Abstract
Pendrin shares with nearly all SLC26/SulP anion transporters a carboxy-terminal cytoplasmic segment organized around a Sulfate Transporter and Anti-Sigma factor antagonist (STAS) domain. STAS domains of divergent amino acid sequence exhibit a conserved fold of 4 β strands interspersed among 5 α helices. The first STAS domain proteins studied were single-domain anti-sigma factor antagonists (anti-anti-σ). These anti-anti-σ indirectly stimulate bacterial RNA polymerase by inactivating inhibitory anti-σ kinases, liberating σ factors to direct specific transcription of target genes or operons. Some STAS domains are nucleotide-binding phosphoproteins or nucleotidases. Others are interaction/transduction modules within multidomain sensors of light, oxygen and other gasotransmitters, cyclic nucleotides, inositol phosphates, and G proteins. Additional multidomain STAS protein sequences suggest functions in sensing, metabolism, or transport of nutrients such as sugars, amino acids, lipids, anions, vitamins, or hydrocarbons. Still other multidomain STAS polypeptides include histidine and serine/threonine kinase domains and ligand-activated transcription factor domains. SulP/SLC26 STAS domains and adjacent sequences interact with other transporters, cytoskeletal scaffolds, and with enzymes metabolizing transported anion substrates, forming putative metabolons. STAS domains are central to membrane targeting of many SulP/SLC26 anion transporters, and STAS domain mutations are associated with at least three human recessive diseases. This review summarizes STAS domain structure and function.
Key Words: Sulfate transporter and anti-sigma factor antagonist domain, SLC26, SulP, Guanine nucleotides, Acyl carrier protein, Carbonic anhydrase, Bicarbonate, Stressosome
Introduction
Pendrin (SLC26A4) is a member of the SLC26 anion transporter family [1, 2]. Pendrin shares with all vertebrate SLC26 proteins and with nearly all SulP anion transport proteins of lower organisms [3, 4, 5, 6, 7] a carboxy-terminal cytoplasmic segment constructed around a central Sulfate Transporter and Anti-Sigma factor antagonist (STAS) domain. This domain is present throughout phylogeny from eubacteriae onward as a conserved fold encoded by highly divergent amino acid sequences [8]. STAS domain proteins include small STAS domain-only polypeptides and larger, multidomain polypeptides. The intramolecular partner domains present in these bacterial multidomain STAS proteins are remarkably diverse. Moreover, for each example of such fusion proteins, the database indicates examples of related but distinct polypeptides encoded as independent open reading frames within the same predicted operon, strongly suggesting coordinated regulation and function.
In bacteria, STAS domain function has been intensively studied in the context of the regulation of the large family of sigma factors (ρ) that bind to RNA polymerase to confer transcriptional target gene specificity [9]. Additional investigations have focused on STAS domain function in various signaling contexts such as blue light phototransduction. More recently, STAS domains of SulP anion transport proteins from bacteria and plants have been studied genetically and structurally, leading to new insights on STAS function. The first structure of an engineered mammalian SLC26 STAS domain [10] and growing numbers of defined SLC26 STAS domain-interacting proteins has provided new contexts in which to evaluate STAS domain function. Most importantly, connections between intrinsic activities or interactions of STAS domains and the regulation of anion transport function in SulP and SLC26 anion transporter polypeptides are emerging.
This review will briefly summarize and update STAS domain structure and function by examining three classes of STAS domains: 1) bacterial anti-sigma factor antagonists of e/, 2) bacterial anti-sigma factor antagonists of ρB in the context of the stressosome and LOV-STAS phototransduction, and 3) several examples of STAS domains in SulP anion transporters of bacteria and plants and in human SLC26 anion transport proteins.
STAS domains as anti-sigma factor antagonists (anti-anti-ρ): SpoIIAA
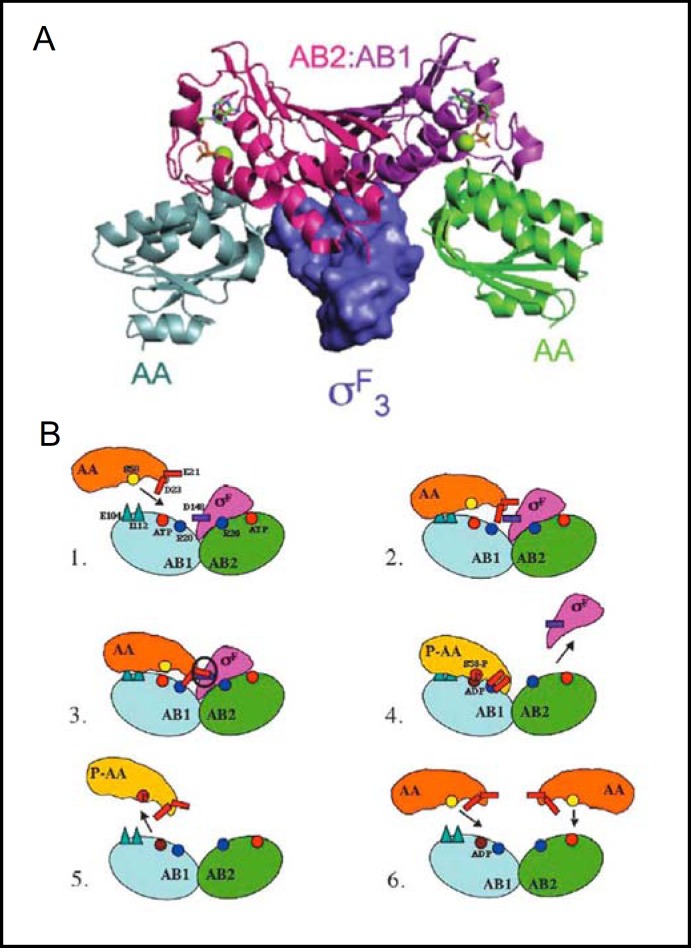
One of the most extensively studied models of biological stress response is the sporulation of Bacillus subtilis. The small forespore is the product of a stress-induced asymmetric division which also yields the larger mother cell with a distinct developmental fate. The sporulation program is initiated by spoIIAC sigma factor gene product ρF, leading to a cascade of downstream activation of forespore-specific gene expression. ρF exerts this initial control by conferring essential target gene specificity for transcriptional activation of the single core bacterial RNA polymerase. Anti-sigma factors (anti-ρ) bind and inhibit their cognate sigma factors. ρF is regulated by anti-ρ SpoIIAB through interactions with three structural domains of ρF. Anti-ρ are themselves inhibited by the anti-sigma factor antagonists (anti-anti-sigma factors, or anti-anti-ρ), which are STAS domain proteins. Thus, SpoIIAB is regulated by STAS domain protein anti-anti-ρ SpoIIAA. The structures of SpoIIAA and other components of the ρF complex have been determined by X-ray crystallography and NMR [11, 12, 13]. A composite structure of the intermediate complex of the SpoIIAB homodimer, two SpoIIAA monomers, and the ρF3 domain of ρF [9] is shown in Fig. 1A.
Fig. 1.
A. X-ray crystal structure of the complex of B. subtilis SpoIIAB anti-ρ homodimer kinase (comprising protomers AB1 (purple) and AB2 (magenta), with the aF domain of holo-sigma factor 0F superposed with the complex of SpoIIAB homodimer and two SpoIIAA anti-anti-ρ monomers (gray and green). Nucleotides bound to each SpoIIAB protomer are shown in green stick and active site Mg2+ as green balls. Reproduced from [9]. B. SpoIIAB catalytic cycle. Residues important for binding and dissociation are shown in (1): AB1 protomer of SpoIIAB (blue) is targeted by SpoIIAA (orange), as its docking surface (R20 in particular) is more accessible than in AB2 (green). (2) SpoIIAA binds to initial sites on SpoIIAB1 (E104, I112). (3). Bound SpoIIAA D23 interacts with SpoIIAB1 R20, leading to steric clash between SpoIIAA E21 and oF D148. (4) The steric clash promotes dissociation of oF from ADP-bound SpoIIAB. SpoIIAA then adopts a conformation that allows S58 phosphorylation (yellow circle changes to red) by SpoIIAA kinase. (5) Phospho-SpoIIAA (yellow) dissociates from ADP-bound SpoIIAB. (6) Unphosphorylated SpoIIAA can bind to SpoIIAB, forming an inhibitory complex that by blocking oF binding maintains oF in its active conformation. Reproduced from [13].
Fig. 1B outlines 6 stages of the regulatory cycle controlling ρF availability to target the activity of RNA polymerase (with important amino acid residues identified in panel 1). When ρF is bound to the SpoIIAB homodimer, its RNA polymerase recognition sites are unavailable, but one of the two ρF-bound SpoIIAB protomers is in a more “open” state. The SpoIIAA anti-anti-ρ monomer targets (1) and binds (2) to the more accessible SpoIIAB anti-ρ protomer (AB1) of the ATP-loaded SpoIIAB homodimer complex with ρF. Slower, additional binding interactions promote steric/electrostatic clash of SpoIIAA with ρF (3), leading to aF dissociation (4) in a form that can regulate RNA polymerase. Tightly bound anti-anti-ρ SpoIIAA is phosphorylated by the kinase activity of anti-ρ SpoIIAB (4), leading in turn to its dissociation (5). Unphosphorylated SpoIIAA can form a tight complex with ADP-loaded SpoIIAB, preventing rebinding of ρF, and prolonging its regulation of RNA polymerase. ATP-loaded SpoIIAB can rebind either SpoIIAA or ρF [13]. SpoIIAA binds and hydrolyzes GTP and, to a lesser degree, ATP. Mutation of SpoIIAA phosphorylation site Ser 58 to Ala reduces but does not abolish GTPase activity [14]. However, the role of GTP binding and hydrolysis to SpoIIAA binding to SpoIIAB and displacement of ρF remains unclear.
