Abstract
The human microbiome project (HMP) reflects the fact that we are supraorganisms composed of human and microbial components. This international effort emanates from a confluence of ongoing technical and computational advances in the genome sciences, an evolving focus of microbiology on the properties and operations of microbial communities, and the notion that rapid, and marked, transformations in human lifestyles are not only affecting the health of the biosphere, but possibly our own health as a result of changes in our microbial ecology. HMP is designed to understand the microbial components of our genetic and metabolic landscape, and how they contribute to our normal physiology and disease predisposition. It is a global and interdisciplinary project that promises to break down the artificial barriers between medical and environmental microbiology. Here, we discuss some the challenges that HMP faces and options for addressing them.
Introduction
Prior to completion of the human genome sequencing project, some predicted that we would find ~100,000 genes. For many, feelings of surprise and perhaps humility were associated with the announcement that our genome only contains ~20,000 protein-coding genes, a number not greatly different from that of the fruit fly. However, by expanding our view of ourselves, we can see that the number 100,000 is likely an underestimate. The microbes that live inside and on us (the microbiota) outnumber our somatic and germ cells by an estimated 10-fold. The collective genomes of our microbial symbionts (the microbiome) provide us with traits we have not had to evolve on our own1. If we consider ourselves to be a composite of microbial and human species, our genetic landscape a summation of the genes embedded in our human genome and microbiome, and our metabolic features a coalescence of human and microbial traits, the self-portrait that emerges is one of a ‘human supraorganism’. Thus, understanding the range of human genetic and physiologic diversity means that we must characterize our microbiome and the factors that influence the distribution and evolution of our microbial partners. The outcome may provide an additional perspective about contemporary human evolution, as we assess whether and how our rapidly advancing technology, with its transformative changes in our lifestyles and biosphere, is influencing our ‘micro’-evolution, and thus our health and predisposition to diseases.
The human microbiome project (HMP) is an imposing yet logical conceptual and experimental extension of the human genome project. HMP is not a single project, but rather a summation of multiple projects that are now being launched, concurrently, in multiple areas of the world, including the USA (as part of the next phase of NIH’s Roadmap Initiative), the European Union, and Asia. It is a manifestation of a thematic expansion in microbiology. Newly introduced, highly parallel DNA sequencers and mass spectrometers are propelling microbiology into a new era where it extends its focus from the properties of single types of organisms in isolation to the operations of entire communities. The new field of metagenomics embraces genome-anchored characterizations of these communities2.
HMP will address some of the most inspiring, vexing and fundamental questions in 21st-century science. Importantly, it promises to break down the artificial barriers between medical and environmental microbiology. The hope is that in addition to providing new ways for defining health and disease predilection (Box 1), HMP will provide us with the parameters needed to design, implement and monitor strategies for intentionally manipulating our microbiota so as to optimize its performance in the context of an individual’s physiology. This article provides a context for HMP, and discusses some of the conceptual and experimental challenges that it faces, as well as the rewards it promises. We focus on the gut to illustrate a number of our points, since this body habitat harbors the largest collection of our microbial partners.
Box 1. A few examples of functional contributions of the gut microbiota, based on studies of gnotobiotic animals, and their implications for human health.
-
Harvest of otherwise inaccessible nutrients/energy from the diet; synthesis of vitamins1,19,42–4444
Implications: The nutrient/energetic value of food is not an absolute term but rather one that is influenced in part by the digestive capacity of the consumer’s microbiota. This has implications for identifying individuals at risk for malnourished states and obesity, and treating them based on more personalized view of nutrition that considers host microbial ecology.
-
Xenobiotic metabolism and other ‘metabotypes’
Implications: The microbiota is a largely under-explored regulator of drug metabolism/bioavailability. Bioremediation functions carried out by the microbiota, such as detoxification of ingested carcinogens, may affect susceptibility to various neoplasms within and outside of the gut. Oxalate metabolism by the microbiota has been linked to predisposition to nephrolithiasis (renal stones; ref. 45). Microbial modification of bile acids affects host lipid metabolism44. Metabotypes (metabolic phenotypes) ascribable to the microbiota should expand our repertoire of personalized biomarkers of health and disease susceptibility.
Gut epithelial cell renewal (slower in germ-free animals) Implications: This phenotype is influenced in part by microbiota-immune cell interactions, and could have a range of effects ranging from susceptibility to neoplasia46 to the capacity to repair a damaged mucosal barrier47. Comparisons of microbial communities physically associated with neoplasms with those having varying degrees of remoteness from the tumors, may provide new mechanistic insights about pathogenesis, including adaptive microbial and host responses.
