Abstract
Glutamate acting on N-methyl-D-aspartate (NMDA) receptors plays an important role in neurodegenerative diseases and neuronal injury following stroke, through activation of poly(ADP-ribose) polymerase-1 and generation of the death molecule poly(ADP-ribose) (PAR) polymer. Here we identify Iduna, a novel NMDA receptor-induced survival gene that is neuroprotective against glutamate NMDA receptor mediated excitotoxicity both in vitro and in vivo and against stroke through interfering with PAR polymer induced cell death (parthanatos). Iduna’s protective effects are independent and downstream of PARP-1 activity. Iduna is a PAR polymer binding protein and mutations at the PAR polymer binding site abolishes the PAR binding activity of Iduna and attenuates its protective actions. Iduna is protective in vivo against NMDA-induced excitotoxicity and middle cerebral artery occlusion (MCAO)-induced stroke in mice. These results define Iduna as the first endogenous inhibitor of parthanatos. Interfering with PAR polymer signaling offers a new therapeutic strategy for the treatment of neurologic disorders.
INTRODUCTION
Glutamate is the major excitatory neurotransmitter regulating normal physiologic activity in the brain. Excessive glutamate release leads to excitotoxicity, which plays a prominent role in many disorders of the nervous system including trauma and ischemic brain injury 1 and dysfunctional glutamate neurotransmission contributes to seizures and neurodegenerative disorders 2. Glutamate excitotoxicity is mediated largely through in flux of calcium through the NMDA receptor leading to activation of PARP-1 and generation of PAR polymer, a newly described death signal that kills cells through apoptosis inducing factor (AIF). Genetic deletion of PARP-1 or drug inhibition result in profound neuroprotection. This form of cell death has recently been designated parthanatos to distinguish it from apoptosis, autophagy and necrosis 3,4. Parthanatos is prominently implicated in models of diabetes, inflammation, MPTP toxicity, myocardial infarction and cerebral ischemia 5.
Under physiologic conditions normal bursts of excitatory activity result in synaptic transmission and the expression of molecular substrates of long-term plasticity, growth and survival 6. The activation of NMDA receptors in glutamatergic neurons plays a prominent role in inducing these long-lasting synaptic changes through multiple downstream signaling molecules and changes in gene expression 7–10. NMDA receptor stimulation may also be important for long-term changes that lead to neuronal survival 11. We report here the identification and characterization of Iduna [MGI: 1915281 (RNF 146)], a novel NMDA-induced cell survival molecule that protects against NMDA excitotoxicity and stroke through binding poly (ADP-ribose) (PAR) polymer and blocking parthanatos.
RESULTS
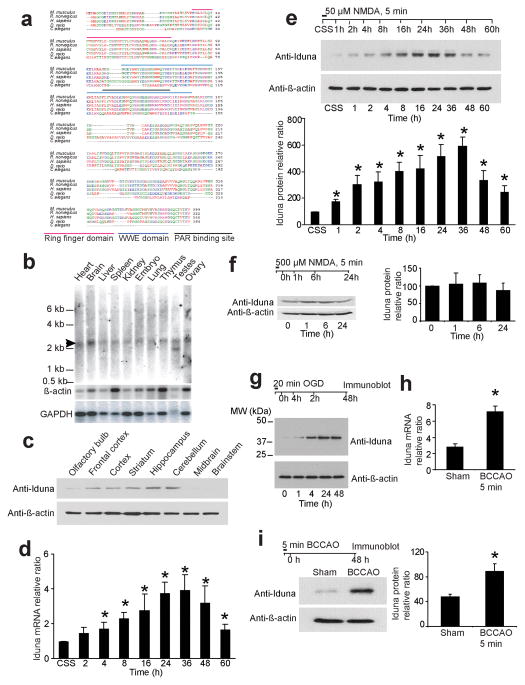
NMDA-induced plasticity late response genes (PLINGS) were identified from cortical neurons by differential analysis of primary cDNA library expression (DAzLE) 12. Of the many genes identified, here we report the characterization of clone 932, named Iduna for the Norse goddess of protection and eternal youth. Iduna encodes for a protein of 359 amino acids with a predicted molecular weight of 39.8 kDa (Fig. 1a). There is a high degree of homology with the human, rat and mouse Iduna proteins. The evolutionary conserved regions with zebra fish and nematode are associated with two domains of Iduna, the Really Interesting New Gene (RING) finger (RF) domain (aa 35–77) and the WWE domain (aa 91–167) (Fig. 1a).
Figure 1.
Iduna is an NMDA induced neuroprotective protein. (a) Amino acid sequences of Iduna proteins are conserved among different species. (b) Northern analysis of Iduna (arrow) in mouse tissue. β-actin and GAPDH are loading controls. (c) Immunoblot analysis of Iduna protein expression in different regions of brain. β-actin is a loading control. Data were repeated with similar results. (d) Induction of Iduna mRNA by 50 μM NMDA detected by RT-PCR in primary neuronal cultures over time. Data are the mean ± SEM from two experiments. (e) Immunoblot of Iduna induced by 50 μM NMDA (upper panel). These data were normalized to β-actin and quantified by optical density (bottom panel). Data are the mean ± SEM from three experiments. (f) Immunoblots of Iduna expression following an excitotoxic 500 μM dose of NMDA in primary cortical neurons. The data were normalized to β-actin and quantified by optical density (side panel). Data are the mean ± SEM from two experiments. (g) Immunoblot of Iduna expression over time following induction by 20 min of oxygen glucose deprivation (OGD) in primary cortical cultures. Experiments were repeated three times. (h) Iduna mRNA expression in mouse forebrain detected by RT-PCR 48 hr after reperfusion following 5 min bilateral carotid artery occlusion (BCCAO). Data are the mean ± SEM, n=4. (i) Immunoblot of Iduna expression in forebrain 48 hr after reperfusion following 5 min BCCAO. Data were normalized to β-actin and quantified by optical density (right panel). Data represents mean ± SEM, n=4. Experimental schedule is indicated above panels and treatment conditions are indicated by horizontal bars. Significance determined by by ANOVA with Tukey-Kramer’s posthoc test.