STAS domain proteins of the stressosome
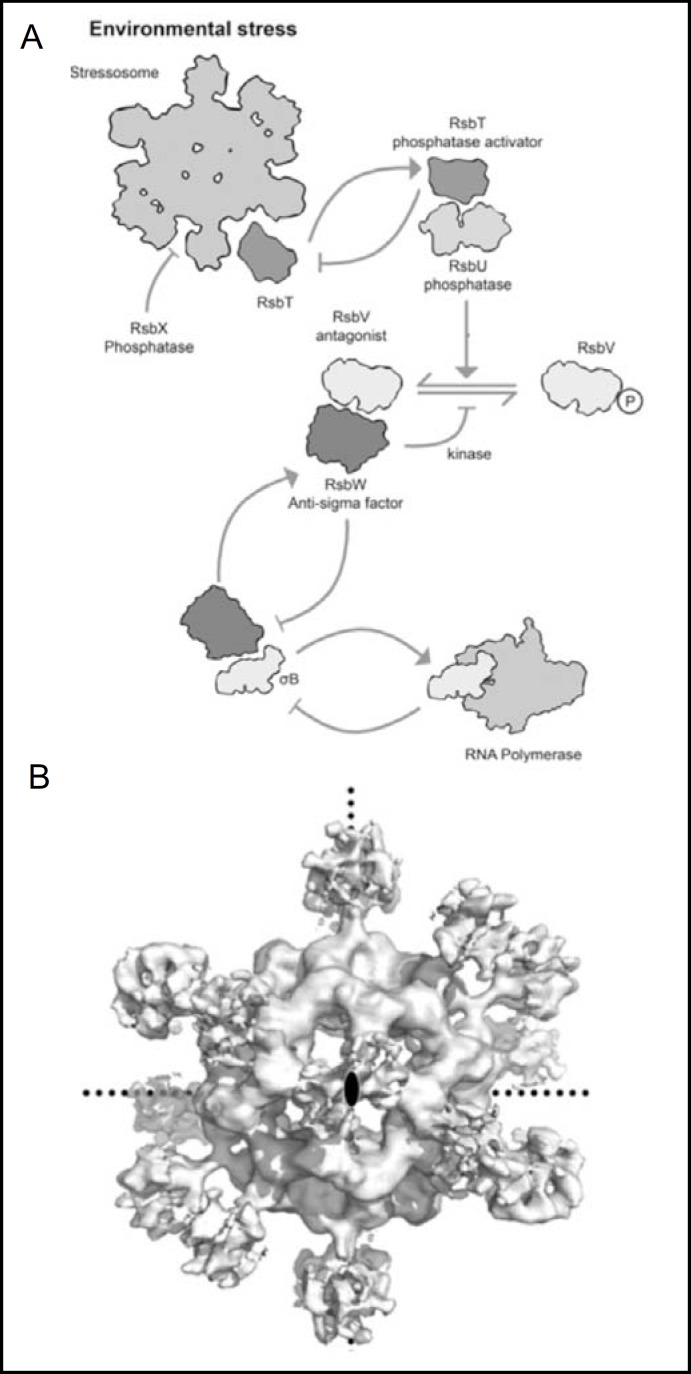
Extreme stress triggers sporulation in B. subtilis, but less extreme, more frequently encountered stresses are triggered by changes in environmental temperature, pH, osmolarity, ethanol, blue light, or cell wall stress. These stresses activate the alternative regulator of RNA polymerase, ρB, which transcribes a regulon of >150 genes [15, 16]. In the absence of stress ρB is maintained in an inactive state in complex with anti-ρ kinase RsbW (Fig. 2A). Free RsbW can be inhibited by the anti-anti-ρ STAS domain protein, RsbV. RsbW can phosphorylate bound STAS protein RsbV, which can be dephosphory-lated by RsbU phosphatase, restoring RsbV to its anti-anti-ρ activity.
Fig. 2.
A. The crB regulatory pathway of B. subtilis. A. The 1.5 MB stressosome, an ordered 1.5 megadalton complex made up of multiple copies of STAS proteins RsbR and RsbS, serves to sequester kinase RsbT in normal conditions. Under stress, RsbT phosphorylates both STAS proteins, resulting in its release from the stressosome to bind and activate RsbU phosphatase. The RsbT/RsbU copmplex-mediated dephosphorylation of anti-anti-ρ factor STAS protein RsbV allows it to bind anti-ρ factor RsbW, liberating ρB from its inactivating complex with RsbW, and allowing activation of RNA polymerase. B. Cryo-transmission electron microscopy-derived molecular envelope of the RbsR/RbsS stressosome at 8Å resolution, viewed down one of its 2-fold axes of symmetry (central black ellipse). Dotted lines mark the other two symmetry axes. Modified from [16].
RsbU activity is controlled by the 1.5 megadalton stressosome (Fig. 2A, Fig. 2B) comprising 40 copies of multidomain STAS protein RsbR, 20 copies of simple STAS protein RsbS, and 20 sequestered, inactive copies of switch kinase RsbT [16]. The C-terminal STAS domain of RsbR is preceded by an N-terminal domain of novel sequence arrayed as a globin fold, but without bound heme or other cofactor. In contrast to the conserved STAS domain sequence, the N-terminal domain is highly varied among bacterial RbsR proteins. The B. subtilis N-terminal domain dimerizes just as do other heme-liganded globin sensor proteins of bacteria [17]. By analogy with mammalian neuroglobin's hypoxia signaling through inhibition of GTP-GDP exchange by heterotrimeric Go, proteins, the N-terminal domain of RbsR may modify in a stress-dependent fashion the binding of ribosome-associated, low-affinity GTPase Obg [18] (not shown in Fig. 2A) to the kinase RsbT.
In response to stress, RsbT phosphorylates RsbS and RsbR, releasing RsbT from the stressosome to activate RsbU phosphatase. RsbU phosphatase activation ultimately leads via anti-anti-ρ RsbV binding to anti-ρ RsbW to ρB release and activation of RNA polyerase to transcribe the ρB regulon. The stress response is dampened or terminated by RsbX phosphatase-mediated dephosphorylation of stressosome STAS proteins RsbS and RsbR, resetting the stressosome to again seqester and inactivate RsbT kinase (Fig. 2A). The polyvalency of the stressosome allows activation of the ρB stress response with highly positive cooperativity and sensitivity [9, 16]. In some bacterial species, the RsbW anti-ρ kinase and an anti-anti-ρ STAS domain are found within a single polypeptide, potentially yielding an even more highly cooperative activation of the stress response.
The STAS domain in phototransduction
As noted above, B. subtilis activates the ρB pathway in response to blue light. The flavin mononucleoside (FMN)-binding LOV (light-oxygen-voltage sensing) domain of the chimeric LOV-STAS protein YtvA is responsible for this photodetection [19, 20, 21] and transmission of the stress signal via the stressosome to ρB. The dark-adapted LOV domain of YtvA noncovalently binds oxidized FMN (LOV447). Blue light illumination triggers msec-scale formation of a metastable FMN thiol adduct with Cys62 in the Da-Ea loop via decay of the excited FMN triplet state (LOV390). Return to dark conditions is accompanied by slow reversion to LOV447 on the sec-min time scale [22, 23].
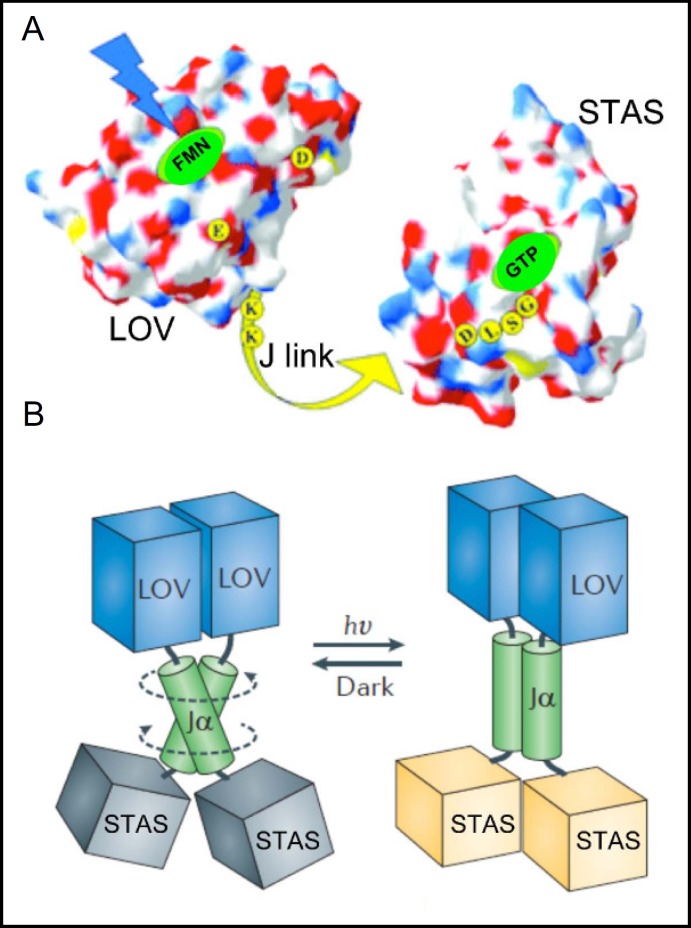
The STAS domain of YtvA has been proposed as the transducer of the photosignal from the LOV-FMN adduct transmitted [21] via conformational change in the short linker connecting the two domains (Fig. 3A). Studies demonstrating YtvA STAS domain-mediated enhancement and red-shift of BODIPYTR-GTP fluorescence in the illuminated state, with further slight enhancement and red-shift during thermally driven recovery in the dark [24, 22] have suggested a guanine nucleotide binding function of the STAS domain with conformational sensitivity to FMN-triggered changes in the LOV domain. A hypothesized STAS domain GTP switch region including the conserved STAS sequence DLSG was identified (Fig. 3A) by assessing the impact of directed mutagenesis on in vivo activation of ρB [20] and by assessment of alterations in 4-bora-3a,4a-diaza-s-indacene (BODIPY)-GTP TR fluorescence enhancement [21]. However, a recent report indicates that some recombinant YtvA LOV constructs lacking the STAS domain similarly enhance BODIPY-GTPT fluorescence in a manner insensitive to competition R by added nucleotide triphosphates, but suggestive of predominant actions of the hydrophobic BODIPY moiety [25]. In addition, GTPase activity of the STAS domain has not yet been detected. Thus, although, the ability of BODIPY-GTPTR to monitor light-sensitive conformational change in YtvA has been confirmed, the specificity of BODIPY-GTP binding to the YtvA STAS domain TR remains in question. The role of the STAS domain in YtvA phototransduction of ρB activation remains unclear, but might involve direct or indirect activation of the RsbT kinase that controls RsbU (Fig. 2A).