-
Maturation and activity of the innate48 and adaptive49 immune systems Implications: Impacts disorders that are manifest within and outside of the gut: e.g., (i) inflammatory bowel diseases, where a there is an apparent dysregulated immune response to the gut microbial community (genome wide association studies of patients with Crohn’s disease have identified a number of human genes involved in both innate and adaptive immunity; ref. 50): (ii) the capacity of the microbiota to influence host expression of antimicrobial compounds produced by components of the innate immune system influences susceptibility to colonization by enteropathogens48,51; and (iii) correlations have been demonstrated between the incidence of asthma, exposure to bacteria52, and treatment with broad-spectrum antibiotics during early childhood53
Cardiac size (smaller as a fraction of body weight in germ-free animals; ref. 54) Implications: The mechanism underlying this phenotype has yet to be defined but the phenomena emphasizes the importance of expanding studies of how much of our physiology is modulated by our microbiomes.
-
Behavior (germ-free mice have greater locomotor activity than their colonized counterparts; ref. 43)
Implications: Has the microbiota evolved ways to benefit itself and its host by influencing human behavior? Is altered production of neuroactive compounds, either directly by the microbiota, or through its modulation of host genes normally involved in biosynthesis/metabolism of these compounds, associated with any neurodevelopmental and/or psychiatric disorders?
Ecology and considerations of scale
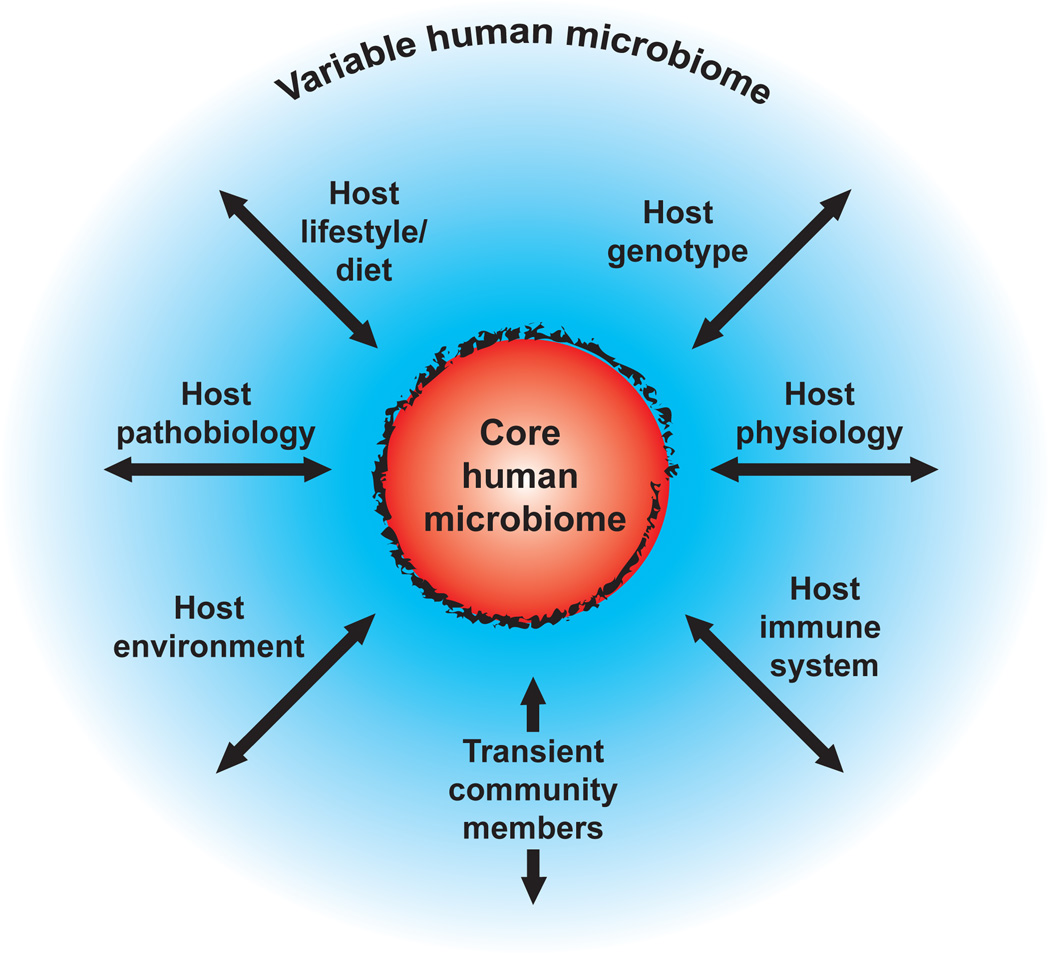
One anticipated outcome of HMP is that it will inform us about whether principles of ecology, gleaned from studies of the macroscopic world, apply to the microscopic world we harbor. To a significant degree, questions about the human microbiome are new only in terms of the system to which they apply: similar questions have inspired and confounded ecologists working on macro-scale ecosystems for decades. How stable and resilient is a microbiota over the course of a day within one individual, and during the course of his or her lifespan? How similar are the microbiomes between members of a family, a human community, and between communities living in different environments? Do all humans share an identifiable ‘core’ microbiome, and if so, how is it acquired and transmitted? What affects the genetic diversity of the microbiome (Figure 1), and how does this diversity impact microbial and host adaptations to markedly different lifestyles, and to various physiological or pathophysiological states?
