Abstract
Background:
Leukocyte differentials are an important component of clinical care. Morphologic assessment of peripheral blood smears (PBS) may be required to accurately classify leukocytes. However, manual microscopy is labor intensive. The CellaVision DM96 is an automated system that acquires digital images of leukocytes on PBS, pre-classifies the cell type, and displays them on screen for a Technologist or Pathologist to approve or reclassify. Our study compares the results of the DM96 with manual microscopy.
Methods:
Three hundred and fifty-nine PBS were selected and assessed by manual microscopy with a 200 leukocyte cell count. They were then reassessed using the CellaVision DM96 with a 115 leukocyte cell count including reclassification when necessary. Correlation between the manual microscopy results and the CellaVision DM96 results was calculated for each cell type.
Results:
The correlation coefficients (r2) range from a high of 0.99 for blasts to a low of 0.72 for metamyelocytes.
Conclusions:
The correlation between the CellaVision DM96 and manual microscopy was as good or better than the previously published data. The accuracy of leukocyte classification depended on the cell type, and in general, there was lower correlation for rare cell types. However, the correlation is similar to previous studies on the correlation of manual microscopy with an established reference result. Therefore, the CellaVision DM96 is appropriate for clinical implementation.
Keywords: Digital pathology, hematopathology, peripheral blood smear
BACKGROUND
Complete blood counts and differentials have been an important and integral component of the clinical management of patients,[1] since its introduction over 100 years go by Ehrlich.[2] Today, automated blood cell counters based on laser-light scatter and flow cytometry principles, such as the Coulter counter have become standard for most blood counts and differentials and are able to provide a five to six part leukocyte differential. However, they are unreliable in the classification of abnormal and immature cells, and do not provide morphological information. Current procedures indicate that when certain criteria are met during the blood count, the blood sample will be flagged for a peripheral blood smear (PBS). These criteria are set by the machine manufacturer and laboratory using the rules such as those published by the International Consensus Group for Hematology Review.[3] Reasons for PBS microscopy include evaluation of immature and abnormal white cell, review of red cell morphology in hemolytic conditions and platelet morphology in thrombocytopenia, and confirmation of abnormal or unexpected blood counts.
Currently at most institutions, PBS is assessed manually by laboratory technologists and pathologists. However, this practice is labor intensive and time consuming. Moreover, there is substantial inter- and intra-observer variability in this process, which negatively impact efficiency.[4,5] since there is increasing demand on the hematopathology services to provide improved turnaround times and 24 h service for clinicians in addition to increasing volumes, there is an eminent need for improved systems with enhanced productivity.
As a result, there has been research and development into automation of morphological PBS assessment. The first automated morphological assessment system introduced with the Cydac Scanning Microscope System (Cydac, Uppsala, Sweden) in 1966.[6] Early systems such as this were not adopted as they were slow and had limited automation and poor accuracy.[1,7,8] In recent years, there has been significant improvement in technology with new machines such as the CellaVision Diffmaster Octavia, the CellaVision DM96, and the next slide digital review network.
Many of these systems, including the CellaVision DM96, operate by scanning barcode-labeled Wright-Giemsa stained slides at low power to locate white blood cells. The system then takes an image of each white blood cell at high power. These images are analyzed by an artificial neural network based on a large database of cells to pre-classify the type of leukocytes into subtypes including: Band neutrophils, segmented neutrophils, lymphocytes, eosinophils, monocytes, promyelocytes, myelocytes, metamyelocytes, and blasts. The cells are then presented on a computer display for someone to review and confirm or reclassify if incorrectly pre-classified. In addition, the systems can review red blood cell morphology and estimate platelet counts.
The objective of our study is to assess the ability of the CellaVision DM96 (DM96) system and software to classify leukocytes by comparing it with the manual PBS examination.