Fig. 3.
A. B. subtilis YtvA electrostatic surface image from LOV domain crystal structure and from STAS domain (modeled on crystal structure of B. subtilis SpoIIAA). Blue light is absorbed by the flavin mononucleotide chromophore of the LOV domain, triggering local conformational change that is believed to be transmitted through the agency of the J linker to the STAS domain, which binds BODIPY-GTP. Yellow residues have been implicated by mutagenesis as important or required for phototransduction. Modified from [21]. B. Light-induced conformational change of YtvA as imagined from holoprotein structure. Modified from [26].
The near complete solution NMR backbone assignment of holo-YtvA in the dark state [19] has demonstrated a dimeric structure with LOV-LOV and STAS-STAS interactions, partially consistent with the proposed mechanism of light-induced conformational change [26] shown in Fig. 3B. However, experiments with small angle X-ray scattering and circular dichroism suggest that the light-induced conformational change may be more modest than depicted in this schematic drawing [25].
STAS domains in bacterial anion transport proteins: BicA, Rv1739c, ychM, and Ye0973
Marine cyanobacteria generate as much as 25% of global primary productivity through efficient inorganic carbon fixation mechanisms. The photosynthesis fueling this productivity is driven by a carbon concentrating mechanism comprising complementary transporters of HCO - and CO2 that generate up to 1000-fold concentration of HCO - over the extracellular environment, with maintenance of the gradient aided by absence of carbonic anhydrase [6]. One of the five known inorganic carbon uptake systems of cyanobacteria such as Synechocystis and Synechococcus is BicA, a STAS domain-containing SulP polypeptide whose expression in intact cells confers high-flux, low-affinity Na+-dependent HCO - uptake [27]. The transmembrane topography of BicA has been studied by progressive C-terminal truncation and fusion with alkaline phosphatase and β-lactamase, yielding an experimentally derived topographical model for SulP and SLC26 anion transporter transmembrane domains [28]. BicA-related transporters have also been noted in genomes of other marine bacteria, in Vibrio cholerae, and in Prochlorococcus species [6]. However, the role of the STAS domain in BicA biosynthesis, stabilization, or function remains unknown.
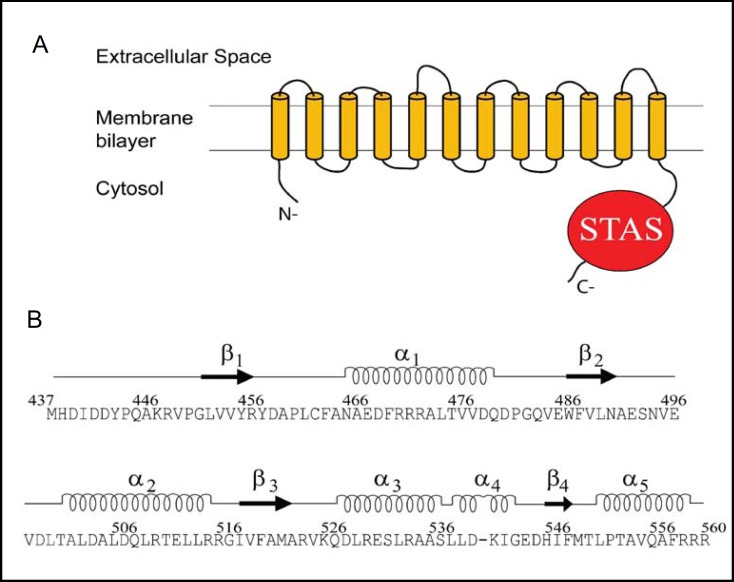
Structral information is available for STAS domains of SulP proteins of Mycobacterium tuberculosis and Escherichia coli. Mycobacteria are characterized by envelopes rich in sulfolipids that include components important for pathogenesis, suggesting that sulfate uptake mechanisms should also be important. Although genetic growth experiments in vitro suggest that mycobacterial sulfate assimilation is mediated entirely by the ABC-type sulfate permease CysTWA, with possible contributions of transport systems for thiosulfate and sulfur-containing amino acids, the sulfate assimilation systems employed by mycobacteria sequestered inside lysosomes are less well understood. Among candidate sulfate or anion transporters in the M. tuberculosis genome are three SulP genes, among which only the Rv1739c gene product was successfully overexpressed and purified [29]. Rv1739c has a SulP-type transmembrane domain and a C-terminal cytoplasmic STAS domain (Fig. 4A). Overexpression of Rv1739c in intact E. coli increased sulfate uptake with K1/2 of 4 μM, and this sulfate uptake was activated at pH 6, inhibited by agents that collapse the electrochemical gradient of the bacterial inner membrane and by several low affinity inhibitors of mammalian sulfate transport [29].
Fig. 4.
A. Schematic of M tuberculosis Rv1739c SulP polypeptide, showing its putatively cytoplasmic N-terminal segment, the transmembrane domain with 12 putative transmembrane oc-helices [7, 74], and the cytoplasmic C-terminal STAS domain. B. Secondary structure of the Rv1739c STAS domain deduced from the three dimensional solution structure, aligned above the STAS domain's 124 amino acid residues (aa 437-560 in the holoprotein). Modified from [30].
The isolated STAS domain of Rv1739c was purified and found to be stable. Purified Rv1739c STAS domain bound guanine nucleotides as assessed by specific photoaffinity labeling with biotinylated azido-GTP and by guanine nucleotide-induced quench of intrinsic Rv1739c STAS fluorescence [30]. Modest GTPase activity also copurified with Rv1739c STAS, but the hypothesis of intrinsic Rv1739c STAS GTPase activity remains untested by mutation of candidate GTP switch or hydrolase residues. Rv1739c STAS was not a phosphoprotein, and did not serve as a phosphorylation substrate for any of the defined recombinant M. tuberculosis Ser/Thr kinases [30].
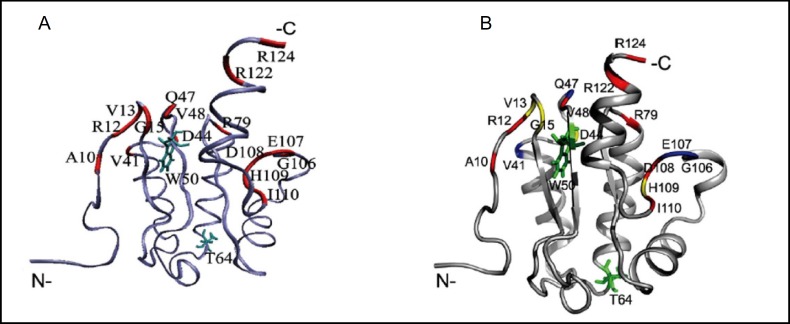
We solved the NMR solution structure of Rv1739c STAS, which showed the interspersed β-strands and a-helices characteristic of the STAS domains of bacterial anti-anti-ρ proteins (Fig. 4B), with the pattern β1-c1-β2-α2-β3-α3-α4-β4-α5 [30]. Discrete, overlapping sets of amino acid residues underwent chemical shift perturbation (CSP) in the presence of nominally saturating concentrations of GDP or of GTP (Fig. 5A), providing more focused evidence of nucleotide binding-induced conformational change in the Rv1739c STAS domain. Of particular interest was the proximity of W50 to some of these chemically shifted residues, providing a partial basis for the ability of nucleotides to quench intrinsic STAS fluorescence [30].
Fig. 5.
A. M. tuberculosis Rv1739c STAS domain backbone structure (blue) with red-highlighted amino acid residues that showed significant chemical shift perturbation (CSP) in the presence of GDP. Side chains of residues W50 and T64 are in cyan. Figure drawn in VMD [74]. B. Rv1739c STAS residues showing GDP-induced CSP mapped onto average STAS structure. Colors denote STAS backbone 15N-1H relaxation parameters. CSP residues in red show increased R2 and J(0) along with decreased NOE and J(<&N) values, indicating high flexibility likely due to increased chemical exchange. CSP residues in yellow deviated from the red pattern in one or two relaxation parameters. CSP residues in blue are those for which at least two relaxation parameters were unavailable due to experimental limitations. W50 and T64 are shown in green. Figure drawn in PyMOL (www.pymol.org). Modified from [30] and from Sharma et al. (manuscript in revision).
Rv1739c STAS backbone 1H-15N dynamics revealed nucleotide-induced internal fluctuations which increased overall rotational tumbling time. This slowed ligand-dependent STAS domain tumbling was dominated by select residues exhibiting slow (on the NMR time scale) conformational exchange. These residues exhibited higher values of the reduced spectral density parameter, J(0), as mapped onto the Rv1739c STAS domain backbone structure in Fig. 5B. Residues with the greatest local changes in relaxation parameters paritially overlap with chemically shifted residues, suggesting a complex relationship between backbone dynamics and steady-state backbone conformation. These backbone dynamics results suggest that nucleotide binding may induce a molecular shape change or a small shift in oligomeric equilibrium away from the predominantly monomeric unliganded state. (Sharma et al., manuscript in revision).
