Abstract
Both inhibitory and activating forms of natural killer (NK) cell receptors are found in mammals. The activating receptors play a direct role in the recognition of virally infected or transformed cells and transduce activating signals into the cell by partnering with an adaptor protein, which contains a cytoplasmic activation motif. Activating NK receptors encoded by the mammalian leukocyte receptor complex (e.g., killer cell immunoglobulin-like receptors) and the natural killer complex (e.g., Ly49s) partner with the adaptor protein DAP12, whereas NK receptors encoded in the CD94/NKG2 complex partner with the adaptor protein DAP10. Novel immune-type receptors (NITRs) found in bony fish share several common features with immunoglobulin-type NK receptors. Nitr9 is a putative activating receptor in zebrafish that induces cytotoxicity within the context of human NK cells. One isoform of Nitr9, Nitr9L, is shown here to preferentially partner with a zebrafish ortholog of Dap12. Cross-linking the Nitr9L–Dap12 complex results in activation of the phosphytidylinositol 3-kinase→AKT→extracellular signal-regulated kinase pathway suggesting that the DAP12-based activating pathway is conserved between bony fish and mammals.
Keywords: Natural cytotoxicity, DAP10, ERK, AKT, DAP12, NITR
Introduction
The cells of the mammalian immune system rely on an intricate network of signaling pathways to differentiate between “self” and “nonself.” The activation or inhibition of these signaling pathways relies on specific cell-surface receptors that engage specific ligands. Receptors can be classified as inhibitory or activating based on the functional outcome of ligand recognition. Natural killer (NK) cells represent a particularly well-characterized example of a cell lineage that utilizes this mode of regulation. When an activating NK cell receptor binds its ligand, the NK cell is activated to kill the target cell; in contrast, when an inhibitory NK receptor binds its ligand, NK cell-mediated killing is repressed.
Primates and rodents utilize different families of NK receptors to effect NK function. Primates typically utilize the killer cell immunoglobulin (Ig)-like receptor (KIR) family of Ig-type receptors, whereas rodents utilize the Ly49 family of lectin-type receptors (Bashirova et al. 2006; Ortaldo and Young 2005; Trowsdale et al. 2001). Cytoplasmic signaling utilized by these disparate receptors is well conserved (Billadeau and Leibson 2002; Cerwenka and Lanier 2000; Djeu et al. 2002). To initiate cell signaling, activating NK receptors physically associate through a positive charge in their transmembrane domain with a negative charge in the transmembrane domain of an adaptor protein. Adaptor proteins possess cytoplasmic signaling motifs necessary for activating a cellular response. The majority of mammalian-activating NK receptors (KIRs and Ly49s) utilize the adaptor protein, DAP12. In contrast, NKG2D, a lectin-type-activating NK receptor, utilizes DAP10 (Hyka-Nouspikel and Phillips 2006; Takaki et al. 2006), indicating a certain degree of specificity in the transmembrane interactions.
Considering the recent and rapid evolution of mammalian NK receptors (Hao and Nei 2005; Parham 2005; Sambrook et al. 2005), it is reasonable to expect that the functionally equivalent receptors in nonmammalian vertebrate species will be structurally divergent from the mammalian NK receptors. In this regard, novel immune-type receptors (NITRs) are members of diversified multigene families of activating/inhibitory receptors. Their overall organization is somewhat similar to KIRs; however, NITRs consist of either one or two ectodomains. All NITRs posses one N-terminal ectodomain of the IgV type, which may contain a J sequence, and many NITRs possess a second C-terminal ectodomain of the Ig-intermediate (I) type. Thus far, NITR genes have been identified only in bony fish and have been characterized most extensively in zebrafish (Evenhuis et al. 2007; Hawke et al. 2001; Piyaviriyakul et al. 2007; Strong et al. 1999; Yoder et al. 2001; Yoder et al. 2002; Yoder et al. 2004). Approximately 36 different NITR genes, which encompass 12 different families of receptors, are located within the NITR gene cluster on chromosome 7 in zebrafish (Litman et al. 2001; Yoder et al. 2001; Yoder et al. 2004); two additional families are located on a different chromosome (Yoder and Litman, unpublished observations). Although most of the NITRs in zebrafish possess cytoplasmic immunoreceptor tyrosine-based inhibition motifs and are predicted to exhibit inhibitory functions, a single gene, nitr9, encodes a positively charged residue within the transmembrane domain and is predicted to encode an activating receptor (Yoder et al. 2004). Nitr9 is expressed in multiple isoforms, all of which include the charged transmembrane residue and are likely to partner with adaptor proteins such as DAP12 or DAP10. Recently, the zebrafish orthologs of DAP12 and DAP10 have been identified (Yoder et al. 2007) permitting the evaluation of their roles in Nitr9 signaling.