METHODS
The study was performed at five sites at Calgary Laboratory Services. There were three academic adult hospitals including one Tertiary Care Academic Centre, one academic children’s hospital, and one community laboratory servicing the metropolitan area.
Three hundred and fifty nine PBS slides were included in the study. They were selected from PBS performed by Calgary Laboratory Services as part of routine clinical service. Smears were selected to include examples of leukocyte abnormalities including, atypical lymphocytes, blasts, and left shifted blood counts. For the slides whole venous blood samples were collected in K3EDTA vaccutainer tubes (BD diagnostics Franklin Lakes, NJ USA). Samples were processed utilizing an automated hematology analyzer-LH750 (Beckman-coulter, Brea, California). These instruments were equipped with automated slide maker and stainer. PBS were stained with the Wright-Giemsa stain.
Manual differential counts of 400 white blood cells were performed at various participating sites by two (200 cell count each) experienced laboratory technologists trained in PBS morphology. The PBS slides were then analyzed by the DM96 slide scanning unit with CellaVision Blood Differential Software (CellaVision, AB, Lund, Sweden). This system automatically selects 115 leukocytes for image analysis. The actual white blood cells counted may have been lower when non-leukocytes were mistakenly counted by the machine. The same technologists who performed the manual differential also performed the CellaVision reclassification; however to minimize the influence of prior exposure of PBS slide morphology on DM96 analysis; the slides were randomly selected, and analysis was performed at different time points. Statistics were performed with Microsoft Excel 2010 (Microsoft, Redmond, WA) and SPSS Statistics 19 (IBM, Armonk, NY). The white blood cell differential percentage for each cell type was compared between manual counts by the technologists and automated counts by the DM96 by calculation of Pearson’s correlation coefficient and represented visually by scatter plots. A P ≤ 0.05 was selected as the level of significance.
RESULTS
Three hundred and fifty nine blood smears were included in the study. The percentage cell type breakdown by manual count and as counted by the DM96 is presented in Table 1. Figure 1 shows scatterplots for each leukocyte cell type. The correlation coefficients (r2) range from a high of 0.99 for blasts to a low of 0.72 for metamyelocytes. Due to low cell counts, metamyelocytes, myelocytes, and promyelocytes were grouped together as “immature granulocytes” for analysis.
Table 1.
Characteristics of peripheral blood slides included in this study
Figure 1.
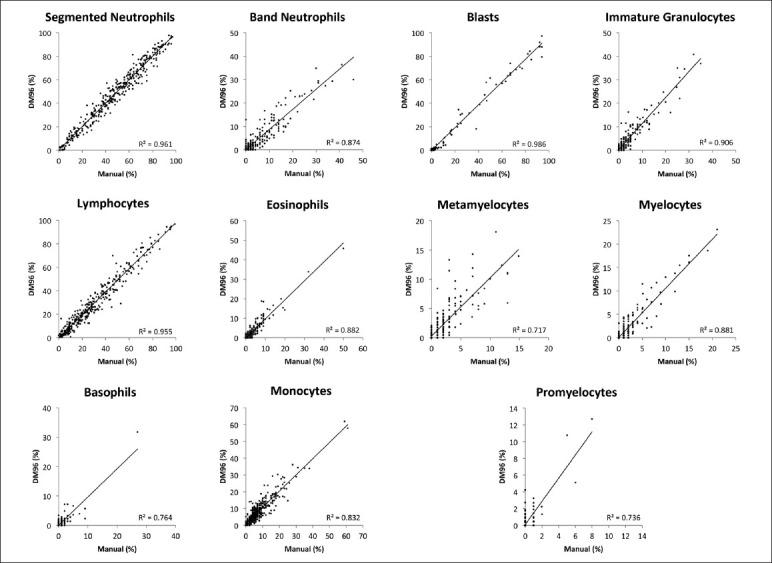
Comparison between manual differential counts and DM96 differential counts for each leukocyte cell type
CONCLUSIONS
Examination of PBS is a labor intensive and time consuming, but clinically necessary, activity in today’s hematopathology laboratory. We evaluated the performance of the DM96 automated morphologic PBS analysis system in classifying the leukocytes in 359 cases and compared the results to a manual assessment of the PBS. Our data showed excellent correlation (r2 > 0.90) between the DM96 and manual microscopy for segmented neutrophils, lymphocytes, and blasts. Lower correlations were seen with eosinophils, monocytes, basophils, and bands. Metamyelocytes, myelocytes, and promyelocytes also showed lower correlations. However, when these were grouped together as immature granulocytes there was excellent correlation.
