Intercalating Interactions between CAT and DNA
Noncovalent interactions with DNA can be adopted by either intercalation (such as daunorubicin and doxorubicin) or groove binding (such as distamycin A and berenil) [23]. Our experiments described above confirmed AT-repeated, sequence-specific, and concentration-dependent interactions between CAT and double-helical DNA. However, additional experiments are required to distinguish between intercalation and groove binding. Since intercalators cause unwinding and lengthening of DNA, increasing ratios of intercalator/DNA increase the viscosity of DNA-containing solutions. As viscosity is proportional to the cube of length, viscosity measurements are very sensitive to changes in the length of DNA [24,25]. The viscosities of increasing concentrations of CAT alone and CT-DNA with increasing concentrations of CAT were measured as flow time using an Ubbelohde viscometer at a temperature of 25 ± 0.5°C. The flow time which is representative of viscosity is plotted as a function of the concentration of CAT (fig. 5a). The flow time for increasing concentrations of CAT remains constant at around 45–46 s all through the measuring range from 0 to 3 × 10−4M. In contrast, the flow time of CT-DNA [3 × 10−4M(bp)] with increasing concentrations of CAT increases from around 60 s to more than 70 s through the measuring range from 0 to 2.4 × 10−4M. This result suggests that CAT can intercalate into linear double-stranded DNA. It has been reported [24,25] that if a circular plasmid is used, the viscosity increases and then declines with increasing concentrations of compound. This is indicative of the DNA changing from a negative supercoiled state to a relaxed and finally a positive supercoiled state. Similar experiments were carried out with pEGFP-N1 plasmid (4.7 kbp) [26] with a concentration of 1.5 × 10−4M(bp) calculated in DNA base pairs. As the ratio of CAT/DNA [M/M(bp)] increased, the flow time (representative of viscosity) of the plasmid solution increased. It reached its maximum at 57.43 s, corresponding to a CAT concentration of 2.68 M. After that, it decreased to its original level (fig. 5b). These viscosity changes correspond to a decrease in negative supercoiling, creating a fully relaxed circular molecule and followed ultimately by the formation of a positively supercoiled molecule [24]. This type of behavior is caused by ligand-induced unwinding of the plasmid DNA. These results suggest that CAT intercalates into circular double-stranded DNA.
Fig. 5.
Viscosities, measured as flow times, as functions of the CAT concentration using an Ubbelohde viscometer at a temperature of 25 ± 0.5°C. a Viscosities, measured as flow times, of increasing concentrations of CAT alone and CT-DNA [3 × 10−4M(bp)] with increasing concentrations of CAT plotted against the CAT concentration. b Viscosities, measured as flow times, of solutions containing CAT/plasmid complexes at a plasmid concentration of 1.5 × 10−4M(bp), plotted against the CAT concentration.
Discussion
In this paper, we described a new anticancer phenanthroindolizidine alkaloid which is originally extracted from T. atrofolliculata and T. ovata. In vivoevaluations showed that CAT has potent anticancer effects. Direct interactions of CAT with DNA were studied in detail, and concentration-dependent interactions were found by both CD spectroscopy and fluorescence spectroscopy. To our surprise, CAT was found to preferably bind to AT-repeated double-helical DNA sequences, and the interactions between CAT and DNA were found to be intercalating.
The CD spectrum is known to be very sensitive to the environment of DNA in solution. The appearance of the ICD of optically inactive molecules or changes in the intrinsic CD of chiral molecule reflects the binding geometry and binding mode of the molecule as well as the arrangements of neighboring DNA bases [12]. Our results suggest that CAT can interact in a concentration-dependent manner with double-helical DNA, and the interactions were later proved to be intercalating by viscosity measurements. Such interactions may inevitably interfere with the cellular processes of replication and transcription as in the case of actinomycin D. Actinomycin D binds to DNA by intercalating its phenoxazone ring at a GpC step with the drug's 2 cyclic pentapeptides located in the DNA minor groove, and strong hydrogen bonds are found between actinomycin D and the N2 amino group of adjacent bases [22,23]. Inspired by the example of actinomycin D, we postulate an interaction mode between CAT and DNA here. The hydrophobic methoxyl-substituted phenanthrene ring in CAT intercalates into the base pairs of double-helical DNA, and the indolizidine ring with a hydrophilic nitrogen atom sticks out into the DNA grooves to interact with protein factors functioning in replication and transcription, such as DNA polymerases, RNA polymerase, topoisomerase, etc. This hypothesis needs further evidence from NMR and X-ray crystallography.