We have shown that a recombinant form of zebrafish Nitr9, when cross-linked, can induce an activating signal within the context of mammalian NK cells (Yoder et al. 2004). To elucidate the function of Nitr9, a recombinant construct of the Nitr9 isoform, Nitr9L, was transfected into cultured human cells and shown to preferentially associate with epitope-tagged zebrafish Dap12. After antibody-induced cross-linking of the Nitr9L-Dap12 complex, phosphorylation of extracellular signal-regulated kinase (ERK) and AKT is increased. These data suggest that the Dap12 cytoplasmic signaling pathway is a common feature of activating receptor function that is conserved between bony fish and mammals.
Materials and methods
Reverse transcriptase–polymerase chain reaction
Kidneys from adult zebrafish were dissected, and myeloid and lymphoid cell lineages were purified cytometrically based on forward and side scatter and complementary deoxyribonucleic acids (cDNAs) generated as described (Yoder et al. 2007). One microliter of cDNA was subjected to thermal cycling with gene-specific primers and Titanium Taq DNA polymerase (BD Biosciences, San Jose, CA) in a 20-µl polymerase chain reaction (PCR) reaction, and 10 µl was analyzed by agarose gel electrophoresis. PCR cycles used for detecting nitr9, dap10, and dap12, and actin transcripts were: 40 (annealing at 65°C), 30 (annealing at 65°C), 40 (annealing at 65°C), and 25 (annealing at 65°C), respectively. Myeloperoxidase transcripts were detected using 25 PCR cycles (annealing at 70°C), and T cell receptor (TCR) α transcripts were detected using 40 PCR cycles (annealing at 60°C). Forward and reverse primers span at least one intron and are listed in Table 1. The identity of amplicons was confirmed by subcloning and DNA sequencing.
Table 1.
Oligonucleotide primer sequence
| Purpose | Primer name | Primer sequence |
|---|---|---|
| RT-PCR Dap10 | ZFDAP10-F3 | AGGGCTTCTAGTGTTTCTCCTCTC |
| ZFDAP10-R3 | GCCAAAGATCTTCATGTTTTGCAG | |
| RT-PCR Dap12 | ZFDAP12-F3 | GGATACTGTTTATAATGTTCCCCG |
| ZFDAP12-R3 | TTCCGATACTGCTGAAGATCACTG | |
| RT-PCR Nitr9 | Nitr9-AUG-For | ATGATCAACTTTTGGATTTTTGGACTTTTC |
| Nitr9-TAA-Rev | TTACTGCTGGTTAGAAACCGAGTTAATCAT | |
| RT-PCR TCRα | TCRα For | GGAACACAAGTTCATGTTGAAAC |
| TCRα Rev | GCTCATCCACGCTTTGAAAGTCA | |
| RT-PCR myeloperoxidase | mpx-For | CCAGAACCAGTGAGCCTGAGACACG |
| mpx-Rev | CAGTCTAACCATGGGCAGCGCTGCAC | |
| RT-PCR β-actin | Actin For | GGTATGGAATCTTGCGGTATCCAC |
| Actin Rev | ATGGGCCAGACTCATCGTACTCCT | |
| Myc-Nitr9L | Nitr9-for-blunt | CACCCAAATGCACCACCTGTGTTTGTTAAAC |
| Nitr9-rev-NotIa | gactgcggccgcTTACTGCTGGTTAGAAAC | |
| FLAG-DAP10 | ZFDAP10-For-bluntYSCF | TACTCATGTTTTGGAGTGAGC |
| ZFDAP10-rev-NotIa | acgtgcggccgcTCATGTTTTGCAGTTTGC | |
| FLAG-DAP12 | ZFDAP12-For-blunt | AATCAAGACTGCAGTTCCTGTTACC |
| ZFDAP12-Rev-NotIa | gcatgcggccgcTTATTTCCGATACTGCTGAAG | |
| FLAG-Dap12D42V | ZFDAP12_D→V-topb | GGGATCATCACGTGTGTTATTATTCTCACGCTC |
| ZFDAP12_D→V-botb | GAGCGTGAGAATAATAACACACGTGATGATCCC | |
| FLAG-Dap12Y96A | ZFDAP12_Y→A-topb | GAAGTAGAATCACCTGCCCAGGAGCTTTACGGA |
| ZFDAP12_Y→A-botb | TCCGTAAAGCTCCTGGGCAGGTGATTCTACTTC |
Overhang (5′) sequences are in lower case text and include a Not I site.