Our results are consistent with several previous studies of the DM96,[9–12] and a study of the next slide digital review network[13] [Table 2]. Notably, our eosinophil correlation was higher compared to previous findings (r2 = 0.50 [9] to 0.85 [12). The reasons for this are not clear but it is important to note that we had generally higher numbers of eosinophils in our selected samples compared to the previous studies.
Table 2.
Correlation coefficients between DM96 and manual microscopy in the classification of leukocytes. Correlation for the nextslide digital review network and correlation between technologists and an expert reference are included for comparison
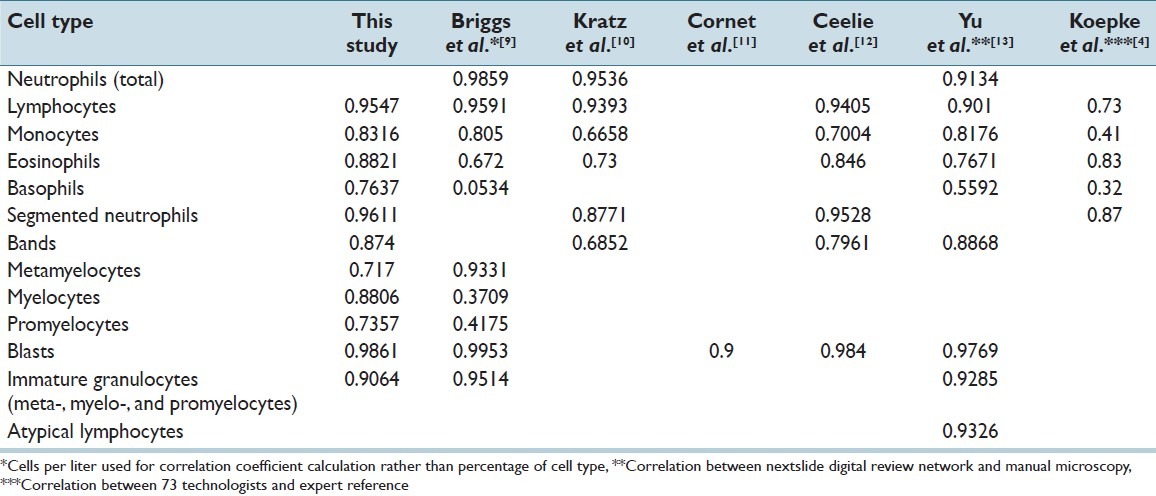
Our results for myelocytes and promyelocytes were significantly better than the only previous study that assessed these cells (myelocytes: r2 = 0.88 for our study, vs. r2 = 0.37 in Briggs et al.;[9] promyelocytes: r2 = 0.74 for our study, vs. r2 = 0.42 in Briggs et al.[9]). Their correlation for metamyelocytes was higher than ours (r2 = 0.72 for our study, vs. r2 = 0.93 in Briggs et al.[9]). These results may be due to the low numbers of immature cells in their study as they included a large number of normal PBS (45/136) while our data does not include normal smears. The aggregation of metamyelocytes, myelocytes, and promyelocytes provided an improved correlation compared to the individual cell subtypes in our study as well as in Briggs et al.[9] (r2 = 0.91 and r2 = 0.95 respectively). This is likely due to the ability of the DM96 and technologists to easily identify immature granulocytes (of which the majority would be of neutrophilic lineage), but difficulty due to subjectivity in subclassifying their maturity.