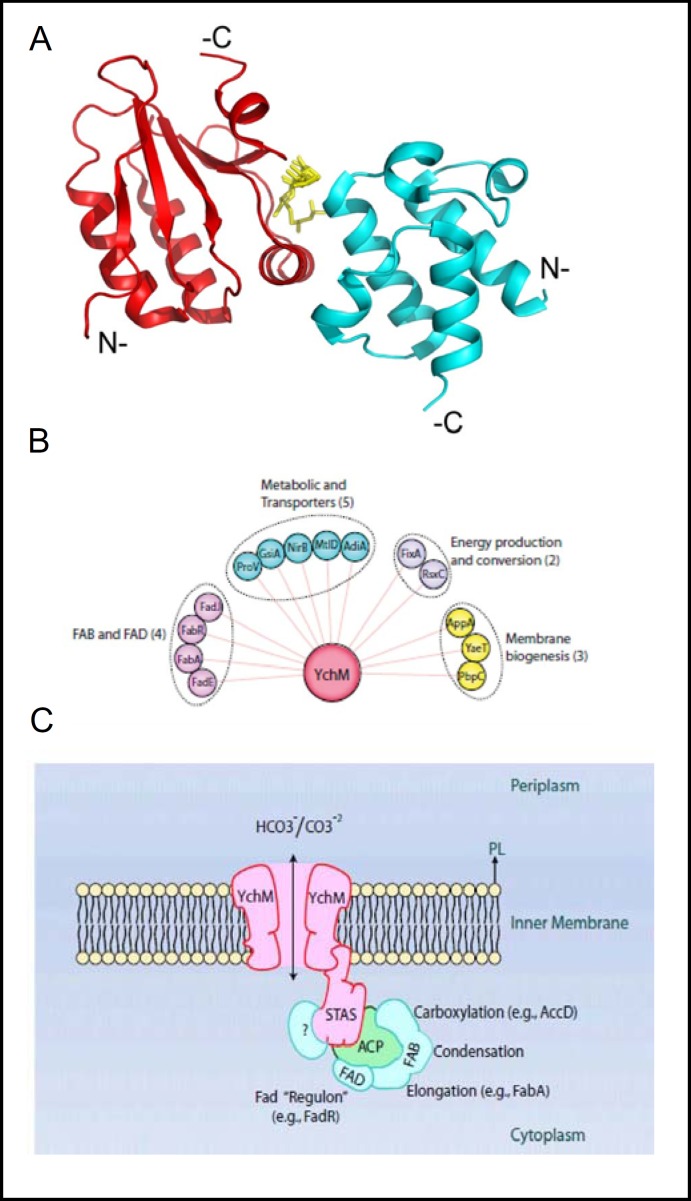
Another SulP polypeptide that can be overexpressed in abundance is the non-essential E coli gene product, YchM. Genetic deletion of YchM or deletion of the YchM STAS domain decreased steady-state Na+-dependent 14C-HCO - accumulation by 30-50% in intact bacteria [31]. YchM differed significantly in its secondary structure as compared to Rv1739c and the anti-anti-ρ proteins, with relative reversal of a1 and β2 in the linear sequence and absence of helix a5, yielding the secondary structural sequence β1-β2-α1-β3-α2-β4-α3-β5-α4. The recombinant STAS domain of YchM was found to copurify and cocrystallize in a 1:1 stoichiometry with bound acyl carrier protein (ACP), an essential component of the fatty acid biosynthetic apparatus. The ACP in the crystal was covalently attached to a malonyl-coA moiety which was situated at the ACP-STAS interface (Fig. 6A). The additional demonstration by both genetic and direct biochemical analyses of a large family of YchM-binding proteins (Fig. 6B) led to the novel suggestion [31] that YchM takes up HCO - for direct channeling to a fatty acid biosynthesis regulon scaffolded around the STAS domain/ACP complex (Fig. 6C). These findings encourage re-evaluation of the role of CO2 and HCO - in a wide spectrum of bacterial functions.
Fig. 6.
A. Crystal structure of STAS domain from E. coli SulP protein YchM (red) in complex with acyl carrier protein (ACP, cyan), as described by [31]. Shown in yellow is the malonyl-4'-phosphopantheine prosthetic group covalently linked through the phosphate to the hydroxyl group of conserved S36 of ACP. The structure (PDB:3NY7) was rendered in PyMOL. B. Experimental evidence supports the existence of numerous YchM-interacting proteins, grouped here by function (reproduced from [31]). C. Schematic model of YchM as a bicarbonate transporter delivering substrate to STAS domain-bound ACP for sequential transfer to multiple components of the fatty acid biosynthesis pathway assembled around the STAS-ACP complex (reproduced from [31]).
A fraction of E. coli YchM STAS domain purified as an uncomplexed monomer [31], and the unliganded M. tuberculosis Rv1739c STAS domain was also monomeric. However, the SulP-STAS holoprotein YE0973 from Yersinia enterocolitica was overexpressed and purified through sequential detergent solutions of dodecylmaltoside and Foscholine-12 was shown by small angle neutron scattering combined with contrast variation to form a dimer stabilized by its transmembrane domains. No evidence was found for involvement in dimer formation of the STAS domains, which were oriented away from each transmembrane homodimer and from each other [32], in contrast to the STAS-STAS interaction observed in the YtvA dimer [19].
Additional putative functions for STAS domains in multidomain bacterial gene products
Bacterial STAS domains are found not only as anti-anti-ρ proteins or C-terminal to SulP anion transporters. The Conserved Domain Architecture Retrieval Tool (cDART) of the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) reveals a wide range of functional domains in STAS domain-containing polypeptides. Thus, a few SulP-STAS transporters exhibit a C-terminal pro-carbonic anhydrase domain, whereas a larger number including M. tuberculosis Rv3273 exhibit a cytoplasmic β-carbonic anhydrase domain in place of the more commonly found STAS domains. These proteins are very likely HCO3−transporters, but their overexpression in quantities required for further study has not been easily achieved. However, the β-carbonic anhydrase domain of putative anion transporter Rv3273 has been overexpressed, purified, documented to be an active carbonic anhydrase, and studied pharmacologically [33]. SulP-STAS domain proteins have been noted within complex multigene antibiotic biosynthesis operons in several species of Streptomyces, including orf29 of the BLM bleomycin biosynthesis operon in S. verticillus [34]. Many similar SulP-STAS open reading frames are evident in antibiotic biosynthesis operons and in other genomic regions of S. coelicolor, S. avermitilis and other antibiotic-producing commercial strains of Streptomyces. This is particularly intriguing in that the YchM-binding protein ACP is required in multiple steps of polyketide antibiotic biosynthesis [35].
As predicted by the NCBI Conserved Domains Database, some SulP-STAS domain proteins include a rhodanese domain thought to encode a thiosulfate:cyanide sulfotransferase activity, and suggesting an intramolecular thiosulfate or cyanide transport metabolon. Other STAS domain proteins may be involved in nutrient transport. These are situated adjacent to domains predicted to encode glutaminase, glycosyltransferase, N-acylglucosamine epimerase (part of the sailic acid synthetic pathway), periplasmic sugar binding, Na+-alanine cotransport activity, prenyl metabolism, or vitamin-K-dependent carboxylase activity, and may be involved in nutrient or cofactor uptake and/or processing. STAS domains in still other proteins are physically adjacent to domains involved in detoxification of alkenes, organic solvents, and aminoglycosides. In addition to the LOV/PAS/PAC light and oxygen sensors and the neuroglobin-like signaling domains described above, STAS domains likely involved in signaling are situated adjacent to domains predicted to encode PAS/PAC oxygen/light sensors, histidine kinases, His kinase-related Rec signaling domains, Ser-Thr kinase G protein regulators, PX phosphoinositide binding domains, CAP-ED/CRP, NTR, and GAF cyclic nucleotide binding domains, a lacI-related transcription regulator domain, and AmiR and NasR transcriptional anti-termination domains.
Plant Sultr sulfate transport proteins
In the plant Arabidopsis thaliana, at least 14 distinct SulP/Sultr genes in 5 classes encode plasmalemmal and intracellular membrane sulfate transporters with C-terminal STAS domains. These transporters are expressed in distinct subcellular membranes in specific tisses and with developmental stage-specific patterns. Although most are sulfate transporters, some (such as SHST1 from Stylosanthes hamata) preferentially exhibit nM affinity uptake of molybdate [36], and at least one has turned out to regulate cellular levels of phytic acid [37]. Plastids utilize sulfate transporters of the ABC superfamily [38]. The two best studied of the SulP proteins are Sultr1;2 and Sultr1;1. Sultr1;2 is the principal plasma membrane sulfate transporter of the roots, but sulfate limitation increases abundance of Sultr1;1 transcripts to a greater degree, suggesting that Sultr1;2 can be acutely regulated by rhizosphere sulfate and cellular energetics. Both plant cell experiments and yeast complementation experiments suggest a mechanism of 3H+/SO42− cotransport for plasmalemmal Sultr transporters of A. thaliana [39]. Unlike the bacterial SulP STAS domains, the Sultr1;2 STAS domain homodimerizes. The STAS domain of Sultr1;2 is also essential to proper plasma membrane targeting of holoprotein, and contributes to protein stability and transport kinetics [40].
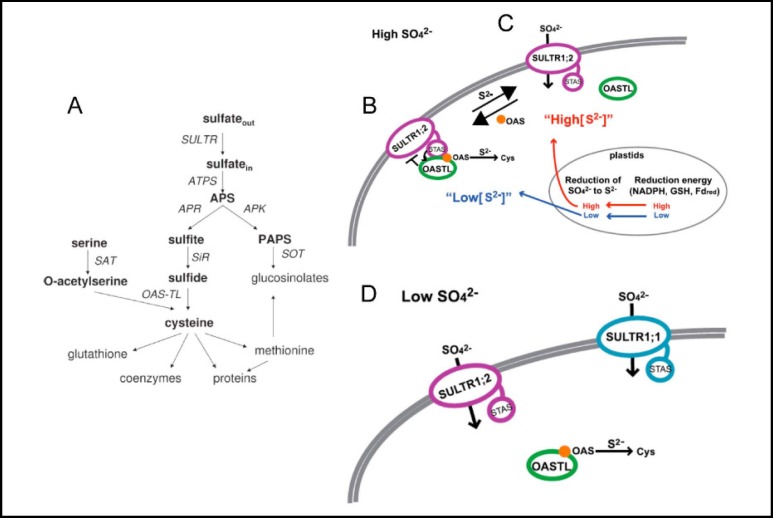
In addition to generation of the metabolic intermediate phosphoadenosine phosphosulfate (PAPS), plant cells reduce sulfate to sulfite and on to sulfide (Fig. 7A) in the plastids (Fig. 7B). Sulfide then is condensed with serine-derived O-acetylserine (OAS) to synthesize cysteine by O-acetylserine-lyase/Cysteine synthase (OASTL) (Fig. 7A). A yeast two-hybrid screen of the Sultr1;2 STAS domain revealed binding with OASTL [41]. The interaction was validated by reciprocal pulldown experiments in vitro, and by reconstitution of a split fluorescent protein in tobacco cells. Sulfate uptake into Sultr1;2-rescued Saccharomyces cerevisiae deficient in endogenous sulfate transport is downregulated by co-expression of OASTL through a mechanism that does not require OASTL catalytic activity and does not reduce Sultr1;2 surface expression. Conversely, OASTL cysteine synthase activity is enhanced by interaction with Sultr1;2 STAS domain [41]. Thus Sultr1;2 and OASTL may comprise a sulfate metabolon assembled on the STAS domain. In a low-energy state root epidermal cell exposed to high sulfate (Fig. 7B), low cytoplasmic sulfide and elevated cytoplasmic OAS promotes OASTL binding to Sultr1;2 STAS, reducing sulfate uptake while increasing OASTL activity. In a high energy state cell exposed to high sulfate (Fig 7C), sulfide synthesis matches OAS levels, OASTL is released from Sultr1;2 STAS, and Sultr1;2 continues to import sulfate. In cells exposed to low sulfate (Fig. 7D), unbound OASTL does not reduce Sultr1;2-mediated sulfate uptake, and Sultr1;1 transcription and biosynthesis are increased to maintain cell sulfate homeostasis [41].