Figure 1. The concept of a core human microbiome.
The core (orange) is viewed as a set of shared genes found in a given habitat (e.g. gut, mouth, skin) in all humans. The core is surrounded by a set of variably represented genes (blue): this variation could be influenced by a combination of factors (arrows) including transient populations of microbes that are not able to persistently colonize (allochthonous organisms), lifestyle (including diet), various environmental exposures (place of residency or work), host genotype, host physiologic status including the properties of the innate and adaptive immune system, and disease. The hazy line surrounding the core indicates the possibility that over the course of human ‘micro-evolution’ new genes may be added to the core microbiome while others may be lost.
Temporal and spatial scales need to be considered when sampling the microbiota. For example, our surface microbial communities have a complex biogeography that can be defined over a dynamic range of distances: at the micron scale (the distribution of microbes on undigested food particles in the distal gut, or across a mucosal barrier); at the centimeter scale (the distribution of communities around different teeth); and over a distance of meters (the cephalocaudal axis of a gut).
Scale has an additional meaning; the concept of a core microbiome represents whatever may be common among the microbiomes of all humans. These commonalities may not manifest as sets of genes or sets of organisms, but rather as functional relationships between the microbiome and the host. There are currently 6.7 billion humans living on Earth. Due to a variety of constraints, we will have to characterize our microbiome(s) by comparing limited types of data collected from a limited set of individuals. If human body habitats, such as the gut, are viewed as islands in space and time, then island biogeography theory, developed from studies of macro-ecosystems3, may be useful for understanding the observed microbial diversity. This theory posits that community composition can depend strongly on the order in which species initially enter a community (a phenomenon known as multiple stable states4). The importance of the initial inoculating microbial community on later community composition has been demonstrated in animal models. For example, in the mouse gut microbiota, the effects of maternal transmission (kinship) are manifest over several generations in animals belonging to the same inbred strain5. Similarly, reciprocal microbiota transplantation experiments, involving conventionally-raised mouse or zebrafish donors and germ-free mouse and zebrafish recipients, demonstrate that legacy effects (the microbial community available to colonize the gut at the time of birth), together with features of the gut habitat itself, conspire to select a microbiota6. We can start by characterizing a few specific islands (humans) in-depth, or we can perform the equivalent of a biogeography experiment in which we perform coarse-grained analysis of a larger number of humans, selected based on demographic, geographical, or epidemiological factors, to infer general trends. These strategies are complementary to one another, and as noted below, both will be needed to fully understand the human microbiome.
What we know about the human microbiome at the present time?
High levels of inter-personal variability in bacterial lineages present
The decreasing cost and increasing speed of DNA sequencing, coupled with advances in the computational approaches used to analyze complex datasets7–11, have prompted a number of groups to embark on bacterial 16S rRNA gene sequence-based surveys of microbial communities that reside on our skin, and in our mouth, esophagus, stomach, colon, and vagina (see refs 12–17 plus other reports in this Insights). The largest reported datasets come from the gut, although the number of humans sampled using these culture-independent surveys is still very limited. The vast majority of the 10–100 trillion microbes in the human gastrointestinal tract live in the colon. More than 90% of all bacterial phylogenetic types (phylotypes) belong to just two of the 70 known divisions (phyla) in the domain Bacteria: the Bacteroidetes and the Firmicutes. Within the colon, differences between individuals are greater than differences between different colonic sampling sites15. Moreover, feces are representative of interpersonal differences5. A recent study of 18,348 fecal 16S rRNA gene sequences collected from 14 unrelated adults over the course of a year demonstrated a high level of ‘between-individual’ differences in microbial community structure and established that community membership in each host was generally stable during this time interval16. How is such high inter-personal diversity sustained? 16S rRNA-based observations about diversity in the human gut microbiota may fit with predictions from the neutral theory of community assembly, which states that most species will share the same general niche, or the biggest niche, and thus are likely to be functionally redundant18. Neutral community assembly predicts highly variable communities as defined by 16S rRNA lineages, but high levels of functional redundancy between community members and assemblages.