Basophil identification also had a significantly better correlation in our study compared to the only previously published result published (r2 = 0.76 for our study, vs. r2 = 0.05 in Briggs et al.[9]). Again, this may be due to the low number of basophils in the older study. Our study potentially had more basophils as we did not include normal PBS. It is interesting to note that in a study by Koepke et al.[4] where the correlation in the classification of PBS cells between 73 technologists and expert references showed an r2 of only 0.32 for basophils. This may be due to improved technologist training in recent years. It may also be related to the fact that in our study, basophils are being identified by the same small group of technologists in both the manual microscopy and in the reclassification of CellaVision cells, while in Koepke et al.,[4] the reference cell identification was performed by a separate group of individuals.
Overall, the correlation between the DM96 and manual microscopy in our study is similar to, and in some cases, better than the range of variance between individual technologists. This is demonstrated in Table 2, comparing our correlation coefficients to those of the study by Koepke et al.[4] Previous precision studies have also confirmed this.[9,12] The impact of this enhanced efficiency and inter-observer correlation on the number of smears referred for pathologist’s review is yet to be seen through case controlled studies in the future.
There are several limitations to our study. First, our selection of PBS is not random. This may have produced a biased result. However, this may also represent a useful aspect of our study as one goal of automation is for the DM96 to classify abnormal smears. The percentage of abnormal smears is low in routine hematopathology practice and a random sample would assess very few abnormal smears. Despite the non-random selection of cases, basophils, and immature granulocytes remain low in numbers for our analysis. Another limitation is that the technologists participating at each laboratory involved in our study only analyzed the slides from their own hospital sites. Within each laboratory, a small group of 2 or 3 technologists manually read the PBS and reclassified the results of the DM96. As a result, there is a large probability that the same technologist manually read the slide and also reclassified the DM96 results. This may artificially improve the correlation between the manual and automated classification methods. Finally, our study, similar to most previous studies, focused on the reclassified cell results rather than the automated pre-classified results. Although, this is how the DM96 will be used in clinical practice, the resultant studies ultimately assess the ability of technologists to identify cells on a computer screen rather than test the ability of the DM96 algorithms to classify cells. As a result, the studies, including our study are unable to test the possibility of full automation of PBS analysis.
Our study shows that the DM96 is a useful tool in the examination of PBS morphology with performance similar to that of manual microscopy. The DM96 will only be useful if it is cost effective, particularly as cost control has become essential in the current era of economic uncertainty. It has previously been shown that the DM96 is faster than manual smear examination an aspect confirmed in our internal departmental validation studies.[9–12,14,15] This is of particular importance as labor is one of the major expenditures in the laboratory. In addition, digitized images provide many advantages compared to manual slide microscopy. Images of individual leukocytes can be stored for educational activities, quality control, and expert consultation. Images may be transmitted from remote locations or areas with a lack of trained technologists to institutions with expertise. Finally, it has been shown that education using images captured by systems such as the DM96 allows for quicker leukocyte recognition among new trainees.[16] Future studies include the assessment of the ability of the DM96 to identify specific PBS morphologic diagnoses.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/1/14/114205