The mutated codons are italicized.
Epitope-tagged expression constructs
The Myc-tagged Nitr9L expression vector was constructed with the pLM plasmid (a derivative of pcDNA3; Invitrogen, Carlsbad, CA), which incorporates an amino-terminal leader sequence and Myc tag, and FLAG-tagged Dap10 and Dap12 expression vectors were constructed with the pLF plasmid (which incorporates a FLAG tag but is otherwise identical to pLM). Epitope-tagged constructs were generated by PCR amplifying the coding sequence of zebrafish nitr9L, dap10, and dap12 (excluding the leader sequence) with Pfu DNA polymerase (Stratagene, La Jolla, CA) employing blunt forward primers and reverse primers that include the endogenous stop codon and incorporate a 3′ NotI site (Table 1). Amplicons were digested with NotI and cloned into the EcoRV/NotI sites of pLM or pLF and confirmed by sequencing.
Site-directed mutagenesis
The plasmid encoding FLAG-tagged Dap12 was mutagenized using the QuickChange® Site Directed Mutagenesis Kit as recommended by the manufacturer (Stratagene). In brief, residue Asp-42 of Dap12 (GenBank EF158446) was mutagenized to a Val in the FLAG-Dap12 expression plasmid using the ZFDAP12_D→V-top and ZFDAP12_D→V-bot primers resulting in a FLAG-Dap12D42V construct. Residue Tyr-96 of Dap12 (GenBank EF158446) was mutagenized to an Ala in the FLAG-Dap12 expression plasmid using the ZFDAP12_Y→A-top and ZFDAP12_Y→A-bot primers resulting in a FLAG-Dap12Y96A construct. Mutations were confirmed by sequencing. Primer sequences are listed in Table 1.
Cell culture and transfections
The human AD-293 cell line (a derivative of the HEK293 cell line, with improved cell adherence) was obtained from the American Type Culture Collection (Manassas, VA) and was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a humidified 5% CO2 incubator. Approximately 1 × 106 cells were seeded into 100-mm dishes ~24 h before transfections. Cells were transfected with 10 µg of recombinant plasmid DNA as indicated in “Results and discussion” using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. The cells were harvested 48 h after transfection and used for further experiments.
Receptor cross-linking
Transfected cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and incubated with 1 µg/ml of anti-Myc antibody (9B11; Cell Signaling Technology, Danvers, MA) on ice for 30 min. After washing with DPBS, cells were incubated with 0.5 µl/ml of affinity purified rabbit anti-mouse IgG (H+L) antibody (Jackson Immunoresearch, West Grove, PA) at 37°C for 10 min. After washing with DPBS, 1 ml of lysis buffer (1% NP-40, 10 mM Tris pH 7.4, 150 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 10 mM iodoacetamide, 50 mM sodium fluoride, 1 mM ethylenediamene tetraacetic acid, 1 mM sodium orthovanadate, 0.25% sodium deoxycholate, 10 µg/ml leupeptin, 10 µg/ml pepsatin, and 10 µg/ml aprotinin, and 10 µl each of protease cocktail I and cocktail II [Sigma-Aldrich, St. Louis, MO]) was added to each 100-mm dish, followed by a 30-min incubation at 4°C. Cells were scraped off the plates and centrifuged for 15 min at 12,000×g and 4°C.
Immunoprecipitation and Western analyses
Transfected cells were washed twice with cold 1× DPBS and placed on ice for 30 min in lysis buffer. Cell lysates were centrifuged at 12,000 × g for 15 min at 4°C to remove nuclei and cell debris. The protein concentrations of the soluble extracts were determined using a Bradford assay (Bio-Rad Laboratories, Hercules, CA). The cleared lysates adjusted to contain equivalent amount of protein were incubated with 5 µg of the anti-Myc antibody and rotated overnight at 4°C, after which 30 µl of protein G-agarose beads (Sigma-Aldrich) were added and mixed for 2 h at 4°C. The precipitates were washed three times with lysis buffer, and proteins were eluted in 2× sodium dodecyl sulfate (SDS) sample buffer. Proteins were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes for Western blotting. Western blots were blocked in PBS–Tween (PBST) containing 4% bovine serum albumin for 1 h and then incubated overnight at 4°C in primary antibodies. The anti-FLAG (M2) antibody was obtained from Sigma-Aldrich; all other antibodies were obtained from Cell Signaling Technology. Blots were washed three times in PBST, followed by incubation for 1 h with horseradish peroxidase-conjugated secondary antibodies in PBST containing 5% nonfat dried milk. The blots were washed three times in PBST, and reaction patterns were visualized using an enhanced chemiluminescence detection system (ECL; GE Healthcare, Piscataway, NJ). In some instances, blots were processed in stripping buffer (Pierce, Rockford, IL) at 37°C for 15 min.