Fig. 7.
A. Sulfate assimilation pathways in plants. Reproduced from [38]. B.-D. Model of proposed regulatory function of A. thaliana Sultr1;2 (modified from [41]) in high extracellular [SO42−] conditions (B. and C.) and in low extracellular [SO42−] conditions (D.). See text for details.
SLC26 anion transporter STAS domains
The human SLC26 STAS domain-associated anion transporter gene family numbers 11 genes encoding at least 10 polypeptides. Among these are three human Mendelian recessive disease genes. The SLC26A3/DRA gene was discovered by positional cloning to be mutated in Congential Chloride-losing Diarrhea [2]. The disease is associated with loss of SLC26A3-mediated Cl−/HCO3−exchange across the microvillar apical membrane of enterocytes, leading to severely reduced intestinal NaCl absorption. An independent positional cloning project led to the realization that the immediately adjacent gene SLC26A4/PDS, likely a product of gene duplication, is responsible for both Pendred Syndrome [1] and for nonsyndromic deafness DFNB4 associated with enlargement of the vestibular aqueduct (reviewed by Ito et al. [42] in this volume). The unlinked SLC26A2/DTDST gene was next linked to diastrophic dysplasia [43] and several related clinical entities encoded by mutations in the same gene. The above human disease phenotypes have been replicated in knockout mice. Autosomal recessive deafness was initially attributed to biallelic mutations in the SLC26A5 gene [44] encoding prestin, the mechanotransducer of the cochlear outer hair cell lateral membrane [45]. However, subsequent studies established that the very few known prestin coding sequence variants found to date are variants also present in individuals with normal hearing [46], despite the severe hearing impairment observed in Slc26a5−/− mice. Mouse-specific pathological phenotypes have been observed in mice genetically deficient in other Slc26 gene products. These include urolithiasis in Slc26a1−/− and Slc26a6−/− mice [47, 48], gastric achlorhydria in Slc26a7−/− and Slc26a9−/− mice [49, 50], distal renal tubular acidosis in Slc26a7−/− mice [50], and male infertility in Slc26a8−/− mice [51].
The SLC26A10 gene is transcribed and mRNA levels exhibit tissue and differentiation-state specificity, but its open reading frame encodes a STAS domain-deficient polypeptide which is inactive as an anion transporter in Xenopus oocytes (unpublished data). However, no data is available on SLC26A10 polypeptide expression, or its potential interactions with other proteins. Moreover, the Slc26a10 gene in most lower organisms for which sequence is available encodes full-length polypeptides with STAS domains. Thus, the designation of SLC26A10 as a pseudogene remains tentative.
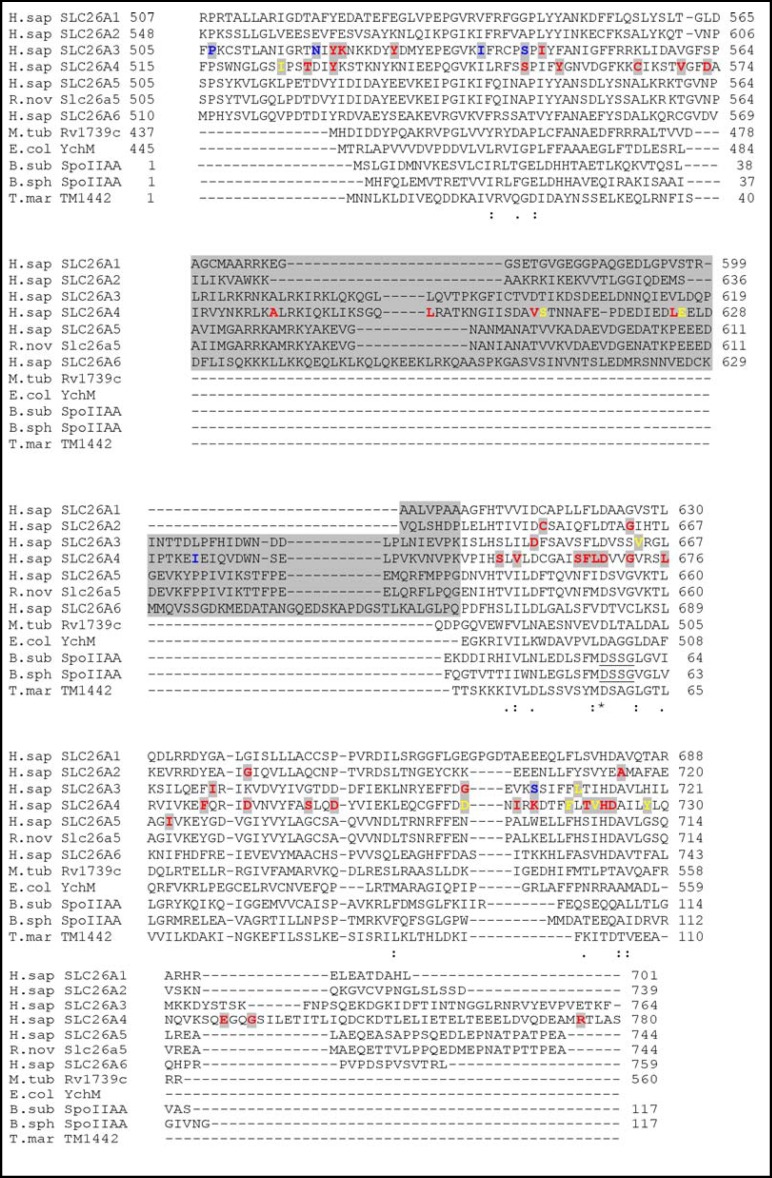
Mammalian and plant SLC26 polypeptides generally encode proteins longer than bacterial SulP proteins, with longer N-terminal cytoplasmic domains, longer loops connecting transmembrane spans, and longer C-terminal regions, including longer STAS domains. Molecular truncation experiments have indicated that STAS domain integrity is required for optimal surface trafficking and functional expression for SLC26A3 [52], as has been shown for A. thaliana Sultr1;2 [40, 53]. Trafficking impairment is noted with most of the examined disease mutations of the SLC26 STAS domains (Fig. 8), which constitute a minority of all SLC26 human disease mutations. The mammalian STAS domains differ from those of anti-anti-ρ proteins and from bacterial SulP polypeptides in their intervening sequences (IVS) inserted into the loop between helix a1 and strand β3 (rat prestin nomenclature). The largest number of distinct disease-associated variants in STAS domains, as well as in holoproteins, is found in pendrin (Fig. 8). Not shown in Fig. 8 are 9 recently discovered novel missense mutations in 8 amino acid residues (B. Wu, pers. comm.). Notably, among IVS sequences of SLC26 human disease genes, only the IVS of pendrin has been shown to date to harbor disease genes (Fig. 8). The IVS sequences are the most divergent regions of SLC26 STAS domains, and a Genbank search of the pendrin IVS revealed no identifiably related amino acid sequences in the database.
Fig. 8.
STAS domain amino acid sequences from the indicated human and rat SLC26 polypeptides, bacterial SulP proteins Rv1739c and YchM, and bacterial anti-anti-ρ pro teins SpoIIAA and TM1442. The anti-anti-ρ proteins are presented in their full lengths. Amino acid numbe ring is from UniProtKB (www.uniprot.org). The underlined DSSG motif of SpoIIAA includes its phosphorylated residue S58. Gray block indicates the intervening sequence (IVS) present in mammalian SLC26 STAS domains but absent from SulP STAS domains and from anti-anti- ρ proteins. SLC26 STAS amino acid residues in bold highlight human disease- associated missense mutations (in red), termination mutations (in yellow), and frameshifts mutations (in blue). Note that among SLC26 STAS domains, only in pendrin have mutations of the IVS been reported. Asterisks under sequences mark positions of complete sequence conservation. Alignment was generated by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Multiple residues of Rv1739c STAS involved in ligand contact or subject to ligand-induced conformational change align with human SLC26 STAS residues implicated in disease. The Rv1739c STAS residues chemically shifted by GTP [30] include (as numbered in Fig. 8 for Rv1739c holoprotein) V477 (corresponding to SLC26A4 V570) and E543 (corresponding to SLC26A3 S706fsX6 and to SLC26A4 K715). One additional Rv1739c STAS residue chemically shifted by GTP but not GDP is V497 (corresponding to SLC26A4 L668P). Rv1739c STAS residues implicated by molecular docking calculations in binding with GTP and GDP [30] include L513 (corresponding to SLC26A3 I675), G516 (corresponding to SLC26A2 G678V and to SLC26A4 D687Y), K540 (corresponding to SLC26A3 G702 and to SLC26A4 D711X), and I541 (corresponding to SLC26A4 I713M). Molecular docking calculations suggest that only GDP may bind to P551 corresponding to the prevalent SLC26A4 deafness mutation H723R. Thus, disease-associated mutations in human STAS domain residues align remarkably well with residues of Rv1739c STAS associated with binding of guanine nucleotides. This interesting relationship between ligand binding residues and disease-associated mutations suggests at minimum, a shared reliance on proper protein folding, and perhaps additional signaling-related functions.
The X-ray crystal and NMR solution structure of the rat prestin STAS domain with an engineered central deletion of the IVS sequence was recently reported [10]. Inclusion of the IVS produced NMR signal broadening and prevented crystallization. Truncation of the C-terminal 9 residues was also required for crystallization. Rat prestin exhibited a secondary structural element sequence of β 1 -β0-β2-α 1 -β3-α2-β4-α3-α4-β5-α5. Although with an extra β strand in its N-terminal region, prestin STAS resembled YchM STAS in the placement of strand β1 before helix a1, and resembled Rv1739c STAS in the presence of a helix a5. The absence from Rv1739c STAS of an antiparallel N-terminal β-strand may reflect inability of Pro residues at positions 8 and 14 to form inter-strand hydrogen bonds. However, secondary structure prediction suggests that inclusion in the Rv1739c construct of upstream residues between the end of the transmembrane domain and the N-terminus of the current STAS construct might elicit formation of this antiparallel N-terminal β-strand. Rat prestin STAS also differs from the STAS domains of YchM and Rv1739c in its higher angle of divergence between helices a1 and a2, possibly a result of the engineered deletion of the nearby IVS region.