By Northern blot analysis Iduna is expressed at relatively high levels in brain, but is also present in spleen, heart, kidney, testis and liver (Fig. 1b), with two Iduna transcripts in the testis. A polyclonal antibody to Iduna, which recognizes a single 40 kDa protein on immunoblot (Supplementary Figure 1a, b), reveals variable expression of Iduna protein in different brain regions, suggesting a regional diversity in Iduna activity (Fig. 1c).
Iduna mRNA as assessed by real time PCR increases following 50 μM NMDA (Fig. 1d), consistent with our microarray screen 12. Iduna protein expression also increases following 50 μM NMDA (Fig. 1e). Both Iduna mRNA and protein follow a similar pattern of expression, peaking at 36 h after NMDA stimulation (Fig. 1d, e). A toxic dose of NMDA (500 μM for 5 min) fails to induce Iduna expression (Fig. 1f). Sublethal exposure to oxygen-glucose deprivation (OGD) which induces tolerance to subsequent lethal insults 13 also induces Iduna (Fig. 1g). A 5 minute bilateral common carotid artery occlusion (BCCAO) in mice results in resistance to subsequent ischemic injury 14 and induces Iduna mRNA and protein (Fig. 1h, i).
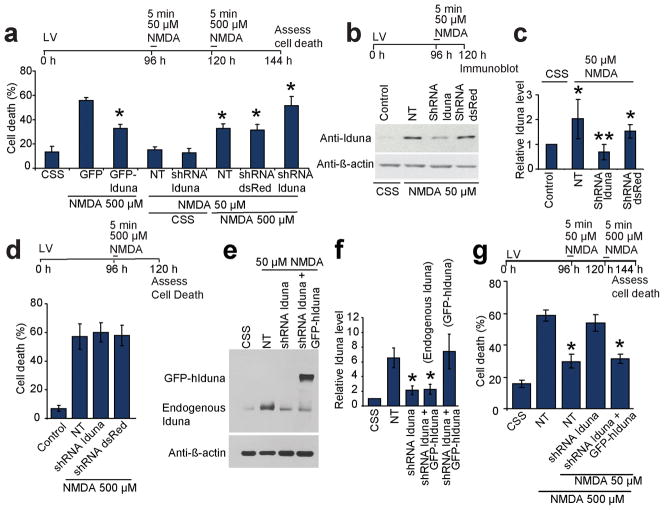
Lentiviral transduction of EGFP-tagged Iduna (GFP-Iduna) in cortical neurons protects against NMDA-induced cell death to a similar degree as the protection afforded by 50 μM NMDA (Fig. 2a). Knockdown of Iduna induction by lentiviral shRNA following treatment with 50 μM NMDA (Fig. 2b, c) abolishes the NMDA-induced protection (Fig. 2a). The control lentiviral shRNA DsRed has no significant effect on NMDA-induced upregulation of Iduna (Fig. 2b, c), NMDA-induced cell survival (Fig. 2a) or cell death (Fig. 2d). Knockdown of Iduna has no effect on cell viability following a toxic 500 μM dose of NMDA (Fig. 2d). The Iduna antibody is equally sensitive to human and mouse Iduna detection (Supplementary Figure 1b). Within the DNA sequence of Iduna between base pairs 556–576 there are five differences between the mouse and human sequence, which is sufficient to render human Iduna resistant to knockdown with the shRNA targeted towards mouse Iduna and thus provide a positive control (Fig. 2e, f). Overexpression of human Iduna is protective in the setting of knockdown of induced mouse Iduna (Fig. 2g), confirming the specificity of the shRNA knockdown of mouse Iduna. These results taken together indicate that Iduna is an NMDA-induced protective protein.
Figure 2.
Iduna is neuroprotective. (a) Primary cortical neurons expressing GFP or Iduna-GFP were exposed to excitotoxic NMDA (500 μM, 5 min). Sister cultures expressing shRNA to Iduna (shRNA-Iduna) or dsRed (shRNA dsRed) were exposed to 50 μM NMDA and then 500 μM NMDA. Abbreviation: NT, not transduced. Data represent mean ± SEM, n = 5 from two experiments; *p < 0.05. (b) Immunoblot of Iduna expression in cortical cultures expressing shRNA Iduna or shRNA dsRed exposed to 50 μM NMDA. (c) Quantification of the data in (b) normalized to β-actin. Data are the mean ± SEM from three experiments, *p < 0.05 vs control, **p < 0.05 vs. NT. (d) Primary cortical cultures expressing shRNA Iduna or shRNA dsRed were challenged with 500 μM NMDA. Data are the mean ± SEM of at least two experiments. (e) Immunoblot of Iduna expression in primary cortical cultures exposed to 50 μM NMDA in the presence of shRNA to mouse Iduna and expression of human Iduna (hIduna). (f) Quantification of (e) normalized to β-actin. Data represent mean ± SEM, n = 4; *p < 0.05. (g) Primary cortical cultures expressing lentivirus shIduna ± human Iduna (hIduna), which is resistant to mouse Iduna shRNA were exposed to NMDA as indicated. Data represent mean ± SEM, n = 4 from 4 experiments; *p < 0.05. Experimental schedule is indicated above panels and treatment conditions are indicated by horizontal bars. Significance determined by by ANOVA with Tukey-Kramer’s posthoc test.