Results and discussion
Zebrafish Nitr9, Dap10, and Dap12 are expressed in lymphocytes
Although the actual associations of adaptors with transmembrane receptors were examined using cell transfection assays, it was essential to first determine if the adaptors and Nitr9 are expressed in the same cell lineage. PCR primers were employed that amplify the entire open reading frame of the previously described long (L) and short (S) transcripts of nitr9 (nitr9L and nitr9S, respectively). Unexpectedly, nitr9L and a nitr9 transcript that was shorter than nitr9S were amplified from zebrafish lymphocytes (Fig. 1a). Sequencing of this smaller transcript revealed a third splice variant of nitr9, encoding Nitr9 “super-short” (Nitr9SS). The entire V domain of Nitr9SS is eliminated by splicing (Fig. 1b and c). It is unknown why transcripts of nitr9S were not detected in lymphocytes: It is possible that nitr9S is not expressed or expressed at levels much lower than nitr9L and nitr9SS in lymphocytes or that expression of nitr9S may be inducible. A reverse transcriptase-PCR strategy was then used to evaluate the expression of nitr9, dap12, and dap10 in purified zebrafish lymphoid and myeloid cells. All three genes were shown to be expressed in lymphocytes and not in the myeloid population (Fig. 1a).
Fig. 1.

Detection of adaptor protein expression. a RT-PCR was used to detect transcripts of the zebrafish nitr9, dap10, and dap12 genes in the zebrafish lymphoid and myeloid lineages. In addition to the nitr9-long (nitr9L) transcript, an alternatively spliced transcript of nitr9, nitr9-super short (nitr9SS) was detected from lymphocytes. RT-PCR of mpx provides a positive control for myeloid cells and RT-PCR of TCRα provides a positive control for lymphocytes. β-Actin is shown as a positive control for both RNA samples. A negative “no template” control was evaluated in parallel with all primer sets. b Visual representations of the three isoforms of Nitr9 are shown: The long and short isoforms were previously described (Yoder et al. 2004). Immunoglobulin (V and I) and transmembrane (TM) domains are indicated. The positive charge within the TM domain is represented by a plus sign, and a cytoplasmic tyrosine (Y) is also shown. c Sequence analyses of the nitr9SS amplicon revealed an open reading frame lacking a V domain. The predicted peptide sequence of Nitr9SS is aligned with the previously described Nitr9L and Nitr9S. Amino acid numbering of the proteins is indicated on the left. Leader sequences and transmembrane domains are boxed (note that this alignment splits the 23 residue leader sequence of NitrSS into two segments). The arginine in the transmembrane domain is indicated with white text on a black shadow. Residues which are highly conserved in immunoglobulin domains are indicated above the alignment and numbered according to the international ImMunoGeneTics (IMGT) information system (Lefranc et al. 2005). Cysteines that are conserved in NITR I domains are denoted by an asterisks above the alignment (Litman et al. 2001)
Nitr9L preferentially associates with Dap12
The zebrafish genes encoding Dap10 and Dap12 have recently been described (Yoder et al. 2007). To evaluate the potential partnering of Nitr9L with zebrafish Dap10 and Dap12, a plasmid encoding an epitope (Myc)-tagged Nitr9L was cotransfected into AD-293 cells with a plasmid encoding an epitope (FLAG)-tagged Dap10 or Dap12 (Fig. 2a). Coimmunoprecipitation experiments demonstrated a stable, physical association of the Nitr9–DAP12 combination (Fig. 2b, lane 8); Nitr9L failed to stably associate with Dap10 (Fig. 2b, lane 6). It has been shown that antibody-induced cross-linking of certain activating receptors can enhance the association of the receptor with an adaptor protein and is sufficient to trigger the activating signal transduction pathway (Campbell et al. 1998). As expected, cross-linking Nitr9L (with an anti-Myc antibody) increases the physical association between Nitr9L and Dap12 (Fig. 2b, compare lanes 8 and 9; and Fig. 3a, compare lanes 8 and 9), whereas cross-linking the Nitr9–Dap10 combination did not result in a detectable receptor–adaptor association (Fig. 2b, lane 7). The association of Nitr9L with Dap12 is specific and enhanced when cross-linked with an antibody.