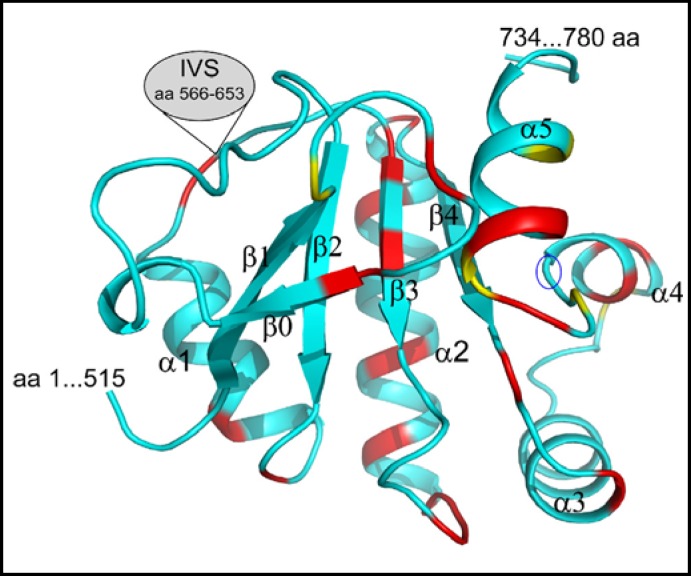
Using the rat prestin STAS domain crystal structure [10] as template in Modeller 9.9 [54], we have modeled the three-dimensional structure of the corresponding regions of human pendrin encompassing aa 515 and 734, but lacking the IVS sequence 566-653 between helix a1 and strand β2 (Fig. 9). The model includes a β1 strand which is the only strand oriented anti-parallel to the other strands. However, secondary structure prediction analysis reveals that in the presence of IVS sequence, the presence of a β1 strand equivalent is not predicted in pendrin or in several other SLC26 STAS domains [30]. Based on mutagenesis studies, Sultr1;2 STAS domain residues A540, Y542, F543 and S588 were proposed to interact with the membrane domain [40], These residues correspond to partially conserved residues P543, Y545, Y546, and V655 of rat prestin STAS, and to residues P560, C562, F563 and T600 of Rv1739c STAS, all of which occupy similar positions on the STAS surface. These residues were proposed to form a juxtamembrane binding surface, or to interact with the SLC26/SulP transmembrane domain [10]. The corresponding pendrin STAS residues P553, F555, Y556, and V671 occupy similar putative positions on the modeled STAS domain structure (Fig. 9). Rv1739c P560 is highly conserved among SLC26 and SulP proteins, whereas the modest conservation at position 563 of Rv1739c includes residues with bulky hydrophobic side chains.
Fig. 9.
Backbone structure of the human pendrin STAS domain encompassing aa 515-734 (excluding the intervening sequence (IVS) region of aa 566-653 between helix a1 and strand β3), as modeled on the structure of rat prestin (PDB ID 3LLO; [10]). The missing portions correspond to regions absent from the rat prestin structure. Red residues are sites of disease-associated missense mutations; yellow residues are sites of disease-associated termination mutations. Blue oval marks nominal strand β5 (2 aa in length) which is absent from the structure generated by PyMOL, but present in that generated by MOLMOL (not shown; [75]).
With the exception of SLC26A5/prestin, identification of proteins that interact directly with mammalian SLC26 polypeptides remains in its initial stages. The interaction of MgcRacGap with the SLC26A8 STAS domain [55], seems highly relevant to guanine nucleotide binding by SpoIIAA and by Rv1739c STAS, but no subsequent reports have explored this interaction. Some but not all SLC26 STAS domain C-terminal regions exhibit PDZ recognition motifs. Consistent with this, SLC26A3 STAS domain binds to NHERF2 [56] and to CAP70/PDZK1 [57]. The SLC26A6 STAS domain binds to NHERF1 and NHERF2 [58]. Conversely, CFTR also binds CAP70/ PDZK1 and NHERF proteins[59], indicating coassembly of scaffolds approximating SLC26 protein with CFTR, and perhaps stabilizing their interactions. The R domain of CFTR also binds STAS domains from SLC26A3, SLC26A6, SLC26A9, with the consequence of mutual, reciprocal activation of anion transport activities in the setting of HEK-293 cell overexpression [60, 61], where R domain phosphorylation was reported to be required. However, purified recombinant SLC26A3 domain interacted with purified recombinant CFTR R domain independent of phosphorylation state [62]. In the context of intact mice, absence of SLC26A6 expression stimulated basal CFTR-dependent HCO3− secretion, but inhibited activated secretion of HCO3− [63]. In yet another set of variations, CFTR coexpression stimulated SLC26A9 anion channel activity in Xenopus oocytes, an interaction inhibited by STAS domain mutation L683P [64], yet the CFTR R domain inhibited both anion exchange and conductance by SLC26A9. Moreover, removal of the SLC26A9 STAS domain eliminated anion conductance but partially preserved anion exchange activity while rendering it CFTR R domain-insensitive [65]. Although pendrin was initially reported to be activated by coexpressed CFTR in HEK-293 cells [60], later studies of iodide secretion in intact parotid duct failed to detect functional interaction of CFTR and pendrin [66]. Thus, the conditions of STAS domain-R domain interaction, the source of STAS domain, and the anion transport properties of the appended transmembrane domain may dictate whether the interaction output is stimulatory or inhibitory. Pendrin STAS-interacting proteins other than CFTR have not been reported. Nor have any binding proteins with specificity for a mammalian STAS domain IVS sequence been reported.
Even SLC26A5/prestin binds to and is potentiated by CFTR coexpression in HEK-293 cells. This interaction with prestin may direct a proportion of CFTR to the outer hair cell lateral membrane, an unprecedented subcellular localization in mammalian cells. However, prestin-mediated non-linear capacitance is not modified in outer hair cells of Cftr−/− mice [67], and cystic fibrosis in humans has not been associated with hearing loss beyond that plausibly attributable to chronic aminoglycoside treatment. MAP1S is a prestin STAS-binding protein discovered through yeast two-hybrid interaction [68]. MAP1S interacts through a linker region between light and heavy chain domains with the proximal region of the STAS domain. The interaction leads to marked stimulation of prestin-mediated nonlinear capacitance in transiently transfected CHO cells in parallel with increased surface expression. MAP1S has a tonotopic distribution that parallels the tonotopic distribution of pendrin through the cochlear spiral [68]. The integral membrane protein “Vesicle-associated membrane protein-associated protein A” (VAPA or VAP-33) was discovered as a prestin-binding protein through a membrane-based yeast two-hybrid screen, and validated by co-immunoprecipitation and immuno-colocalization in outer hair cells. Outer hair cells from Slc26a5~'~ mice expressed VAPA at decreased abundance, suggesting a possible role for VAPA in vesicular trafficking of prestin [69]. Additional, incompletely validated prestin-binding proteins identified in this membrane-based yeast two-hybrid screen [70] include integrin-interacting proteins tetraspanins 6 and 29 (CD9) and β2-microglobulin, glycosylphosphoinositide-linked costimulatory molecule CD52/CAMPATH-1, the small conductance Ca2+-activated K+ channel KCNN2/ SK2, fatty acid binding protein FABp3, Wnt receptor frizzled-3, bone γ-carboxyglutamate protein I, and dynein light chain Tctex-type I [70]. Kcnn2 is expressed in inner hair cells [71], and frizzled-3 is involved in embryonic cochlear hair cell development [72]. Prestin has also been shown to interact indirectly with βV-Spectrin, likely through the cytoskeletal proteins F-actin and protein 4.1 [73]. However, these intermediate proteins have themselves not yet been shown to bind directly to prestin.
Conclusion
The STAS domain is the central structural feature of the C-terminal cytoplasmic domains of SulP and SLC26 anion transport proteins. The phylogenetically ancient protein fold of the STAS domain also constitutes the core of anti-anti-ρ factor function in multiple bacterial ρ factor pathways. The STAS domain secondary structures vary slightly at their N- and C-termini and more extensively in the potentially disordered IVS sequence between helix a1 and strand β3 (rat prestin nomenclature), present only in STAS domains of higher organisms. The STAS domain of B. subilis YtvA responds to a blue light-induced conformational change in the adjacent LOV domain to transduce a transcriptional signal to ρB by a yet unknown pathway. E. coli YchM STAS likely functions in fatty acid biosynthesis through its binding to acyl carrier protein, but its role in bacterial lipid metabolism remains incompletely understood. Since acyl carrier protein is a cofactor in numerous bacterial biosynthetic pathways, including those of polyketide antibiotics, the impact of YchM may extend beyond lipid metabolism. M. tuberculosis Rv1739c STAS binds and may hydrolyze guanine nucleotides, but the functional importance of these activities remains obscure for Rv1739c as well as for anti-anti-ρ SpoIIAA of B. subtilis. Additional putative functions of bacterial STAS domains can be hypothesized based on the nature of adjacent functional domains within the same multidomain polypeptide, or based on adjacency or proximity within a single operon.
Although widespread among bacteria, in higher organisms the STAS domain appears restricted to SulP/SLC26 anion transporter polypeptides. The presence of disease-causing mutations in the STAS domains of SLC26A2/DTDST, SLC26A3/DRA, and SLC26A4/ pendrin attests to the structural importance of the STAS domain, as suggested by recombinant expression and targeting experiments. No intrinsic enzymatic activity has been associated with STAS domains of mammalian SLC26 anion transporters. However the STAS domains have a complex, context-dependent relationship with the R domain of CFTR and with their own adjacent SLC26 anion transport domains that may be modified by scaffolding through adjacent PDZ recognition motifs. The growing number of identified, specific prestin-interacting proteins of the cochlear outer hair cell may presage a similar bounty of proteins that interact with the other, less highly conserved SLC26 STAS domains.