Ecosystem-level functions revealed by metagenomic studies of the microbiome
The first reported application of metagenomic techniques to a human microbiome involved two unrelated healthy adults. Compared to all previously sequenced microbial genomes and the human genome, metabolic reconstructions of their gut (fecal) microbiomes revealed statistically significant enrichment for genes involved in (i) the metabolism of glycans, amino acids, and xenobiotics, (ii) methanogenesis, and (iii) 2-methyl-D-erythritol 4-phosphate pathway-mediated biosynthesis of vitamins and isoprenoids1.
The ability of comparative metagenomics to reveal functional attributes of a microbiome is further underscored by a recent study, which showed that a host phenotype (obesity) can be correlated with the degree of representation of microbial genes involved in certain metabolic pathways19. Community DNA was isolated from the distal gut contents of genetically obese (ob/ob) mice and their lean (+/+ or ob/+) littermates and analyzed with a traditional capillary sequencer, and with a representative of the new generation of highly parallel instruments. In silico metabolic reconstructions, based on microbial community gene content, indicated that the obesity-associated gut microbiome has an increased capacity to harvest energy from the diet; the ob/ob microbiome was enriched for genes involved in importing and metabolizing otherwise indigestible dietary polysaccharides to short chain fatty acids, which are absorbed by the host and stored as more complex lipids in adipose tissue. Biochemical analyses supported these predictions. Moreover, when adult germ-free wild-type mice were colonized with a microbiota harvested from obese (ob/ob) or lean (+/+) donors, adiposity in recipients of the obese microbiota increased to a significantly greater degree than in recipients of a lean microbiota, supporting the conclusion that the obese gut microbiota has an increased (and transmissible) capacity to promote fat deposition19. This marriage of comparative metagenomics and gnotobiotic animal models illustrates one way to proceed from in silico predictions to experimental tests of whole community microbiome function.
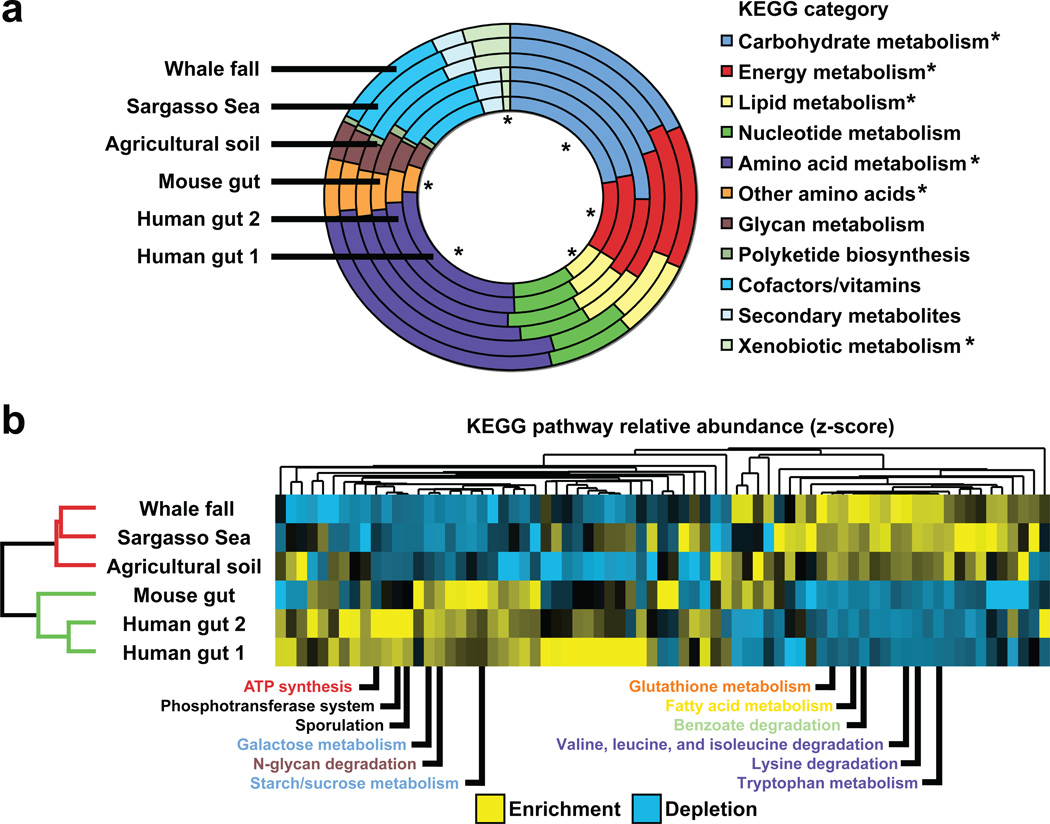
Metagenomic datasets from very different microbial ecosystems can also be compared to reveal the traits that are important to each20. Figure 2 compares the two human and five mouse capillary sequencer-derived gut microbiome datasets described above with datasets obtained from three environmental communities: one from decaying whale carcasses located at the bottom of the ocean (three whale falls), another from an agricultural soil community, and another from a survey of the Sargasso Sea20,21. Unidirectional (forward) DNA sequencing reads were culled from each dataset and matched to annotated genes represented in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database22. The gut microbiomes cluster together and are enriched for predicted genes assigned to KEGG categories and pathways for carbohydrate and glycan metabolism (Figure 2A,B). Given the currently limited sequencing coverage and number of individuals sampled, deeper sequencing of additional human gut microbiomes will be necessary to determine if these features are common traits of the human microbiome (see discussion of sampling issues below).
Figure 2. Functional comparisons of the gut microbiome versus other sequenced microbiomes1,19–21.