Fig. 2.

Nitr9L associates specifically with Dap12. a Predicted protein structures of recombinant zebrafish proteins utilized in cotransfection assays. Myc-tagged Nitr9-long (Nitr9L) is shown on the left, and FLAG-tagged Dap10 and Dap12 are shown on the right. Nitr9L encodes two extracellular Ig domains of the variable (V) and intermediate (I) types (Yoder et al. 2004). The cytoplasmic ITAM of Dap12 and the putative Dap10 signaling motif, YMNV, are indicated (Yoder et al. 2007). b AD-293 cells were cotransfected with plasmids encoding Myc-tagged Nitr9L and FLAG-tagged Dap10 or Dap12 as indicated (lanes 3–9). Cells were then preincubated with an anti-Myc antibody for cross-linking (X-link) before immunoprecipitation. pcDNA3 (lane 1) was used as a vector control, and isotype-matched antibody (lane 2) was used as a negative control for cross-linking. Immunoprecipitates (anti-Myc) were separated by SDS-PAGE and subjected to Western analyses using an anti-FLAG antibody (upper panel). Whole-cell lysates from the same treatments also were separated by SDS-PAGE and Western analyses completed with an anti-FLAG antibody to show the expression level of Dap10 and Dap12 (bottom panel)
Fig. 3.

Nitr9L–Dap12 signaling relies on key residues within Dap12. AD-293 cells were transfected with plasmids encoding Myc-tagged Nitr9L (lanes 6–13) and FLAG-tagged zebrafish Dap12 (lanes 3, 8, and 9) or FLAG-tagged Dap12D42V (lanes 4, 10, and 11) or FLAG-tagged Dap12Y96A (lanes 5, 12, and 13). Nitr9L was then cross-linked with an anti-Myc antibody (lanes 9, 11, and 13), and cells were lysed. a Nitr9L was immunoprecipitated (anti-Myc) and Western analyses completed with an anti-FLAG antibody to evaluate the association of Nitr9L with Dap12, Dap12D42V, and Dap12Y96A (top). Western analysis also was carried out on the whole-cell lysates to verify expression of the Dap12 proteins (bottom). b Whole-cell lysates also were subjected to Western analyses to detect phosphorylated ERK (p-ERK; top) as compared to total ERK (bottom) and for c phosphorylated AKT (p-AKT; top) as compared to total AKT (bottom). Control cells either were exposed to an isotype matched antibody (lane 2) or transfected with the parental plasmid, pcDNA3 (lane 1). Note the increase in phosphorylation of ERK and AKT after cross-linking Nitr9L in the presence of Dap12 (compare lanes 8 and 9)
Elucidating the Nitr9–Dap12 signaling pathway
When activated, mammalian DAP12 signals via SYK tyrosine kinase to engage downstream signaling molecules such as PI3K and ERK (Djeu et al. 2002). PI3K and ERK are rapidly activated in NK cells upon receptor cross-linking and account for redistribution of granules, representing a critical step for NK cell-mediated cytolysis. Cross-linking activating receptor/DAP12 complexes results in increased phosphorylation of PI3K and ERK, which can be monitored by Western blot analyses using antiphosphorylated AKT (p-AKT) and ERK (p-ERK) antibodies (AKT lies downstream of PI3K and is used as an indicator for PI3K activation). The cotransfection and cross-linking experiments described above were used to determine if zebrafish Dap12 utilizes the PI3K-ERK pathway. Coexpression and cross-linking of the Nitr9–Dap12 complex results in an increased level of p-AKT and p-ERK (compare lanes 8 and 9 in Fig. 3b and c). Mutated forms of zebrafish Dap12: Dap12D42V and Dap12Y96A were utilized to determine if the stimulation of this activating pathway specifically relies on Dap12. Substitution of the transmembrane Asp→Val abrogates the capacity of Dap12 to associate with Nitr9L via an intratransmembranous electrostatic interaction whereas the Tyr→Ala abrogates the function of the immunoreceptor tyrosine-based activation motif within Dap12. Although Nitr9L can associate with Dap12D42V, it appears to be weaker (compare lanes 8 and 9 with 10 and 11 in Fig. 3a). Cross-linking of Nitr9L with Dap12D42V fails to result in the phosphorylation of ERK or AKT (compare lanes 8 and 9 with 10 and 11 in Fig. 3b and c). Similarly, Nitr9L associates with Dap12Y96A, but the cross-linking of this complex also fails to result in the phosphorylation of ERK or AKT (compare lanes 8 and 9 with 12 and 13 in Fig. 3b and c). Taken together, these results suggest that both PI3K and ERK are activated after cross-linking the Nitr9–Dap12 protein complex and indicate that Nitr9L and Dap12 not only physically associate but likely utilize a similar signaling pathway to initiate cytotoxic (e.g., NK) cell-equivalent activation in zebrafish.