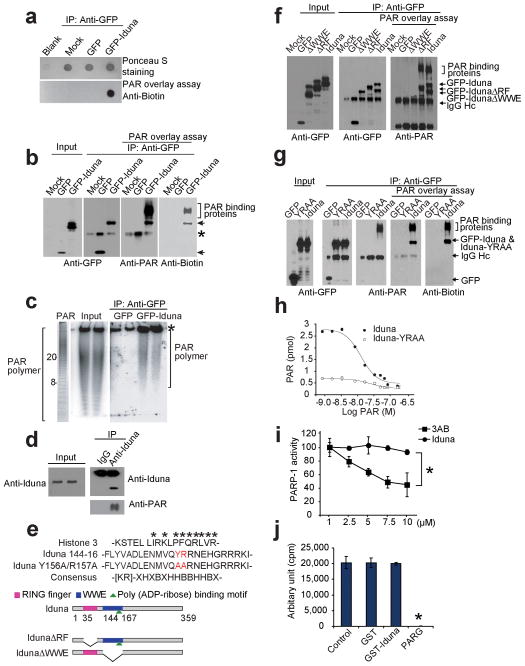
Iduna contains a RING finger domain and a WWE domain. Within the WWE domain there is a putative PAR binding motif (Fig. 1a). Since PAR is a newly discovered death signal 15, the ability of Iduna to bind PAR polymer was determined. Dot blots of immunoprecipitated GFP-Iduna incubated with biotin-labeled PAR polymer and probed with an anti-biotin antibody show GFP-Iduna binds to PAR polymer, whereas GFP alone fails to bind (Fig. 3a). PAR polymer overlay assays with immunoprecipitated GFP-Iduna reveals PAR polymer specifically binds to GFP-Iduna, but it fails to bind to GFP (Fig. 3b). In addition, poly(ADP-ribosyl)ated proteins co-immunoprecipiate with GFP-Iduna (Fig. 3b). In a PAR polymer binding assay GFP-Iduna or histone 3 (H3) (a positive control) bind radiolabeled free PAR polymer but GFP does not (Supplementary Figure 2a). Iduna binds to a range of PAR polymers of varying length as determined by phosphorimager detection of radiolabeled PAR polymer bound to GFP-Iduna following separation by Tris-borate-EDTA PAGE (Fig. 3c). In primary neuronal cultures treated with 50 μM NMDA, immunoblot analysis shows that Iduna co-immunoprecipitates with PAR polymer (Fig. 3d). Since in resting neurons Iduna is expressed at low levels and there is relatively little PAR polymer, there is no detectable interaction between Iduna and PAR polymer under resting conditions. Taken together these results indicate that Iduna is a PAR polymer binding protein.
Figure 3.
PAR binding activity of Iduna. (a) Dot blot of immunoprecipitated GFP-Iduna and GFP with biotin-labeled PAR polymer and detected with anti-biotin antibody. Data were reproduced with similar results. (b) Far western analysis of Iduna PAR binding activity. Arrows indicate GFP-Iduna fusion protein and bracket indicates PAR binding proteins. Asterisk indicates a IgG heavy chain signal recognized by polyclonal antibodies including the GFP or PAR antibodies. Data were reproduced with similar results. (c) [32P]-PAR polymer bound to GFP-Iduna or GFP analyzed in Trisborate-EDTA PAGE. Values represent ADP-ribose units in the PAR polymer. Asterisk indicates non-specific PAR polymer binding. (d) Immunoblot of endogenous Iduna and PAR from cortical neurons treated with 50 μM NMDA. These experiments were repeated at least two times with similar results. (e) Alignment of the PAR binding motif in Iduna and Histone 3. The Iduna Y156A/R157A PAR binding mutant (red) is indicated. Schematic of Iduna functional domains and deletion mutants. Ring finger (RF) domain (AA 35–77) [pink bar], WWE domain (AA 91–167) [blue bar] and the PAR binding domain (144–167) [green triangle] are highlighted. Full-length Iduna, and RF (IdunaΔRF) and WWE (IdunaΔWWE) domain deletion mutants of Iduna are shown. (f) Far western analysis of PAR binding activity of Iduna and Iduna deletion mutants. Arrows indicate GFP-Iduna fusion proteins and bracket indicates PAR binding proteins. IgG heavy chain (IgG Hc) is indicated by arrow. n=2 (g) Far western analysis of PAR binding activity of Iduna and Iduna-YRRA mutant. Arrows indicate GFP-Iduna fusion proteins and bracket indicates PAR binding proteins. (h) Analysis of PAR binding properties of wild type Iduna (●) and Iduna-YRAA mutant (○). (i) Chemiluminescent activity of PARP-1 in the presence of Iduna or the PARP-1 inhibitor 3-aminobenzamide (3-AB). Data represent two separate experiments. *p < 0.05 (j) Quantification of [32P]-PAR polymers synthesized by PARP-1 in the presence of GST, GST-Iduna or PARG (which catalytically degrades PAR), respectively. Data represent mean ± SEM, n=3 *p < 0.05 by ANOVA with Tukey-Kramer’s posthoc test.
PAR binding is specified by a sequence of approximately 20 amino acids containing N-terminal basic amino acids and a C-terminal region containing alternating hydrophobic and basic amino acids (Fig. 3e) 16,17. Iduna contains a predicted PAR polymer binding sequence within amino acids 144–167 of the WWE domain (Fig. 3e, Fig. 1a). The PAR-binding domain was defined by comparing to the consensus sequence for PAR-binding and the known PAR binding domain of Histone 3 (Fig. 3e). HEK293 cells were transfected with GFP, GFP-Iduna lacking the WWE domain (GFP-IdunaΔWWE), GFP-Iduna lacking the RF domain (GFP-IdunaΔRF), and full-length GFP-Iduna, followed by immunoprecipitation with a GFP antibody. The PAR polymer overlay assay of the immunoprecipitate shows PAR polymer binds to GFP-IdunaΔRF and full-length GFP-Iduna and co-immunoprecipitated PAR binding proteins, but it fails to bind to GFP and GFP-IdunaΔWWE (Fig. 3f). Only GFP-Iduna-ΔRF and full-length GFP-Iduna can co-immunoprecipiate PAR binding proteins (Fig. 3f). The critical amino acids in the PAR binding domain, 156Y and 157R, were mutated to 156A and 157A in full length Iduna (Iduna-YRAA). A PAR polymer overlay assay reveals that PAR polymer binds to GFP-Iduna and co-immunoprecipitated PAR binding proteins, but it fails to bind to GFP-Iduna-YRAA and PAR binding proteins fail to co-immunoprecipitate (Fig. 3g). Similar results are obtained with a biotin-tagged PAR polymer followed by detection with an anti-biotin antibody (Fig. 3g). A synthesized peptide fragment of the predicted PAR polymer binding sequence in Iduna between of amino acids 144–167 of the WWE domain (Iduna 144–167) binds to PAR polymer in a manner comparable to full-length Iduna (Supplementary Figure 2b). However, when the peptide fragment was synthesized with amino acids 156A and 157A to disrupt the PAR binding site, (Iduna 144–167 YRAA) it fails to bind PAR polymer (Supplementary Figure 2b). Histone H3 is a known PAR binding protein and was used as a positive control for the PAR overlay assay. Full length GST-Iduna binds to PAR polymer whereas GST alone or GST-Iduna-YRAA fails to bind PAR polymer in the PAR polymer overlay assay (Supplementary Figure 2b). An assay based on electrophorectic mobility shift (EMSA) for PAR binding was developed to monitor PAR polymer binding to Iduna. Iduna retards the mobility shift of PAR polymer whereas GST or GST-Iduna-YRAA has no effect of PAR polymer mobility shift. Histone 3, a positive control also retards the PAR polymer mobility shift (Supplementary Figure 2c).