Unlike actinomycin D, which prefers the GpC site, CAT was proved to preferably interact with AT-repeated sequences. AT-rich sequences are abundant in promoter regions. The AT-rich sequences located at a fixed distance upstream of the transcription start site have been identified in essentially all animals, plants, and fungi that have been examined [27,28,29,30]. The preference of CAT for intercalating into AT-repeated sequences raises the possibility that CAT may interfere with the normal function of transcription through blocking the recognition of the promoter site by protein factors. Many commercially available anticancer drugs, like doxorubicin and other anthracyclines, can intercalate into DNA at certain sites and form DNA-doxorubicin adducts. It is a classic topoisomerase II poison because these adducts inhibit the function of this enzyme which catalyzes the unwinding of DNA for transcription and replication. The intercalation of CAT into DNA may well obstruct the normal interaction and identification between DNA sequences and functional protein factors [31,32,33,34]. Tylocrebrine, another phenanthroindolizidine compound, was previously reported to inhibit RNA synthesis as measured by the amount of 14C-labeled uridine incorporation [8]. Intercalation with AT-repeated DNA and concomitant inhibition of the process of transcription may be an important mechanism for CAT's anticancer effects, though other mechanisms may also work. Some of CAT's derivatives after structural modifications were also determined for their interactions with double-helical DNA by CD spectroscopy. Similar concentration-dependent and AT-sequence-specific interactions were also observed (data not shown). Therefore, the sequence preference may be commonly shared by many, if not all, of the phenanthroindolizidine alkaloids.
Supplementary Material
Supplementary Data
Acknowledgement
This work was supported by the National Natural Science Foundation of China (90713039).
References
- 1.Gellert E. The indolizidine alkaloids. J Nat Prod. 1982;45:50–73. [Google Scholar]
- 2.Chemler SR. Phenanthroindolizidines and phenanthroquinolizidines: promising alkaloids for anti-cancer therapy. Curr Bioact Compd. 2009;5:2–19. doi: 10.2174/157340709787580928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rathnagiriswaran AN, Venkatachalam K. The chemical examination of Tylophora asthmatica and isolation of the alkaloids tylophorine and tylophorinine. Indian J Med Res. 1935;22:433. [Google Scholar]
- 4.Grant P, Sánchez L, Jiménez A. Cryptopleurine resistance: genetic locus for a 40S ribosomal component in Saccharomyces cerevisiae. J Bacteriol. 1974;120:1308–1314. doi: 10.1128/jb.120.3.1308-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao KN, Bhattacharya RK, Venkatachalam SR. Thymidylate synthase activity in leukocytes from patients with chronic myelocytic leukemia and acute lymphocytic leukemia and its inhibition by phenanthroindoli-zidine alkaloids pergularinine and tylophorinidine. Cancer Lett. 1998;128:183–188. doi: 10.1016/s0304-3835(98)00061-5. [DOI] [PubMed] [Google Scholar]
- 6.Rao KN, Venkatachalam SR. Inhibition of dihydrofolate reductase and cell growth activity by the phenanthroindolizidine alkaloids pergularinine and tylophorinidine: the in vitro cytotoxicity of these plant alkaloids and their potential as antimicrobial and anticancer agents. Toxicol In Vitro. 2000;14:53–59. doi: 10.1016/s0887-2333(99)00092-2. [DOI] [PubMed] [Google Scholar]
- 7.Shiah HS, Gao W, Baker DC, Cheng YC. Inhibition of cell growth and nuclear factor-kappaB activity in pancreatic cancer cell lines by a tylophorine analogue, DCB-3503. Mol Cancer Ther. 2006;5:2484–2493. doi: 10.1158/1535-7163.MCT-06-0146. [DOI] [PubMed] [Google Scholar]
- 8.Huang MT, Grollman AP. Mode of action of tylocrebrine: effects on protein and nucleic acid synthesis. Mol Pharmacol. 1972;8:538–550. [PubMed] [Google Scholar]
- 9.You X, Pan M, Gao W, et al. Effects of a novel tylophorine analog on collagen-induced arthritis through inhibition of the innate immune response. Arthritis Rheum. 2006;54:877–886. doi: 10.1002/art.21640. [DOI] [PubMed] [Google Scholar]
- 10.Garbett NC, Ragazzon PA, Bhaires JB. Circular dichroism to determine binding mode and affinity of ligand-DNA interactions. Nat Protoc. 2007;2:3166–3172. doi: 10.1038/nprot.2007.475. [DOI] [PubMed] [Google Scholar]
- 11.Willis B, Arya DP. Recognition of B-DNA by neomycin-Hoechst 33258 conjugates. Biochemistry. 2006;45:10217–10232. doi: 10.1021/bi0609265. [DOI] [PubMed] [Google Scholar]