The intact STAS domains of mammalian SLC26 polypeptides have proven thus far refractory to crystallization, or even to NMR solution structure determination. This difficulty may reflect unstructured or metastable IVS domains, N-terminal linker sequences, and/or C-terminal sequences of STAS domains. The latter two possibilities may require empiric selection of optimal N- and C-terminal boundaries of STAS domain constructs for structure determination. The unanticipated, stoichiometric presence of acyl carrier protein was important in the crystallization of E. coli YchM. The yet unreported proteins hypothesized to interact with mammalian SLC26 STAS domains may similarly prove important for stabilization and structure determination of the putatively disordered IVS regions of intact mammalian SLC26 STAS domains, and perhaps of the corresponding holoproteins as well. They may, in addition, provide information crucial to decoding the mechanisms by which STAS domains regulate SLC26 polypeptide trafficking, stability, and anion transport function.
Acknowledgements
This work was supported by NIH grant DK43495 with core facility support from the Harvard Digestive Diseases Center (NIH DK34854) to SLA.
References
- 1.Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, Sheffield VC, Green ED. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- 2.Hoglund P, Haila S, Socha J, Tomaszewski L, Saarialho-Kere U, Karjalainen-Lindsberg ML, Airola K, Holmberg C, de la Chapelle A, Kere J. Mutations of the down-regulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat Genet. 1996;14:316–319. doi: 10.1038/ng1196-316. [DOI] [PubMed] [Google Scholar]
- 3.Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- 4.Kere J. Overview of the SLC26 family and associated diseases. Novartis Found Symp. 2006;273:2–11. disc. 11-18, 261-264. [PubMed] [Google Scholar]
- 5.Dorwart MR, Shcheynikov N, Yang D, Muallem S. The solute carrier 26 family of proteins in epithelial ion transport. Physiology. 2008;23:104–114. doi: 10.1152/physiol.00037.2007. [DOI] [PubMed] [Google Scholar]
- 6.Price GD, Howitt SM. The cyano-bacterial bicarbonate transporter bicA: Its physiological role and the implications of structural similarities with human SLC26 transporters. Biochem Cell Biol. 2011;89:178–188. doi: 10.1139/o10-136. [DOI] [PubMed] [Google Scholar]
- 7.Felce J, Saier MH., Jr Carbonic anhydrases fused to anion transporters of the sulP family: Evidence for a novel type of bicarbonate transporter. J Molec Microbiol Biotechnol. 2004;8:169–176. doi: 10.1159/000085789. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin EV. The stas domain - a link between anion transporters and antisigma-factor antagonists. Curr Biol. 2000;10:R53–55. doi: 10.1016/s0960-9822(00)00335-3. [DOI] [PubMed] [Google Scholar]
- 9.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: A structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualetto E, Aiello R, Gesiot L, Bonetto G, Bellanda M, Battistutta R. Structure of the cytosolic portion of the motor protein prestin and functional role of the STAS domain in SLC26/SulP anion transporters. J Molec Biol. 2010;400:448–462. doi: 10.1016/j.jmb.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs H, Comfort D, Lord M, Campbell ID, Yudkin MD. Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaf of Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:5067–5071. doi: 10.1073/pnas.95.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seavers PR, Lewis RJ, Brannigan JA, Verschueren KH, Murshudov GN, Wilkinson AJ. Structure of the Bacillus cell fate determinant SpoIIAA in phosphorylated and unphosphorylated forms. Structure. 2001;9:605–614. doi: 10.1016/s0969-2126(01)00623-2. [DOI] [PubMed] [Google Scholar]
- 13.Masuda S, Murakami KS, Wang S, Anders Olson C, Donigian J, Leon F, Darst SA, Campbell EA. Crystal structures of the ADP- and ATP-bound forms of the Bacillus anti-sigma factor SpoIIAB in complex with the anti-anti-sigma SpoIIAA. J Mol Biol. 2004;340:941–956. doi: 10.1016/j.jmb.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 14.Najafi SM, Harris DA, Yudkin MD. The SpoIIAA protein of Bacillus subtilis has GTP-binding properties. J Bacteriol. 1996;178:6632–6634. doi: 10.1128/jb.178.22.6632-6634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecker M, Pane-Farre J, Volker U. Sigma b-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 16.Marles-Wright J, Lewis RJ. The stressosome: Molecular architecture of a signalling hub. Biochem Soc Trans. 2010;38:928–933. doi: 10.1042/BST0380928. [DOI] [PubMed] [Google Scholar]
- 17.Murray JW, Delumeau O, Lewis RJ. Structure of a nonheme globin in environmental stress signaling. Proc Natl Acad Sci USA. 2005;102:17320–17325. doi: 10.1073/pnas.0506599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo S, Demeler B, Haldenwang WG. The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation. J Bacteriol. 2008;190:6625–6635. doi: 10.1128/JB.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurk M, Dorn M, Schmieder P. Blue flickers of hope: Secondary structure, dynamics, and putative dimerization interface of the blue-light receptor YtvA from Bacillus subtilis. Biochemistry. 2011;50:8163–8171. doi: 10.1021/bi200782j. [DOI] [PubMed] [Google Scholar]
- 20.Avila-Perez M, Vreede J, Tang Y, Bende O, Losi A, Gartner W, Hellingwerf K. In vivo mutational analysis of YtvA from Bacillus subtilis: Mechanism of light activation of the general stress response. J Biol Chem. 2009;284:24958–24964. doi: 10.1074/jbc.M109.033316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Cao Z, Livoti E, Krauss U, Jaeger KE, Gartner W, Losi A. Interdomain signalling in the blue-light sensing and GTP-binding protein YtvA: A mutagenesis study uncovering the importance of specific protein sites. Photochem Photobiol Sci. 2010;9:47–56. doi: 10.1039/b9pp00075e. [DOI] [PubMed] [Google Scholar]
- 22.Buttani V, Gartner W, Losi A. NTP-binding properties of the blue-light receptor YtvA and effects of the E105l mutation. Eur Biophys J. 2007;36:831–839. doi: 10.1007/s00249-007-0155-1. [DOI] [PubMed] [Google Scholar]
- 23.Losi A. Flavin-based blue-light photosensors: A photobiophysics update. Photochem Photobiol. 2007;83:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 24.Buttani V, Losi A, Polverini E, Gartner W. Blue news: NTP binding properties of the blue-light sensitive YtvA protein from Bacillus subtilis. FEBS Lett. 2006;580:3818–3822. doi: 10.1016/j.febslet.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Nakasone Y, Hellingwerf KJ. On the binding of BODIPY-GTP by the photosensory protein YtvA from the common soil bacterium Bacillus subtilis. Photochem Photobiol. 2011;87:542–547. doi: 10.1111/j.1751-1097.2011.00919.x. [DOI] [PubMed] [Google Scholar]
- 26.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelden MC, Howitt SM, Price GD. Membrane topology of the cyanobacterial bicarbonate transporter, BicA, a member of the SulP (SLC26A) family. Molec Membr Biol. 2011;27:12–23. doi: 10.3109/09687680903400120. [DOI] [PubMed] [Google Scholar]
- 29.Zolotarev AS, Unnikrishnan M, Shmukler BE, Clark JS, Vandorpe DH, Grigorieff N, Rubin EJ, Alper SL. Increased sulfate uptake by E. Coli overexpressing the Slc26-related SulP protein RV1739c from Mycobacterium tuberculosis. Comp Biochem Physiol. 2008;149:255–266. doi: 10.1016/j.cbpa.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma AK, Ye L, Baer CE, Shanmugasundaram K, Alber T, Alper SL, Rigby AC. Solution structure of the guanine nucleotide-binding STAS domain of Slc26-related SulP protein RV1739c from Mycobacterium tuberculosis. J Biol Chem. 2011;286:8534–8544. doi: 10.1074/jbc.M110.165449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babu M, Greenblatt JF, Emili A, Strynadka NC, Reithmeier RA, Moraes TF. Structure of a Slc26 anion transporter STAS domain in complex with acyl carrier protein: Implications for E. Coli YchM in fatty acid metabolism. Structure. 2010;18:1450–1462. doi: 10.1016/j.str.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 32.Compton EL, Karinou E, Naismith JH, Gabel F, Javelle A. Low resolution structure of a bacterial Slc26 transporter reveals dimeric stoichiometry and mobile intracellular domains. J Biol Chem. 2011;286:27058–27067. doi: 10.1074/jbc.M111.244533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimori I, Minakuchi T, Vullo D, Scozzafava A, Innocenti A, Supuran CT. Carbonic anhydrase inhibitors. Cloning, characterization, and inhibition studies of a new beta-carbonic anhydrase from Mycobacterium tuberculosis. J Med Chem. 2009;52:3116–3120. doi: 10.1021/jm9003126. [DOI] [PubMed] [Google Scholar]
- 34.Du L, Sanchez C, Chen M, Edwards DJ, Shen B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus atcc15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 35.Lai JR, Koglin A, Walsh CT. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochem. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 36.Fitzpatrick KL, Tyerman SD, Kaiser BN. Molybdate transport through the plant sulfate transporter Shst1. FEBS Lett. 2008;582:1508–1513. doi: 10.1016/j.febslet.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 37.Ye H, Zhang XQ, Broughton S, Westcott S, Wu D, Lance R, Li C. A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct Integr Genom. 2011;11:103–110. doi: 10.1007/s10142-011-0209-4. [DOI] [PubMed] [Google Scholar]
- 38.Davidian JC, Kopriva S. Regulation of sulfate uptake and assimilation—the same or not the same? Molec Plant. 2010;3:314–325. doi: 10.1093/mp/ssq001. [DOI] [PubMed] [Google Scholar]
- 39.Buchner P, Takahashi H, Hawkesford MJ. Plant sulphate transporters: Coordination of uptake, intracellular and long-distance transport. J Exp Botany. 2004;55:1765–1773. doi: 10.1093/jxb/erh206. [DOI] [PubMed] [Google Scholar]