The affinity of Iduna and Iduna-YRAA for PAR binding was determined by a competition assay with increasing concentrations of unlabeled PAR polymer against 2.5 nM [32P]-labeled PAR polymer (mean size of 40 ADP-ribose units) (Fig. 3h). From the competitive binding curve, the EC50 of wild type Iduna for PAR polymer is 14.5 ± 0.13 nM (p>0.001) calculated as a function of PAR polymer concentration, and the maximum binding capacity (Bmax) is 3.04 ± 0.16 pmol. The PAR polymer synthesized by in vitro automodification of PARP-1 has a mean length of 40 ADP-ribose residues 18 and accordingly the concentration of PAR is given as a function of polymer molecules with a mean size of 40 ADP-ribose units. These concentrations of PAR polymers are within the range of polymer concentrations found in intact cells during NMDA excitotoxicity and N-methyl-N-nitro-N-nitrosoguanidine (MNNG) toxicity 15,19. PAR polymer fails to bind to Iduna-YRAA (Fig. 3h). The homologous competitive binding curve for wild type Iduna analyzed by the Cheng-Prusoff equation provides a dissociation constant (Kd) for wild type Iduna of 12.0 nM. The observed Kd is 10 times less than the PAR polymer concentration found after NMDA-induced excitotoxicity in cortical neurons 15. These results taken together indicate that Iduna is a high affinity and saturable PAR polymer binding protein at its WWE domain, and that the basic and hydrophobic amino acids YR located at position 156 and 157 are critical for PAR polymer binding.
Since PARP-1 activation plays a prominent role in NMDA excitotoxicity 5,20 we tested whether PARP-1 activity is directly affected by Iduna. PARP-1 activity was assessed by incorporation of biotinylated PAR onto histone proteins. The PARP-1 inhibitor 3-aminobenzamide (3-AB) inhibits PARP-1 activity, but Iduna has no effect (Fig. 3i). Iduna also fails to inhibit PARP-1 catalytic activity as assessed by 32P-NAD incorporation into radiolabeled PAR polymer (Fig. 3j). Thus, Iduna does not inhibit PARP-1 catalytic activity, and is a PAR binding protein that acts downstream of PARP-1 activation.
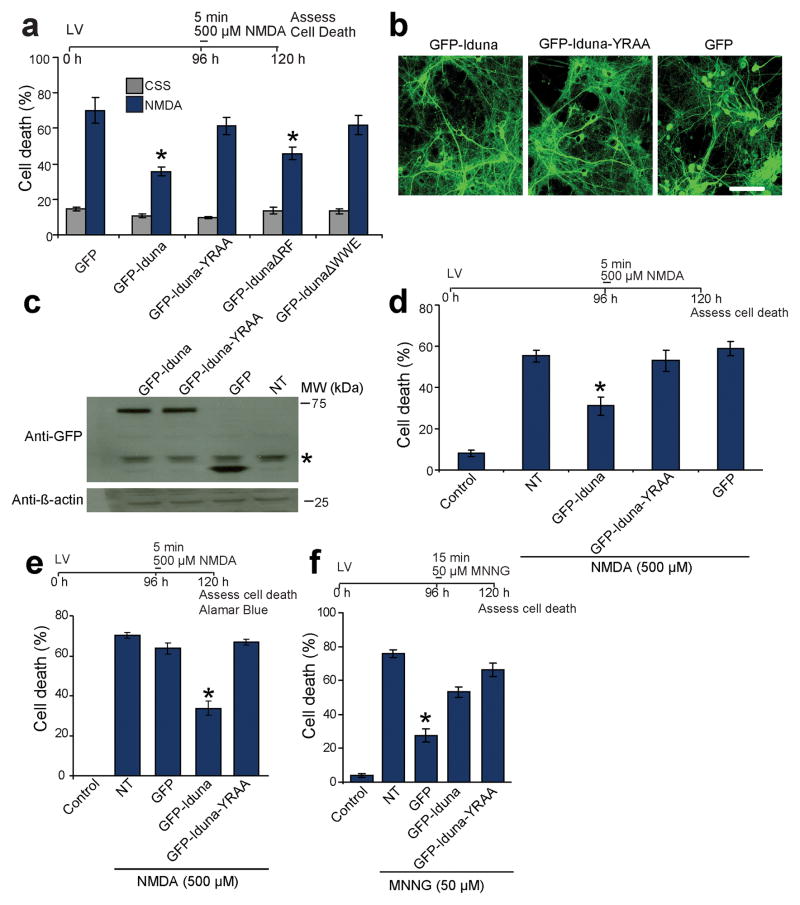
To ascertain whether the binding of Iduna to PAR polymer effects the neuroprotective actions of Iduna against NMDA excitotoxicity and parthanatos, neuronal cultures were transiently transfected with GFP-tagged-Iduna and GFP-tagged-Iduna mutants (Fig. 4a). Both GFP-Iduna and GFP-IdunaΔRF, which contain the PAR-binding domain, prevent NMDA excitotoxicity whereas GFP-IdunaΔWWE and GFP-Iduna-YRAA fail to protect against NMDA excitotoxicity (Fig. 4a). Since transient transfections are effective in only a small population of neurons, lentiviral expression of GFP, GFP-Iduna or GFP-Iduna-YRAA was used in neuronal culture with an efficiency greater than 95% (Fig. 4b and Supplemental Figure 3). Equivalent levels of GFP-Iduna and GFP-Iduna-YRAA protein are expressed (Fig. 4c) with primarily cytoplasmic localizations similar to endogenous Iduna as determined by confocal microscopy and subcellular fractionation (Fig. 4b and Supplementary Figure 4). A small amount of endogenous Iduna and lentiviral expressed GFP-Iduna and GFP-Iduna-YRAA seem to translocate to the nucleus after an excitotoxic dose of NMDA (500 μM) (Supplementary Figure 4). Thus, we cannot exclude the possibility of a contributory effect of Iduna’s translocation to the nucleus. However, since Iduna is a primarily a cytosolic protein, and the protective actions of Iduna do not require an interaction with PARP-1 (see Fig. 3i, j), it is likely the protective effects occur in the cytoplasm. Since PAR exits the nucleus to mediate its toxicity 15,21, the actions of Iduna in the cytoplasm may