It is interesting to note that when the transmembrane domains of activating NITRs are compared to the transmembrane domains of mammalian IgSF receptors, which are know to partner with DAP12, Nitr9L is the only DAP12-associated receptor to possess an Arg (rather than a Lys) within the transmembrane domain (Table 2): The biological significance of this observation remains to be determined.
Table 2.
Transmembrane domains of DAP12-associated, Ig-domain receptors
| Receptor | Transmembrane domain1 | Species shown |
|---|---|---|
| KIR2DS2 | Human | |
| NKp44 | Human | |
| CD300e | Human | |
| PILRβ | Human | |
| SIRPβ1 | Human | |
| TREM1 | Human | |
| TREM2 | Human | |
| Siglec-H | Mouse | |
| CD300d | Mouse | |
| Cd200r3 | Mouse | |
| Cd300R4 | Mouse | |
| TREM3 | Mouse | |
| Nitr9 | Zebrafish | |
| IpNITR22 | Channel catfish | |
| IpNITR32 | Channel catfish | |
| IpNITR42 | Channel catfish | |
| IpNITR10/112 | Channel catfish |
Transmembrane domains predicted by SMART software (http://smart.embl-heidelberg.de/; Letunic et al. 2006). Charged residues are white text on black.
Catfish IpNITR2, IpNITR3, IpNITR4, IpNITR10, and IpNITR11 encode transmembrane domains similar to zebrafish Nitr9 (Hawke et al. 2001) and are predicted to associate with DAP12.
Concluding remarks
These data demonstrate that nitr9, dap12, and dap10 are expressed by zebrafish lymphocytes and strongly suggest that Nitr9L partners with Dap12 in vivo in zebrafish lymphocytes. In addition, Nitr9S and the newly identified Nitr9 variant, Nitr9SS, potentially partner with Dap12 as they all possess an identical transmembrane domain. Based on its overall structure, patterns of diversification, and other data presented here, it is reasonable to hypothesize that Nitr9 functions in zebrafish NK cells as well as cytotoxic T cells. In catfish, multiple NITRs are expressed in clonal lines of NK-like as well as cytotoxic T and macrophage cells (Evenhuis et al. 2007; Hawke et al. 2001). A single inhibitory NITR (poNITR1), cloned from Japanese flounder, was found recently to be expressed in T cells (TCRα+) as well as B cells (IgM+; Piyaviriyakul et al. 2007); however, transcripts of only one catfish NITR gene have been detected in clonal B cell lines (Evenhuis et al. 2007). In humans, KIR receptors are expressed in NK and T cell lineages, whereas other genes encoded in the leukocyte receptor complex (e.g., LILR/LIR/ILT) are expressed in various hematopoietic lineages (Nakajima et al. 2003; Xu et al. 2005). Ongoing efforts are directed at further characterizing the hematopoietic cell types (in vivo) that express and/or rely on the function of NITRs as well as Dap12 and Dap10. It is possible that NITRs as well NK receptors exhibit differential expression and effect alternative functions in different cell lineages (Valiante et al. 1997).
We propose a model incorporating previous and current data on the function of Nitr9L that parallels what is known about NK receptor-DAP12 signaling in mammals (Fig. 4). The major points of this model are that (1) Nitr9L preferentially partners with Dap12, which is enhanced by receptor cross-linking, (2) cross-linking the Nitr9-Dap12 complex results in activating the PI3K→ERK pathway, and that (3) cross-linking of Nitr9, within the context of an NK cell, can induce cytotoxicity (Yoder et al. 2004). NK function likely has a long phylogenetic history. For these various reasons, we have predicted that NITRs may effect this type of function. As the genomes of bony fish are being solved, it is becoming increasingly more likely that NITRs could function in an NK capacity, which is supported by the observations made here on adaptor associations and signaling.
Fig. 4.