(A) Relative abundance of predicted genes assigned to KEGG categories for metabolism. Analyses were performed on the combined mouse gut dataset (n=5 animals), both human gut datasets, and three ‘environmental’ datasets: the combined Whale fall dataset (n=3 samples), agricultural soil, and the combined Sargasso Sea dataset (n=7 samples). Forward sequencing reads were culled from each dataset and mapped onto reference microbial and eukaryotic genomes from the KEGG database22 (version 40; BLASTX best-blast-hit e-value<10−5). Asterisks indicate categories that are significantly enriched or depleted in the combined gut dataset versus the combined environmental dataset (the distribution of ~15,000 KEGG pathway assignments across each of the six datasets was used to construct two combined datasets of ~45,000 KEGG pathway assignments each; Χ2 test using the Bonferroni correction for multiple hypotheses, p<10−4). (B) Hierarchical clustering based on the relative abundance of predicted proteins assigned to KEGG pathways reveals specific differences between gut (green) and environmental (red) microbiomes. The relative abundance of pathways that exceeded a threshold of >0.6% (assignments to a given pathway divided by assignments to all pathways) in at least two environments was transformed into a z-score (yellow=enrichment; blue=depletion), and clustered by environments and pathways20 using a Euclidean distance metric (Cluster 3.040). The results were visualized in Treeview41. Environmental clustering was consistent using multiple distance metrics, including Pearson Correlation (centered/uncentered), Spearman Rank Correlation, Kendall’s tau, and City-block distance. The twelve most discriminating KEGG pathways are listed (based on the ratio of average gut relative abudance versus average environmental relative abundance). Metabolic pathway names are colored based on KEGG category [pathways not colored include sporulation (cell growth/death) and phosphotranferase system (membrane transport)]. The gut microbiome is enriched for pathways involved in importing and degrading polysaccharides and simple sugars (‘starch/sucrose metabolism’, ‘galactose metabolism’, ‘N-glycan degradation’, and ‘phosphotransferase system’). The gut microbiome is also enriched for genes involved in ‘sporulation’, reflecting the high relative abundance of Gram-positive Firmicutes.
Moving forward: some of the things that HMP will need
Additional reference genomes
Metagenomic analyses of complex microbial communities are currently limited by the availability of suitable reference genomes for confident assignment of short sequences generated by the current generation of highly parallel DNA sequencers, and by knowledge of the professions (niches) of their component organismal lineages. An ongoing project to sequence the genomes of 100 cultured representatives of the phylogenetic diversity in the human gut microbiota23 illustrates how reference genomes will help interpret metagenomic studies. Capillary sequencing reads from the human and mouse gut microbiome datasets described above were anchored to previously published microbial and eukaryotic genomes (KEGG database version 40; ref. 22) and seventeen recently sequenced human gut bacterial genomes belonging to the Bacteroidetes, Firmicutes and Actinobacteria (BLASTX homology, e-value<10−5). These analyses revealed that the quality, and percentage of metagenomic read assignments increases significantly with the addition of each individual gut genome.