be important for the regulation of cell viability. Overexpression of GFP-Iduna protects neuronal cultures against NMDA excitotoxicity, whereas GFP-Iduna-YRAA or GFP fail to provide neuroprotection (Fig. 4d, and Fig. 2a). Similar results were obtained using Alamar Blue reduction to assess cell viability 22 (Fig. 4e). Iduna overexpression also protects neuronal cultures against a lower excitotoxic dose of NMDA (100 μM for 5 min) (cell death %: NT 44.2 ± 3.2, Iduna 23.0 ± 1.8). To determine whether Iduna protects against other forms of parthanatos, cortical neurons were exposed to the DNA alkylating agent and PARP-1 activator, MNNG under conditions where MNNG toxicity is PARP-1 dependent (MNNG, 50 μM for 15 min). Iduna protects neuronal cultures against MNNG-induced cell death whereas Iduna-YRAA or GFP fail to provide any protection (Fig. 4f). Expression of GFP-Iduna or GFP-Iduna-YRAA has no effect against apoptotic cell death in neuronal cultures treated with staurosporine (STS, 500 nM) or camptothecin (CPT 20 μM). Both STS and CPT induce caspase-dependent cell death as the pan-caspase inhibitor z-VAD fmk inhibited cell death in neuronal cultures (Supplementary Figure 5). In neuronal cultures treated with 100 μM or 500 μM H2O2. Iduna protects only against 100 μM H2O2, further indicating that Iduna-mediated protection is specific for PAR mediated cell death as the PARP inhibitor DPQ only protected against 100 μM H2O2 toxicity as well (Supplementary Figure 6).
Figure 4.
PAR-binding property of Iduna mediates neuroprotection. (a) Quantification of 500 μM NMDA induced cell death in primary cortical neurons transciently transfected to express GFP, GFP-Iduna, GFP-Iduna-YRAA, GFP-IdunaΔRF or GFP-IdunaΔWWE. Cells with fragmented processes were considered dead. Data represent mean ± SEM, n=6 from two independent experiments, *p < 0.05 by ANOVA with Tukey-Kramer’s posthoc test. (b) Representative photomicrographs of lentiviral expression of GFP, GFP-Iduna or GFP-Iduna-YRAA in primary cortical neurons. n=4, scale bar = 50 μm (c) Immunoblots of lentiviral expression of GFP, GFP-Iduna or GFP-Iduna-YRAA in primary cortical neurons. No signal is seen in control cultures (*) non-specific band. Data were repeated three times with similar results. (d) Quantification of 500 μM NMDA induced cell death in primary cortical neurons with lentiviral expression of GFP, GFP-Iduna or GFP-Iduna-YRAA. Control cultures were treated with control salt solution (CSS) alone. NT, non-transduced. n=12–20 from two experiments. *p < 0.05 (e) Quantification of cell death in cortical neurons treated in an identical manner to panel 4d but cell death was assessed via AlamarBlue® reduction assay. Data represents mean ± SEM, n=5, *p < 0.05 by ANOVA with Tukey-Kramer’s posthoc test. (f) Quantification of cell death due to DNA damage by MNNG in primary neuronal cultures expressing GFP, GFP-Iduna or GFP-Iduna YRAA. Data represent mean ± SEM, n = 5 of two experiments. *p ≤ 0.05 by ANOVA with Tukey-Kramer’s posthoc test.
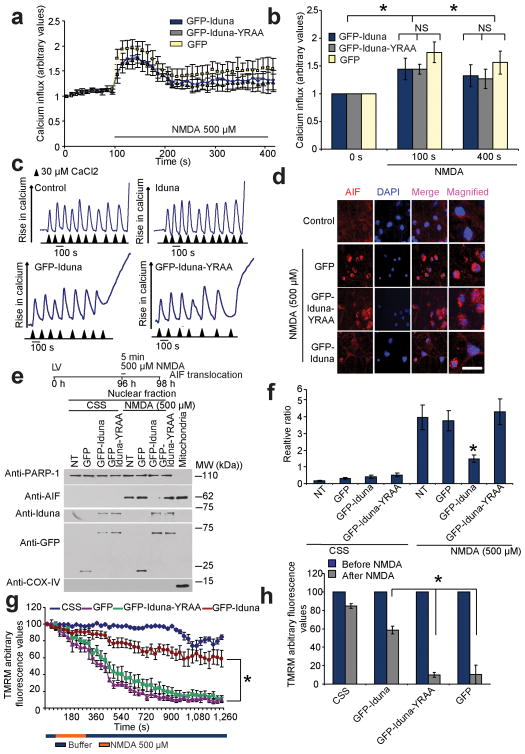
Iduna does not interfere with NMDA-induced intercellular calcium influx determined in neuronal cultures loaded with fluo-5F (Invitrogen). Live-cell calcium imaging captured using a confocal microscope (LSM-710, Carl Zeiss) observed following a 5 min application of NMDA is similar in neurons expressing Iduna, Iduna-YRAA or GFP (Fig. 5a, b, Supplementary Figure 7). Iduna overexpression does not interfere with mitochondrial calcium uptake (Fig. 5c). These results taken together indicate that the neuroprotection elicited by Iduna against NMDA excitotoxcity is not due to interference with NMDA-induced elevations of calcium.