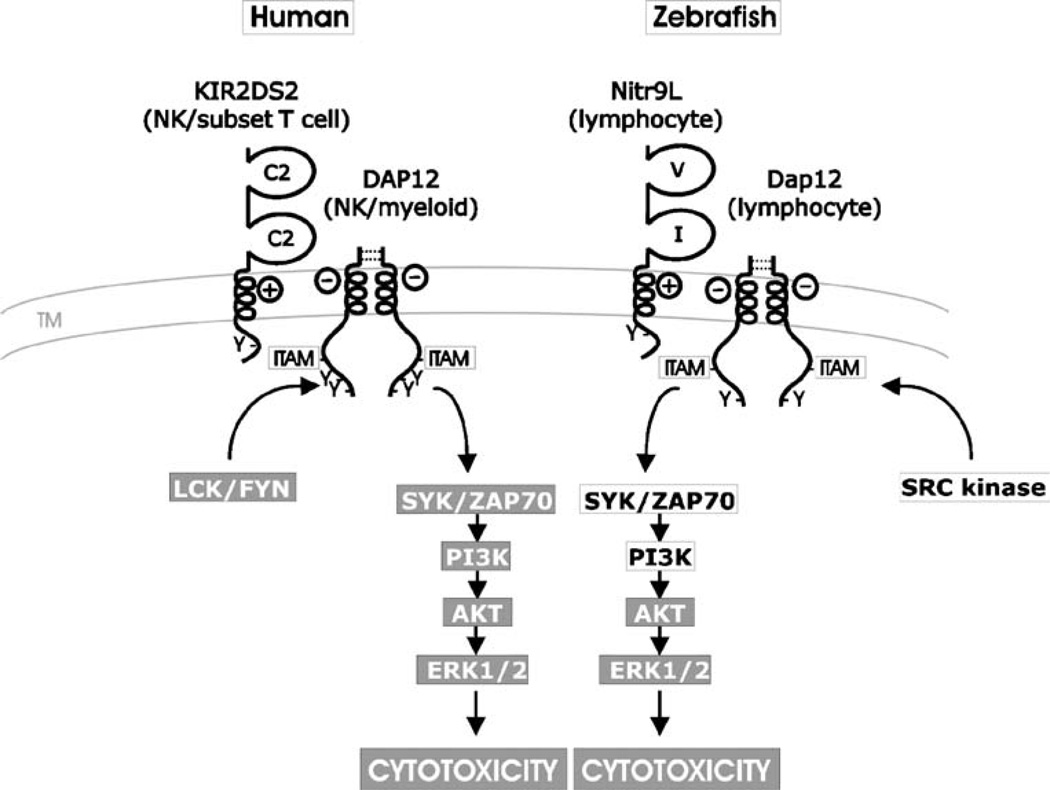
Model for Nitr9L–Dap12 activation of cytotoxic cells. The NK cell-activating pathway in humans is shown in the context of ligand recognition by the activating NK receptor KIR2DS2. DAP12 forms a dimer via two pairs of extracellular cysteines (dotted lines) and associates via complementary charges in the transmembrane domain of the activating NK receptor. Upon cross-linking, LCK/FYN phosphorylates the ITAM within the cytoplasmic tail of DAP12 that initiates the PI3K→ERK signal transduction pathway leading to cytokine release and cytotoxicity. A model for Nitr9L–Dap12 activation of cytotoxic cells parallels this process. Zebrafish Dap12 possesses the requisite cysteines for dimerization as well as the potential to interact with Nitr9L via complementary charges in transmembrane domains (Yoder et al. 2004; Yoder et al. 2007). In this study, it is shown that Nitr9L and Dap12 can physically associate and that cross-linking Nitr9L results in increased phosphorylation of AKT and ERK (Fig. 3). Components of the signaling pathway, which have been shown to be downstream of cross-linking Nitr9L/Dap12, are indicated (gray background). Cytoplasmic signaling components predicted to be associated with Nitr9L/Dap12 signaling are listed in black text
Acknowledgments
We thank P.K. Epling-Burnette for helpful discussions and Barb Pryor for editorial assistance. Sequence data (nitr9 super short isoform) from this article have been deposited with the GenBank database under accession number EF690658. This research was supported by grants awarded by NIH R01 AI057559 (GWL) and R01 AI056213 (SW), NSF MCB-0505585 (JAY), and the All Children’s Hospital Foundation (GWL).
Contributor Information
Sheng Wei, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA.
Jun-min Zhou, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA.
Xinghong Chen, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA.
Radhika N. Shah, Department of Molecular Biomedical Sciences and Center for Comparative Medicine and Translational Research, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA Immunology Program, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA.