- 12.Hwangbo HJ, Yun BH, Cha JS, Kwon DY, Kim SK. Enantioselective binding of S- and R-ofloxacin to various synthetic polynucleotides. Eur J Pharm Sci. 2003;18:197–203. doi: 10.1016/s0928-0987(02)00279-8. [DOI] [PubMed] [Google Scholar]
- 13.Xi Z, Zhang R, Yu Z, Ouyang D, Huang R. Selective interaction between tylophorine B and bulged DNA. Bioorg Med Chem Lett. 2005;15:2673–2677. doi: 10.1016/j.bmcl.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Norden B, Kurucsev T. Analysing DNA complexes by circular and linear dichroism. J Mol Recognit. 1994;7:141–156. doi: 10.1002/jmr.300070211. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov VI, Minchenkova LE, Schyolkina AK, Poletayev AI. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- 16.Lyng R, Hard T, Norden B. Induced CD of DNA intercalators: electric dipole allowed transitions. Biopolymers. 1987;26:1327–1345. doi: 10.1002/bip.360260809. [DOI] [PubMed] [Google Scholar]
- 17.Olsthoorn CS, Bostelaar LJ, Derooij JF, Vanboom JH, Altona C. Circular dichroism study of stacking properties of oligodeoxy-adenylates and polydeoxyadenylate. Eur J Biochem. 1981;115:309–321. doi: 10.1111/j.1432-1033.1981.tb05240.x. [DOI] [PubMed] [Google Scholar]
- 18.Norden B, Tjerneld F. Structure of methylene blue-DNA complexes studied by linear and circular dichroism spectroscopy. Biopolymers. 1982;21:1713–1734. doi: 10.1002/bip.360210904. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Kumar A, Boykin DW, Wilson WD. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys Chem. 2007;131:1–14. doi: 10.1016/j.bpc.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhadra K, Maiti M, Kumar GS. Molecular recognition of DNA by small molecules: AT base pair specific intercalative binding of cytotoxic plant alkaloid palmatine. Biochim Biophys Acta. 2007;1770:1071–1080. doi: 10.1016/j.bbagen.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Vives M, Tauler R, Gargallo ER. Spectroscopic study of the interaction of actinomycin D with oligonucleotides carrying the central base sequences -XGCY- and -XGGC-CY- using multivariate methods. Anal Bioanal Chem. 2007;387:311–320. doi: 10.1007/s00216-006-0946-8. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Liu X, Patel DJ. DNA bending and unwinding associated with actinomycin D antibiotics bound to partially overlapping sites on DNA. J Mol Biol. 1996;258:457–479. doi: 10.1006/jmbi.1996.0262. [DOI] [PubMed] [Google Scholar]
- 23.Yang XL, Wang AH. Structural studies of atom-specific anticancer drugs acting on DNA. Pharmacol Ther. 1999;83:181–215. doi: 10.1016/s0163-7258(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 24.Pilch DS, Waring MJ, Sun JS, et al. Cha racterization of a triple helix-specific li -gand: BePI (3-methoxy-7H-8-methyl-11-[(3′-amino)propylamino] -benzo [e] pyrido [4,3-b] indole) intercalates into both double-helical and triple-helical DNA. J Mol Biol. 1993;232:926–946. doi: 10.1006/jmbi.1993.1440. [DOI] [PubMed] [Google Scholar]
- 25.Palchaudhuri R, Hergenrother PJ. DNA as a target for anti cancer compounds: methods to determine the mode of binding and the mechanism of action. Curr Opin Biotechnol. 2007;18:497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Glinka EM, Andryushchenko AS, Sapozhnikov AM, Zatsepina OV. Construction of the plasmid for expression of ETA-EGFP fusion protein under control of the cytomegalovirus promoter and its effects in HeLa cells. Plasmid. 2009;62:119–127. doi: 10.1016/j.plasmid.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Tan S, Wang C, Lu C, et al. Quercetin is able to demethylate the p16INK4a gene promoter. Chemotherapy. 2009;55:6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- 28.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 30.Takeda S, Nakai T, Ikeda F, Hatano K. Overproduction of a metallo- ß-lactamase by a strong promoter causes high-level imipenem resistance in a clinical isolate of Pseudomonas aeruginosa. Chemotherapy. 2008;54:181–187. doi: 10.1159/000140362. [DOI] [PubMed] [Google Scholar]
- 31.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 32.Swift LP, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 2006;66:4863–4871. doi: 10.1158/0008-5472.CAN-05-3410. [DOI] [PubMed] [Google Scholar]
- 33.Eskiocak U, Işeri OD, Kars M, Biçer A, Gunduz U. Effect of doxorubicin on telomerase activity and apoptotic gene expression in doxorubicin-resistant and -sensitive MCF-7 cells: an experimental study. Chemotherapy. 2008;54:209–216. doi: 10.1159/000140464. [DOI] [PubMed] [Google Scholar]
- 34.Thongphasuk P, Stremmel W, Chamulitrat W. 2,3-dehydrosilybin is a better DNA topoisomerase I inhibitor than its parental silybin. Chemotherapy. 2009;55:42–48. doi: 10.1159/000175466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data