Figure 5.
Iduna does not interfere with NMDA-induced changes in Ca2+ or mitochondrial Ca2+ loading, but prevents AIF translocation and reductions in mitochondrial membrane potential (Δψm). (a) Ca2+ influx imaged in primary cortical neurons expressing GFP, GFP-Iduna or GFP-Iduna-YRAA assessed by the Ca2+-sensitive fluorochrome fluo-5F (2.0 μM) over time. Intensity gain in these neurons was reduced to avoid saturation effects because of the spectral overlap between GFP and fluo-5F. (b) Graphic representation of Ca2+ influx before and after 500 μM NMDA. *p < 0.05 by ANOVA with Tukey-Kramer’s posthoc test. (c) Assessment of mitochondrial Ca2+ uptake in isolated mitochondria incubated with or without recombinant Iduna protein (top panels) or digitonin permeabilized MCF7 cells expressing GFP-Iduna or GFP-Iduna-YRAA (bottom panels), using Calcium green-5N as an indicator of free Ca2+. Experiments were repeated twice with similar results (d) Representative confocal photomicrographs of NMDA-induced AIF translocation in cortical neurons expressing GFP, GFP-Iduna or GFP-Iduna-YRAA. AIF immunoreactivity (red), DAPI (blue) scale bar = 20 μm (e) Immunoblot analysis of subcellular fractionations from cortical cultures treated as indicated in (d) for AIF. PARP-1, nuclear fraction, COX IV post-nuclear mitochondrial fraction. Data were repeated three times with similar results. (f) Quantification of AIF immunoblot analysis in (e). Data are the mean ± SEM from three experiments, *p < 0.05 vs NT after NMDA treatment by ANOVA with Tukey-Kramer’s posthoc test. (g) Analysis of Δψm, using TMRM live imaging in primary cortical neurons expressing GFP, GFP-Iduna or GFP-Iduna-YRAA. Neurons were treated with either 500 μM NMDA or CSS, *p < 0.05 (h) Graph shows loss of Δψm (TMRM fluorescence) before and after 20 min of NMDA application. *p < 0.05. Significance determined by ANOVA with Tukey-Kramer’s posthoc test.
Parthanatos involves the translocation of apoptosis inducing factor (AIF) from the mitochondria to the nucleus following NMDA excitotoxicity 5,15,19,21. AIF translocation following excitotoxic NMDA treatment was monitored by immunohistochemistry and confocal microscopy (Fig. 5d) and by immunoblot analysis of nuclear and mitochondrial subcellular fractions (Fig. 5e). AIF translocates to the nucleus in GFP and Iduna-YRAA transduced neurons following NMDA excitotoxicity, whereas Iduna reduces the translocation of AIF (Fig. 5f) comparable to the degree of neuroprotection afforded by Iduna overexpression. During parthanatos, cytochrome c is released from mitochondria long after AIF translocates to the nucleus, after 1–2 hours 19,20. Consistent with the protective effects of Iduna against parthanatos, we observe a reduction of cytochrome c translocation from mitochondria to the cytoplasm with Iduna overexpression compared to Iduna-YRAA or GFP (Supplementary Figure 8).
Mitochondrial membrane potential (Δψm) reduction accompanies the translocation of AIF during NMDA excitotoxicity 20. Overexpression of Iduna prevents NMDA induced loss of Δψm as monitored by TMRM fluorescence compared to Iduna-YRAA or GFP (Fig. 5g, h, Supplementary Figure 9) similar to the reduction in AIF nuclear translocation and cell death. Taken together these results indicate that Iduna prevents AIF translocation and reductions in Δψm in a PAR binding dependent manner.
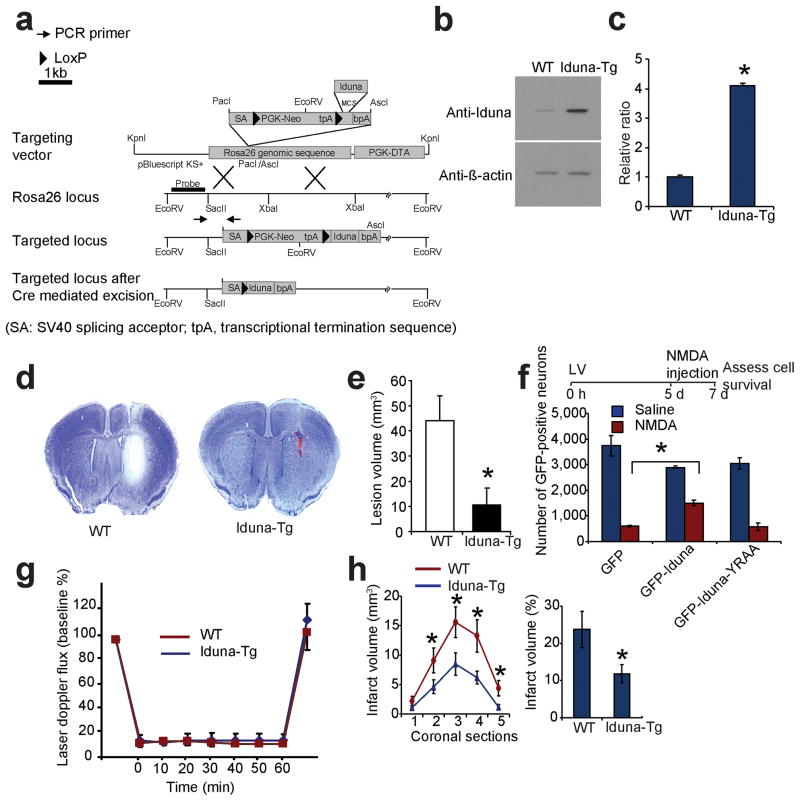
To determine whether Iduna is protective in vivo, transgenic mice overexpressing Iduna were generated by knocking in Iduna into the ROSA26 genomic locus resulting in a four-fold expression over wild type littermate mice (Fig. 6a–c). NMDA induced lesions are reduced by approximately 80% in the Iduna transgenic mice compared to littermate wild type control animals following an intrastriatal injection of NMDA (20 nmoles) (Fig. 6d, e).