Jinhong Liu, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA.
Timothy M. Orcutt, Department of Molecular Biomedical Sciences and Center for Comparative Medicine and Translational Research, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA
David Traver, Division of Biological Sciences, Section of Cell and Developmental Biology, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Julie Y. Djeu, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA
Gary W. Litman, Immunology Program, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Avenue, Tampa, FL 33612, USA Department of Molecular Genetics, All Children’s Hospital, 801 Sixth Street South, St. Petersburg, FL 33701, USA; Department of Pediatrics, University of South Florida College of Medicine, University of South Florida/All Children’s Hospital Children’s Research Institute, 830 First Street South, St. Petersburg, FL 33701, USA.
Jeffrey A. Yoder, Email: Jeff_Yoder@ncsu.edu, Department of Molecular Biomedical Sciences and Center for Comparative Medicine and Translational Research, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA; Immunology Program, College of Veterinary Medicine, North Carolina State University, 4700 Hillsborough Street, Raleigh, NC 27606, USA.
References
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Billadeau DD, Leibson PJ. ITAMs versus ITIMs: striking a balance during cell regulation. J Clin Invest. 2002;109:161–168. doi: 10.1172/JCI14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Cella M, Carretero M, Lopez-Botet M, Colonna M. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev. 2000;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- Djeu JY, Jiang K, Wei S. A view to a kill: signals triggering cytotoxicity. Clin Cancer Res. 2002;8:636–640. [PubMed] [Google Scholar]
- Evenhuis J, Bengten E, Snell C, Quiniou SM, Miller NW, Wilson M. Characterization of additional novel immune type receptors in channel catfish, Ictalurus punctatus. Immunogenetics. 2007;59:661–671. doi: 10.1007/s00251-007-0230-x. [DOI] [PubMed] [Google Scholar]
- Hao L, Nei M. Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC Class I genes. Gene. 2005;347:149–159. doi: 10.1016/j.gene.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Hawke NA, Yoder JA, Haire RN, Mueller MG, Litman RT, Miracle AL, Stuge T, Shen L, Miller N, Litman GW. Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci USA. 2001;98:13832–13837. doi: 10.1073/pnas.231418598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyka-Nouspikel N, Phillips JH. Physiological roles of murine DAP10 adapter protein in tumor immunity and autoimmunity. Immunol Rev. 2006;214:106–117. doi: 10.1111/j.1600-065X.2006.00456.x. [DOI] [PubMed] [Google Scholar]
- Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clement O, Chaume D, Lefranc G. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2005;33:D593–D597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Hawke NA, Yoder JA. Novel immune-type receptor genes. Immunol Rev. 2001;181:250–259. doi: 10.1034/j.1600-065x.2001.1810121.x. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Asai A, Okada A, Ping L, Hamajima F, Sata T, Isobe K. Transcriptional regulation of ILT family receptors. J Immunol. 2003;171:6611–6620. doi: 10.4049/jimmunol.171.12.6611. [DOI] [PubMed] [Google Scholar]
- Ortaldo JR, Young HA. Mouse Ly49 NK receptors: balancing activation and inhibition. Mol Immunol. 2005;42:445–450. doi: 10.1016/j.molimm.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Piyaviriyakul P, Kondo H, Hirono I, Aoki T. A novel immunetype receptor of Japanese flounder (Paralichthys olivaceus) is expressed in both T and B lymphocytes. Fish Shellfish Immunol. 2007;22:467–476. doi: 10.1016/j.fsi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, Rast JP, Amemiya CT, Litman GW. A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci USA. 1999;96:15080–15085. doi: 10.1073/pnas.96.26.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki R, Watson SR, Lanier LL. DAP12: an adapter protein with dual functionality. Immunol Rev. 2006;214:118–129. doi: 10.1111/j.1600-065X.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- Xu J, Vallejo AN, Jiang Y, Weyand CM, Goronzy JJ. Distinct transcriptional control mechanisms of killer immunoglobulin-like receptors in natural killer (NK) and in T cells. J Biol Chem. 2005;280:24277–24285. doi: 10.1074/jbc.M500727200. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc Natl Acad Sci USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Nichols KM, Ristow SS, Thorgaard GH, Ota T, Litman GW. Cloning novel immune-type inhibitory receptors from the rainbow trout, Oncorhynchus mykiss. Immunogenetics. 2002;54:662–670. doi: 10.1007/s00251-002-0511-3. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Ota T, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW. Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci USA. 2004;101:15706–15711. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–164. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