Additional reference genome sequences, including multiple isolates of selected species-level phylotypes, should also help answer questions regarding genetic variation within and between the major phylogenetic lineages in a given habitat such as the gut. For example, comparisons of multiple members of the Firmicutes and Bacteroidetes should provide insights as to the extent of genetic redundancy and/or specialization between these two divisions. Given the extraordinary density of colonization in the distal gut (1011–1012 organisms/ml of luminal content), these additional genomes also provide an opportunity to determine more accurately the role of lateral gene transfer in the evolution of gut microbes within and between hosts24, and the extent to which the gene content of these organisms reflects their phylogenetic history.
Developing new methods for retrieving previously unculturable organisms will be critical for obtaining reference genome sequences. Recently, fluorescence in situ hybridization with phylogenetic markers, flow cytometry, whole-genome amplification, and shotgun-sequencing have been used to obtain a partial genome assembly for a member of the TM7 phylum, providing a first look at a group of organisms with no currently available cultured representatives25. Additionally, methods such as gel microdroplet technology aim to allow high-throughput culture of microorganisms in a simulated natural environment26.
Linking short gene fragments to organisms
Another major challenge is to link genes to organisms, or at least to broader taxonomic classifications, because metagenomic datasets consist of largely unassembled sequence data. Several approaches currently exist27–29, but no tools have been developed for the automated analysis of large datasets containing mostly short sequence reads without relying on phylogenetic marker genes. Thus, an accurate and scalable way to perform phylogenetic classification on vast numbers of short reads is essential.
The two general marker-independent approaches to phylogenetic assignment are to use Markov models based on the frequency of short nucleotide sequences, or ‘words’, in the reads, and to use homology searching to place each fragment in the context of a phylogenetic tree. The former approach is likely to be relatively insensitive, especially for short sequences and for sequences from heterogeneous genomes, because of statistical sampling issues. The latter approach is expected to be more accurate, and provides the additional advantage of placing each sequence within the context of a multiple alignment and a phylogenetic tree for downstream studies. However, sequences without identifiable homologs cannot be analyzed in this way. Some combination of these two general strategies will likely provide the best approach for understanding the functions present in each metagenome. The key issues are to (i) understand how accurate the phylogenetic classification obtained from each method can be, especially in the face of lateral gene transfer, (ii) find better, faster, and more scalable heuristics for generating huge phylogenetic trees containing millions of sequences, and (iii) identify the best way of accounting for the influence of both the genome and the function of each protein on the overall composition of each sequence. In particular, heterogeneous rates of evolution in different protein families pose substantial problems for search-based methods: significant similarities at the primary sequence level may not persist over time and the structures of the proteins are typically unknown, thus preventing the use of structure-based alignment techniques.
Towards a global comparison of human microbial communities
Understandably, there will be great pressure at early stages of the HMP to focus on disease states. However, time, resources, and discipline are needed to define ‘normal’ before we can determine the impact of the microbiota on disease predisposition and pathogenesis.
Several issues need to be considered when designing ways to generate an initial set of reference microbiomes from healthy individuals: (i) the degree of genetic relatedness among those that are being sampled (e.g. should the initial focus be on mono-and dizygotic twins-pairs and their mothers?); (ii) place in family structure; (iii) age; (iv) demographics (e.g. rural versus urban); (v) ethical, legal and logistical barriers that need to be overcome in order to obtain, without exploitation, samples (and metadata) from subjects living in very diverse cultural and socio-economic contexts; (vi) the types of comparisons that are needed [within sample (alpha) diversity; between sample (beta) diversity; between-habitat comparisons within a given individual (e.g., skin, nasal passage, mouth, hypopharynx, intestine, urogenital tract), and between habitat comparisons among family members]; plus (vii) the protocols that could or should be used for sampling surface-associated microbial communities (a major unresolved technical issue is how to reproducibly retrieve sufficient and representative quantities of microbes, largely free of human cells, from a body surface such as the skin for metagenomic analysis, and over what temporal and spatial scales this sampling should occur).