Figure 6.
Iduna is neuroprotective in vivo. (a) Targeting strategy for ROSA26-Iduna conditional transgenic (Tg) mouse. (b) Immunoblot of Iduna expression in wild type (WT) and Iduna-Tg mice. (c) Quantification of (b). Data represent mean ± SEM, n = 6; *p < 0.05 by Student’s t-test. (d) Representative coronal sections stained with nissl to reveal lesions in control (left) and Iduna-Tg (right) mice 48 h after intrastriatal injections with NMDA (20 nmoles). (e) Quantification of the lesion-volume. Data represent mean ± SEM, n = 4; *p < 0.05 by Student’s t-test. (f) Stereological counts of GFP-positive neurons from mouse brain injected with GFP, GFP-Iduna or GFP-Iduna-YRAA lentivirus followed by NMDA (20 nmoles) or normal saline. Quantification of GFP-positive surviving neurons. Data represent mean ± SEM, n=4 from two experiments; *p < 0.05 by ANOVA with Tukey-Kramer’s posthoc test. (g) Laser-Doppler flux measured over the lateral parietal cortex in the core of the ischemic region in WT (n=10) and Iduna-Tg (n=11) mice. Values are mean ± SEM, expressed as a percent of the pre-ischemic baseline values. (h) Brain infarct volume after 60 min of middle cerebral artery occlusion in WT (n=10) and Iduna Tg (n=11) mice. Left panel, *two-way analysis of variance indicated a significant overall effect of genotype among the five coronal levels (level 1 is most anterior), and the Holm-Sidak multiple comparison procedure indicated significant differences at coronal levels 3 and 4 where infarct volume was greatest. Mean ± S.E.M. Right panel, total infarct volume expressed as a percent of the entire ischemic hemisphere. * p < 0.05 from WT Student’s t-test. The time course of the various experiments are indicated at top of the panels.
Mice were injected stereotactically with GFP-Iduna, GFP-Iduna-YRAA or GFP lentiviruses, followed by intrastriatal injection of NMDA (20 nmoles) 5 days after viral injection. Cell survival was assessed by stereological cell counting of GFP-positive neurons in mouse brain sections 48 h after the NMDA injection. In GFP-injected animals, NMDA injection leads to a 90% loss of GFP positive cells whereas GFP-Iduna protects approximately 51% of the neurons against NMDA lesions (Fig. 6f), similar to protection observed in vitro and in the Iduna transgenic mice. GFP-Iduna-YRAA is not able to protect against NMDA excitotoxic injections (Fig. 6f and Supplementary Figure 10). These data indicate that either constitutive or acute overexpression of Iduna is neuroprotective in vivo. Moreover, Iduna mediated protection is dependent on PAR binding.
Neuroprotection against ischemic injury was determined by subjecting wild type littermates and Iduna transgenic mice to transient occlusion of the middle cerebral artery. Over the 60-min period of occlusion, cortical perfusion monitored by laser-Doppler flowmetry was reduced equivalently in wild type mice (10 ± 1% of baseline; ± SE) and Iduna transgenic mice (12 ± 2%). The reduction was stable throughout the occlusion period and recovered to pre-ischemic levels immediately upon removal of the filament in both groups (Fig. 6g). Despite the similar intensity of the ischemic insult, infarct volume was reduced by 50% in Iduna transgenic mice compared to their wild type counterparts (Fig. 6h). Moreover, the reduction in infarct size was not skewed to a particular coronal level (Fig. 6h). Likewise, Iduna-Tg mice showed improved neurological function following stroke. There was no baseline neurobehavioural differences in Iduna-Tg and WT mice (Supplementary Figure 11). Thus, Iduna overexpression protects against stroke induced neuronal injury.
DISCUSSION
We report the discovery of Iduna, a neuroprotective protein. Iduna protects against parthanatos, NMDA receptor mediated glutamate excitotoxicity both in vitro and in vivo, and ischemia due to middle cerebral artery occlusion. Iduna is normally expressed at low levels in the nervous system, but expression substantially increases following a low dose of NMDA, a sub-lethal exposure to OGD, or a brief exposure to BCCAO, all of which induce neuroprotection 13,14,23. Thus, Iduna plays a role in the protective response to NMDA and ischemia and the subsequent development of tolerance to lethal insults. Consistent with this notion is our observation that shRNA mediated knockdown of Iduna completely abolishes the protective effects of the neuroprotective dose of NMDA and overexpression of Iduna is neuroprotective. The PAR polymer binding activity of Iduna is essential for its neuroprotective function. Emerging evidence reveals that PAR polymer binds to a variety of proteins in a saturable and highly specific manner 24–29. A recent unbiased proteomic screen for PAR-binding proteins identified a number of proteins including AIF 16. Mutation of the PAR-binding domain in AIF prevents the translocation of AIF from the mitochondria and promotes cell survival 4. That Iduna blocks the translocation of AIF from the mitochondria to the nucleus is consistent with these observations. How PAR binding to Iduna regulates subsequent PAR dependent signaling events, including AIF release, is not yet known. However, the ability to interfere with PAR dependent signaling events positions Iduna as the first endogenous functional antagonist of PAR polymer death signaling.