REFERENCES
- 1.Tatsumi N, Pierre RV. Automated image processing. Past, present, and future of blood cell morphology identification. Clin Lab Med. 2002;22:299–315, viii. doi: 10.1016/s0272-2712(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich P. Farbenanalytische Untersuchungen Zur Histologie Und Klinik Des Blutes: Gesammelte Mittheilungene. Berlin, Germany: Hirschwald; 1891. [Google Scholar]
- 3.Barnes PW, McFadden SL, Machin SJ, Simson E international consensus group for hematology. The international consensus group for hematology review: Suggested criteria for action following automated CBC and WBC differential analysis. Lab Hematol. 2005;11:83–90. doi: 10.1532/LH96.05019. [DOI] [PubMed] [Google Scholar]
- 4.Koepke JA, Dotson MA, Shifman MA. A critical evaluation of the manual/visual differential leukocyte counting method. Blood Cells. 1985;11:173–86. [PubMed] [Google Scholar]
- 5.Rümke CL. Statistical reflections on finding atypical cells. Blood Cells. 1985;11:141–4. [PubMed] [Google Scholar]
- 6.Prewitt JM, Mendelsohn ML. The analysis of cell images. Ann N Y Acad Sci. 1966;128:1035–53. doi: 10.1111/j.1749-6632.1965.tb11715.x. [DOI] [PubMed] [Google Scholar]
- 7.Riley RS, Ben-Ezra JM, Massey D, Cousar J. The virtual blood film. Clin Lab Med. 2002;22:317–45. doi: 10.1016/s0272-2712(03)00077-5. [DOI] [PubMed] [Google Scholar]
- 8.Beksaç M, Beksaç MS, Tipi VB, Duru HA, Karakás MU, Cakar AN. An artificial intelligent diagnostic system on differential recognition of hematopoietic cells from microscopic images. Cytometry. 1997;30:145–50. doi: 10.1002/(sici)1097-0320(19970615)30:3<145::aid-cyto5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 9.Briggs C, Longair I, Slavik M, Thwaite K, Mills R, Thavaraja V, et al. Can automated blood film analysis replace the manual differential. An evaluation of the CellaVision DM96 automated image analysis system? Int J Lab Hematol. 2009;31:48–60. doi: 10.1111/j.1751-553X.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- 10.Kratz A, Bengtsson HI, Casey JE, Keefe JM, Beatrice GH, Grzybek DY, et al. Performance evaluation of the CellaVision DM96 system: WBC differentials by automated digital image analysis supported by an artificial neural network. Am J Clin Pathol. 2005;124:770–81. doi: 10.1309/XMB9-K0J4-1LHL-ATAY. [DOI] [PubMed] [Google Scholar]
- 11.Cornet E, Perol JP, Troussard X. Performance evaluation and relevance of the CellaVision DM96 system in routine analysis and in patients with malignant hematological diseases. Int J Lab Hematol. 2008;30:536–42. doi: 10.1111/j.1751-553X.2007.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceelie H, Dinkelaar RB, van Gelder W. Examination of peripheral blood films using automated microscopy; evaluation of Diffmaster Octavia and Cellavision DM96. J Clin Pathol. 2007;60:72–9. doi: 10.1136/jcp.2005.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Ok CY, Hesse A, Nordell P, Connor D, Sjostedt E, et al. Evaluation of an automated digital imaging system, Nextslide Digital Review Network, for examination of peripheral blood smears. Arch Pathol Lab Med. 2012;136:660–7. doi: 10.5858/arpa.2011-0285-OA. [DOI] [PubMed] [Google Scholar]
- 14.Billard M, Lainey E, Armoogum P, Alberti C, Fenneteau O, Da Costa L. Evaluation of the CellaVision DM automated microscope in pediatrics. Int J Lab Hematol. 2010;32:530–8. doi: 10.1111/j.1751-553X.2009.01219.x. [DOI] [PubMed] [Google Scholar]
- 15.Rollins-Raval MA, Raval JS, Contis L. Experience with CellaVision DM96 for peripheral blood differentials in a large multi-center academic hospital system. J Pathol Inform. 2012;3:29. doi: 10.4103/2153-3539.100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiuchi Y, Tabe Y, Idei M, Bengtsson HI, Ishii K, Horii T, et al. The use of CellaVision competency software for external quality assessment and continuing professional development. J Clin Pathol. 2011;64:610–7. doi: 10.1136/jcp.2011.089888. [DOI] [PubMed] [Google Scholar]