As is the case throughout ecology, we must choose between extensive sampling of a small number of sites (individual people/body habitats), and the broad sampling that will allow us to uncover general principles that govern community structure and function. Deep sampling of body habitats from a few individuals is needed to estimate the distribution of species and gene abundances: these estimates, in turn, will allow modeling of the results of different trade-offs between deeper sampling of fewer individuals and shallower sampling of more individuals. Unlike the situation with the HapMap30, we have no baseline expectation of the amount of diversity in different communities, and the development of careful sampling models will be essential for optimizing use of resources. Also, given the rapid pace of development of new and more massively parallel sequencing technologies, systematic testing will be required to identify ways for maximizing sequencing coverage at affordable cost, while maintaining the ability to analyze and assemble genome fragments.
Ultimately, we will need to connect differences in communities to differences in metabolic function and/or disease. Thus, another key challenge in the HMP is to define the concept of ’distance’ between communities, and relate these distances to host biology and a variety of meta-data. UniFrac11,31,32 and other phylogenetic techniques provide an answer to this problem for 16S rRNA gene datasets, and may be extensible to metagenomic data. With distances defined, statistical techniques will have to be developed and refined to integrate multivariate datasets into a unified framework that allows us to identify components of the microbiome that could impact human health and disease.
HMP will also require us to move beyond comparative genomics to an integrated ‘systems metagenomics’ approach that accounts for microbial community structure (the ‘microbiota’), gene content (the ‘microbiome’), gene expression (the ‘meta-transcriptome’ and ‘meta-proteome’), and metabolism (the ‘meta-metabolome’). Some progress has been made in generating ‘functional gene arrays’ with the goal of interrogating microbiomes for the relative abundance of specific genes or transcripts33–35. More work is needed to improve array sensitivity, and to apply these approaches to complex communities such as the human microbiome. cDNA library construction and sequencing present an alternative approach, and have already been applied to microbial and eukaryotic mRNA from environmental samples36,37. However, high-throughput methods for eliminating highly abundant transcripts (e.g. rRNAs) are needed. Proteomic tools are available for analyzing complex samples [e.g. Elucidator (Rosetta) and SEQUEST]. Comprehensive microbial protein sequence databases are undergoing continuous development (e.g. NCBI Protein Clusters). In addition, custom databases can be created from metagenomic datasets, and used to interpret mass spectrometry datasets38. Given the limited knowledge about the biotransformations that human microbial communities support, ‘meta-metabolomics’ will likely prove to be very challenging. Despite highly accurate instrumentation (e.g., Fourier transform ion cyclotron resonance mass spectrometers have a mass accuracy of <1–10 ppm), tools and databases for metabolite identification need to be developed. This situation should be helped by efforts currently underway to catalog all human-associated metabolites, and generate a searchable database39. Together, these complementary measurements will allow a far richer characterization of human microbial communities, and will allow us to identify the variation that is typical of a healthy state so that we can then search for deviations associated with disease.
Data deposition and distribution
The vast amounts of information that will emanate from an international HMP, together with the avalanche of data from metagenomic surveys of the environment, necessitates new procedures and expanded capabilities for depositing, storing, and mining different types of data. Goals include, but are not limited to: a minimal set of standards for annotation; a flexible, simple and open format for depositing meta-data (taking a lesson from clinical studies because the important relevant parameters are presently unknown); widely useful and efficient analysis tools for the general user (including tools for meta-analyses of varied types of data); and the development of an adequate cyber-infrastructure to support the computing needs of the broad community.
Model systems
Although HMP is by definition human-focused, model organisms and other experimental systems are needed to define how communities operate and interact with their hosts, to characterize the determinants of their robustness and to identify biomarkers of community composition/performance. Gnotobiotic animals, both wild-type and genetically engineered, colonized at various stages of their life cycles with intentionally designed simplified communities composed of a few sequenced members, or with more complex enumerated communities, should be very useful. In vitro models, including micro-fluidic-based techniques for single cell sorting and measurements, should be informative for defining the biological properties of microorganisms and the consequences of microbial-microbial interactions.
A scenario for staging the HMP
Based on these considerations, Table 1 outlines one potential scenario for staging HMP. The search for data will be global in many senses of the word: it embraces the planet and its (human) inhabitants; it requires the participation of individuals from the clinical, biological and physical-engineering sciences with expertise in disciplines ranging from mathematics, to statistics, computer science, computational biology, microbiology, ecology, evolutionary biology, comparative genomics and genetics, environmental and chemical engineering, chemistry and biochemistry, human systems physiology, anthropology, sociology, and law/ethics, among others; it mandates coordination among scientists, governments, and funding agencies; and it represents one element of a world-wide effort to document, understand and respond to the consequences of our human activities - not only as they relate to our own health but to the sustainability of our biosphere. We hope that just as microbial observatories have been established to monitor world-wide changes in terrestrial and oceanic ecosystems, an early manifestation of HMP will be to establish ‘human observatories’ to monitor our microbial ecology in different settings.