Induction of neuroprotective proteins including Iduna is likely a result of multiple different signaling events. Low concentrations of NMDA (50 μM) or non-lethal OGD induces long-lasting neuroprotection that appears similar to that induced by dis-inhibition of GABAergic neurons by bicuculline administration 13,30–33, to activate calcium, nitric oxide and MEK dependent pathways 13,34, as well as, CREB dependent signaling 30–32. Different induction paradigms likely activate divergent cell survival pathways by preferential activation of synaptic and extrasynaptic receptors 11,35–37. Research into neuroprotective mechanisms has at its heart the goal of developing new therapeutic strategies to treat patients 23. Although induction strategies might have limited use for acute injuries such as stroke or trauma, they may be extremely useful in treating patients undergoing cardiac bypass surgery, neurosurgery or other surgical cohorts where ischemia is a risk. Patients with subarachnoid hemorrhage, transient ischemic attacks, soldiers at risk for blast injury or perhaps even patients suffering from chronic neurodegenerative diseases could also benefit from enhancing neuronal survival. It is intriguing that a cohort of diverse signaling events have been linked to neuroprotection 23, which suggests that in the brain there is a complex epistasis where multiple parallel pathways are capable of producing a neuroprotective phenotype. As technology for genome-wide detection of transcriptional targets and high throughput proteomics becomes more robust, we will be in a better position to address this very interesting and important question and understand how these divergent signals integrate to regulate neuronal survival.
In summary, Iduna represents a novel protein, which confers protection against parathanatos in a manner analogous to that in which Bcl-2 and IAPs prevent apoptosis. The mechanism by which Iduna protects against parthanatos unveils a previously unrecognized endogenous protective process that involves interference with PAR polymer mediated toxicity downstream of PARP-1 activation. Due to the prominent role of PARP-1 activation in many neurologic diseases and ischemia reperfusion injury in organs 5,8,38, therapies aimed at blocking PAR polymer induced cell death by activating Iduna or mimicking the effects of Iduna could represent novel therapeutic targets to prevent the toxic effects of PARP-1 activation and cell death.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH/NINDS NS039148, NS067525 NIDA DA000266, and NS051764 and the McKnight Endowment for the Neurosciences. SSA is an American Heart Research Postdoctoral Fellow. GGP was supported from a Canadian Institutes Health Research grant and holds a Research Chair in Proteomics. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
AUTHOR CONTRIBUTIONS
S.A.A., H.C.K., designed and performed in vitro and in vivo experiments.
Y-IL., Performed the RT-PCR and some WB for Figure 1.
J.Z. and R.C.K. conducted and analyzed the MCAO experiments.
Z.C., A.B., AB initially purified Iduna Lentiviral particles and performed some cell death assays. ZC supplied OGD samples for Figure 1g.
J-F.H. and G.G.P. Purified PAR antibodies and conducted in vitro PAR activity assays
T.M.D. and V.L.D. formulated the hypothesis, initiated and organized the study and wrote the manuscript. S.A.A., H.C.K., R.C.K., G.G.P., T.M.D. and V.L.D. contributed to finalized manuscript.
References
- 1.Aarts MM, Tymianski M. Molecular mechanisms underlying specificity of excitotoxic signaling in neurons. Curr Mol Med. 2004;4:137–147. doi: 10.2174/1566524043479202. [DOI] [PubMed] [Google Scholar]
- 2.Waxman EA, Lynch DR. N-methyl-D-aspartate Receptor Subtypes: Multiple Roles in Excitotoxicity and Neurological Disease. Neuroscientist. 2005;11:37–49. doi: 10.1177/1073858404269012. [DOI] [PubMed] [Google Scholar]
- 3.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu SW, Wang H, Dawson TM, Dawson VL. Poly(ADP-ribose) polymerase-1 and apoptosis inducing factor in neurotoxicity. Neurobiol Dis. 2003;14:303–317. doi: 10.1016/j.nbd.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 6.West AE, et al. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnegy ME. Ca2+/calmodulin signaling in NMDA-induced synaptic plasticity. Crit Rev Neurobiol. 2000;14:91–129. [PubMed] [Google Scholar]
- 8.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25:259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 10.Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 11.Hardingham GE, Bading H. The Yin and Yang of NMDA receptor signalling. Trends Neurosci. 2003;26:81–89. doi: 10.1016/S0166-2236(02)00040-1. [DOI] [PubMed] [Google Scholar]
- 12.Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM. Identification and analysis of plasticity-induced late-response genes. Proc Natl Acad Sci U S A. 2004;101:2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Zulueta M, et al. Requirement for nitric oxide activation of p21(ras)/extracellular regulated kinase in neuronal ischemic preconditioning. Proc Natl Acad Sci U S A. 2000;97:436–441. doi: 10.1073/pnas.97.1.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faraco G, et al. Brain ischemic preconditioning does not require PARP-1. Stroke. 2010;41:181–183. doi: 10.1161/STROKEAHA.109.567826. [DOI] [PubMed] [Google Scholar]
- 15.Andrabi SA, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagne JP, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, et al. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 21.Yu SW, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deb C, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009;4:e6077. doi: 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dirnagl U, Becker K, Meisel A. Preconditioning and tolerance against cerebral ischaemia: from experimental strategies to clinical use. Lancet Neurol. 2009;8:398–412. doi: 10.1016/S1474-4422(09)70054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 26.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 27.Gottschalk AJ, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 29.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 30.Hara T, et al. CREB is required for acquisition of ischemic tolerance in gerbil hippocampal CA1 region. J Neurochem. 2003;86:805–814. doi: 10.1046/j.1471-4159.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee HT, et al. cAMP response element-binding protein activation in ligation preconditioning in neonatal brain. Ann Neurol. 2004;56:611–623. doi: 10.1002/ana.20259. [DOI] [PubMed] [Google Scholar]
- 32.Mabuchi T, et al. Phosphorylation of cAMP response element-binding protein in hippocampal neurons as a protective response after exposure to glutamate in vitro and ischemia in vivo. J Neurosci. 2001;21:9204–9213. doi: 10.1523/JNEUROSCI.21-23-09204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papadia S, Stevenson P, Hardingham NR, Bading H, Hardingham GE. Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J Neurosci. 2005;25:4279–4287. doi: 10.1523/JNEUROSCI.5019-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng S, et al. NMDA-induced neuronal survival is mediated through nuclear factor I-A in mice. J Clin Invest. 2010;120:2446–2456. doi: 10.1172/JCI33144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- 36.Soriano FX, et al. Preconditioning doses of NMDA promote neuroprotection by enhancing neuronal excitability. J Neurosci. 2006;26:4509–4518. doi: 10.1523/JNEUROSCI.0455-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang SJ, et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173:2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.