- 40.Shibagaki N, Grossman AR. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem. 2006;281:22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- 41.Shibagaki N, Grossman AR. Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J Biol Chem. 2010;285:25094–25102. doi: 10.1074/jbc.M110.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito T CB, King KA, Zalewski CK, Muskett J, Chattaraj P, Shawker T, Reynolds JC, Butman JA, Brewer CC, Wangemann P, Alper SL, Griffith AJ. SLC26A4 and phenotypes associated with enlargment of the vestibular aqueduct (EVA) Cell Physiol Biochem. 2011;28:545–552. doi: 10.1159/000335119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hastbacka J, Kerrebrock A, Mokkala K, Clines G, Lovett M, Kaitila I, de la Chapelle A, Lander ES. Identification of the finnish founder mutation for diastrophic dysplasia (DTD) Eur J Hum Genet. 1999;7:664–670. doi: 10.1038/sj.ejhg.5200361. [DOI] [PubMed] [Google Scholar]
- 44.Liu XZ, Ouyang XM, Xia XJ, Zheng J, Pandya A, Li F, Du LL, Welch KO, Petit C, Smith RJ, Webb BT, Yan D, Arnos KS, Corey D, Dallos P, Nance WE, Chen ZY. Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum Molec Genet. 2003;12:1155–1162. doi: 10.1093/hmg/ddg127. [DOI] [PubMed] [Google Scholar]
- 45.Zheng J, Shen W, He DZ, Long KB, Madison LD, Dallos P. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 46.Minor JS, Tang HY, Pereira FA, Alford RL. DNA sequence analysis of SLC26A5, encoding prestin, in a patient-control cohort: Identification of fourteen novel DNA sequence variations. PloS One. 2009;4:e5762. doi: 10.1371/journal.pone.0005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson PA, Russell CS, Lee S, McLeay SC, van Dongen JM, Cowley DM, Clarke LA, Markovich D. Urolithiasis and hepatotoxicity are linked to the anion transporter Sat1 in mice. J Clin Invest. 2010;120:706–712. doi: 10.1172/JCI31474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Z, Asplin JR, Evan AP, Rajendran VM, Velazquez H, Nottoli TP, Binder HJ, Aronson PS. Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nat Genet. 2006;38:474–478. doi: 10.1038/ng1762. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Song P, Miller ML, Borgese F, Barone S, Riederer B, Wang Z, Alper SL, Forte JG, Shull GE, Ehrenfeld J, Seidler U, Soleimani M. Deletion of the chloride transporter Slc26a9 causes loss of tubulovesicles in parietal cells and impairs acid secretion in the stomach. Proc Natl Acad Sci USA. 2008;105:17955–17960. doi: 10.1073/pnas.0800616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J, Song P, Nakamura S, Miller M, Barone S, Alper SL, Riederer B, Bonhagen J, Arend LJ, Amlal H, Seidler U, Soleimani M. Deletion of the chloride transporter Slc26a7 causes distal renal tubular acidosis and impairs gastric acid secretion. J Biol Chem. 2009;284:29470–29479. doi: 10.1074/jbc.M109.044396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toure A, Lhuillier P, Gossen JA, Kuil CW, Lhote D, Jegou B, Escalier D, Gacon G. The testis anion transporter 1 (Slc26a8) is required for sperm terminal differentiation and male fertility in the mouse. Hum Molec Genet. 2007;16:1783–1793. doi: 10.1093/hmg/ddm117. [DOI] [PubMed] [Google Scholar]
- 52.Chernova MN, Jiang L, Shmukler BE, Schweinfest CW, Blanco P, Freedman SD, Stewart AK, Alper SL. Acute regulation of the SLC26A3 congenital chloride diarrhoea anion exchanger (DRA) expressed in Xenopus oocytes. J Physiol. 2003;549:3–19. doi: 10.1113/jphysiol.2003.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouached H, Berthomieu P, El Kassis E, Cathala N, Catherinot V, Labesse G, Davidian JC, Fourcroy P. Structural and functional analysis of the C-terminal STAS (sulfate transporter and anti-sigma antagonist) domain of the Arabidopsis thaliana sulfate transporter Sultr1.2. J Biol Chem. 2005;280:15976–15983. doi: 10.1074/jbc.M501635200. [DOI] [PubMed] [Google Scholar]
- 54.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by modeller. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 55.Toure A, Morin L, Pineau C, Becq F, Dorseuil O, Gacon G. Tat1, a novel sulfate transporter specifically expressed in human male germ cells and potentially linked to Rho GTPase signaling. J Biol Chem. 2001;276:20309–20315. doi: 10.1074/jbc.M011740200. [DOI] [PubMed] [Google Scholar]
- 56.Lamprecht G, Heil A, Baisch S, Lin-Wu E, Yun CC, Kalbacher H, Gregor M, Seidler U. The down regulated in adenoma (DRA) gene product binds to the second PDZ domain of the NHE3 kinase regulatory protein (E3KARP), potentially linking intestinal Cl-/HCO3-exchange to Na+/H+ exchange. Biochem. 2002;41:12336–12342. doi: 10.1021/bi0259103. [DOI] [PubMed] [Google Scholar]
- 57.Rossmann H, Jacob P, Baisch S, Hassoun R, Meier J, Natour D, Yahya K, Yun C, Biber J, Lackner KJ, Fiehn W, Gregor M, Seidler U, Lamprecht G. The Cftr associated protein Cap70 interacts with the apical Cl-/HCO3- exchanger DRA in rabbit small intestinal mucosa. Biochem. 2005;44:4477–4487. doi: 10.1021/bi048828b. [DOI] [PubMed] [Google Scholar]
- 58.Lohi H, Lamprecht G, Markovich D, Heil A, Kujala M, Seidler U, Kere J. Isoforms of SLC26A6 mediate anion transport and have functional PDZ interaction domains. Am J Physiol Cell Physiol. 2003;284:C769–779. doi: 10.1152/ajpcell.00270.2002. [DOI] [PubMed] [Google Scholar]
- 59.Li C, Naren AP. Analysis of CFTR interactome in the macromolecular complexes. Methods Mol Biol. 2011;741:255–270. doi: 10.1007/978-1-61779-117-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko SB, Shcheynikov N, Choi JY, Luo X, Ishibashi K, Thomas PJ, Kim JY, Kim KH, Lee MG, Naruse S, Muallem S. A molecular mechanism for aberrant Cftr-dependent HCO3- transport in cystic fibrosis. EMBO J. 2002;21:5662–5672. doi: 10.1093/emboj/cdf580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ko SB, Zeng W, Dorwart MR, Luo X, Kim KH, Millen L, Goto H, Naruse S, Soyombo A, Thomas PJ, Muallem S. Gating of Cftr by the STAS domain of SLC26 transporters. Nat Cell Biol. 2004;6:343–350. doi: 10.1038/ncb1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorwart MR, Shcheynikov N, Baker JM, Forman-Kay JD, Muallem S, Thomas PJ. Congenital chloride-losing diarrhea causing mutations in the STAS domain result in misfolding and mistrafficking of SLC26A3. J Biol Chem. 2008;283:8711–8722. doi: 10.1074/jbc.M704328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Soyombo AA, Shcheynikov N, Zeng W, Dorwart M, Marino CR, Thomas PJ, Muallem S. Slc26a6 regulates Cftr activity in vivo to determine pancreatic duct HCO3- secretion: Relevance to cystic fibrosis. EMBO J. 2006;25:5049–5057. doi: 10.1038/sj.emboj.7601387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avella M, Borgese F, Ehrenfeld J. Characterization of the l683P mutation of SLC26A9 in Xenopus oocytes. Biochim Biophys Acta. 2011;1810:577–583. doi: 10.1016/j.bbagen.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 65.Chang MH, Plata C, Sindic A, Ranatunga WK, Chen AP, Zandi-Nejad K, Chan KW, Thompson J, Mount DB, Romero MF. SLC26A9 is inhibited by the R-region of the cystic fibrosis transmembrane conductance regulator via the STAS domain. J Biol Chem. 2009;284:28306–28318. doi: 10.1074/jbc.M109.001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shcheynikov N, Yang D, Wang Y, Zeng W, Karniski LP, So I, Wall SM, Muallem S. The Slc26a4 transporter functions as an electroneutral Cl-/I-/HCO3- exchanger: Role of Slc26a4 and Slc26a6 in I- and HCO3- secretion and in regulation of Cftr in the parotid duct. J Physiol. 2008;586:3813–3824. doi: 10.1113/jphysiol.2008.154468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Homma K, Miller KK, Anderson CT, Sengupta S, Du GG, Aguinaga S, Cheatham M, Dallos P, Zheng J. Interaction between Cftr and prestin (Slc26a5) Biochim Biophys Acta. 2010;1798:1029–1040. doi: 10.1016/j.bbamem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai JP, Surguchev A, Ogando Y, Song L, Bian S, Santos-Sacchi J, Navaratnam D. Prestin surface expression and activity are augmented by interaction with MAP1S, a microtubule-associated protein. J Biol Chem. 2010;285:20834–20843. doi: 10.1074/jbc.M110.117853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sengupta S, Miller KK, Homma K, Edge R, Cheatham MA, Dallos P, Zheng J. Interaction between the motor protein prestin and the transporter protein VAPA. Biochim Biophys Acta. 2010;1803:796–804. doi: 10.1016/j.bbamcr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng J, Anderson CT, Miller KK, Cheatham M, Dallos P. Identifying components of the hair-cell interactome involved in cochlear amplification. BMC Genomics. 2009;10:127. doi: 10.1186/1471-2164-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcotti W, Johnson SL, Kros CJ. A transiently expressed SK current sustains and modulates action potential activity in immature mouse inner hair cells. J Physiol. 2004;560:691–708. doi: 10.1113/jphysiol.2004.072868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Guo N, Nathans J. The role of frizzled3 and frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J Neurosci. 2006;26:2147–2156. doi: 10.1523/JNEUROSCI.4698-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Legendre K, Safieddine S, Kussel-Andermann P, Petit C, El-Amraoui A. Alpha2-betaV spectrin bridges the plasma membrane and cortical lattice in the lateral wall of the auditory outer hair cells. J Cell Sci. 2008;121:3347–3356. doi: 10.1242/jcs.028134. [DOI] [PubMed] [Google Scholar]
- 74.Humphrey W, Dalke A, Schulten K. Vmd: Visual molecular dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-38. [DOI] [PubMed] [Google Scholar]
- 75.Koradi R, Billeter M, Wuthrich K. Molmol: A program for display and analysis of macromolecular structures. J Molec Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29-32. [DOI] [PubMed] [Google Scholar]