Table 1.
A Scenario for Staging the Human Microbiome Project
| Tier 1 - Initial data acquisition and analysis |
| [Pillar 1] - Deep draft assemblies of reference genomes. |
|
| [Pillar 2] - Reference microbiome datasets |
|
| [Pillar 3] - Shallower 16S rRNA and community metagenomic datasets from moderate number of samples |
|
| Tier 2 - Choice of individuals that represent different clusters for additional deep sequencing |
|
| Tier 3 - Global human microbiome diversity project |
|
Many outcomes of HMP can be envisioned, ranging from new diagnostic biomarkers of health, to a 21st century pharmacopeia that includes members of our microbiota and the chemical messengers they produce, to enzymes manufactured by our microbiota that are able to process substrates in ways that have important industrial applications. We anticipate that one important outcome will be a deeper understanding of the nutritional needs of humans, and the generation of viable options for matching changes in our agricultural practices and food supply, mandated by changes in climate and economic growth, with an understanding of our microbial ecology.
Acknowledgements
We apologize that due to space constraints we are not able to cite many excellent studies by our colleagues in this emerging field.
Footnotes
The authors declare no competing financial interests.
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Committee on Metagenomics: Challenges and Functional Applications, National Research Council. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet. Washington, D.C: The National Academies Press; 2007. p. 174. [PubMed] [Google Scholar]
- 3.MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton, N.J: Princeton University Press; 1967. p. 203. [Google Scholar]
- 4.Ambramsky Z, Rosenzweig ML. The productivity diversity relationship: Tilman’s pattern reflected in rodent communities. Nature. 1984;309:150–151. doi: 10.1038/309150a0. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, et al. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole JR, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schloss PD, Handelsman J. DOTUR, a computer program for defining operational taxonomic units estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis TZ, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone C, Hamady M, Knight R. UniFrac - an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z, et al. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei Z, et al. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bik EM, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbial ecology linked to obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 17.Hyman RW, et al. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. USA. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbell SP. Neutral theory and the evolution of ecological equivalence. Ecology. 2006;87:1387–1398. doi: 10.1890/0012-9658(2006)87[1387:ntateo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 20.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 21.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon JI, et al. Extending Our View of Self: the Human Gut Microbiome Initiative (HGMI) 2005 http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/HGMISeq.pdf.
- 24.Xu J et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podar M, et al. Targeted access to the genomes of low abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zengler K, et al. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teeling H et al. TETRA: a web-service and stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McHardy AC, et al. Accurate phylogenetic classification of variable-length DNA fragments. Nature Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 29.von Mering C, et al. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 30.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, et al. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 2001;67:5780–5790. doi: 10.1128/AEM.67.12.5780-5790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentry TJ, et al. Microarray application in microbial ecology research. Microb. Ecol. 2006;52:159–175. doi: 10.1007/s00248-006-9072-6. [DOI] [PubMed] [Google Scholar]
- 35.Gao H, et al. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl. Environ. Microbiol. 2007;73:563–571. doi: 10.1128/AEM.01771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poretsky RS, et al. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 2005;71:4121–4126. doi: 10.1128/AEM.71.7.4121-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant S, et al. Identification of eukaryotic open reading frames in metagenomic cDNA libraries made from environmental samples. Appl. Environ. Microbiol. 2006;72:135–143. doi: 10.1128/AEM.72.1.135-143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ram RJ, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- 39.Wishart DS, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 41.Saldanha AJ. Java Treeview - extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 42.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin FJ, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidhu H, Allison MJ, Chow JM, Clark A, Peck AB. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J. Urol. 2001;166:1487–1491. [PubMed] [Google Scholar]
- 46.Chu FF, et al. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- 47.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc. Natl. Acad. Sci. USA. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 49.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun-Fahrlander C, et al. Environmental exposure to endotoxin its relation to asthma in school-age children. N. Engl. J. Med. 2002;347:869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 53.Kozyrskyj AL, Ernst P, Becker AB. Increased risk of childhood asthma from antibiotic use in early life. Chest. 2007;131:1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 54.Wostmann BS, Bruckner-Kardoss E, Pleasants JR. Oxygen consumption and thyroid hormones in germfree mice fed glucose-amino acid liquid diet. J. Nutr. 1982;112:552–559. doi: 10.1093/jn/112.3.552. [DOI] [PubMed] [Google Scholar]