Synopsis
Steroid hormone receptor maturation is a multi-step process that involves several TPR proteins that bind to the maturation complex via the C-termini of hsp70 and hsp90. We produced a random T7 peptide library to investigate the roles played by the C-termini of the two heat shock proteins in the TPR/hsp interactions. Surprisingly, phages with the MEEVD sequence, found at the C-terminus of hsp90, were not recovered from our biopanning experiments. However, two groups of phages were isolated that bound relatively tightly to HsPP5 TPR. Multiple copies of phages with a C-terminal sequence of LFG were isolated. These phages bound specifically to the TPR domain of HsPP5 although mutation studies produced no evidence that they bound to the domain’s hsp90-binding groove. However, the most abundant family obtained in the initial screen had an aspartate at the C-terminus. Two members of this family with a C-terminal sequence of VD appeared to bind with approximately the same affinity as the hsp90 C-12 control. A second generation pseudo-random phage library produced a large number of phages with a LD C-terminus. These sequences acted as hsp70 analogues and had relatively low affinities for hsp90 specific TPR domains. Unfortunately, we failed to identify residues near hsp90’s C-terminus that impart binding specificity to individual hsp90/TPR interactions. The data suggest that the C-terminal sequences of hsp70 and hsp90 act primarily as non-specific anchors for TPR proteins.
Keywords: Phage display, TPR domains, PP5, immunophilin, steroid receptor, heat shock protein
INTRODUCTION
The maturation process of steroid receptors is a complex and dynamic multi-protein pathway [1]. Many of the proteins that interact with steroid receptor complexes contain tetratricopeptide repeat (TPR) motifs. These motifs consist of degenerate 34 amino acid sequences and 1–16 adjacent motifs form a TPR domain. The domains contain a binding groove, which has the potential to bind specifically to partner proteins [2]. Consequently, TPR proteins mediate many important protein-protein interactions within the cell [3]. Heat shock protein-organizing protein (Hop) is the TPR protein whose role in steroid receptor maturation is best understood. Hop’s function is to replace the receptor bound heat shock protein 70 (hsp70) with heat shock protein 90 (hsp90) during the early stages of receptor maturation. This exchange is mediated by Hop’s TPR domains which include a hsp70 specific TPR1 and a hsp90 specific TPR2A domain [4]. The role played by Hop’s TPR2B domain in steroid receptor maturation is unclear, but it may play a role in HOP binding to hsp70 [5]. Hop then dissociates from the maturation complexes and is not normally found in mature steroid receptor complexes [1].
Several TPR proteins compete for the same binding site in the complex during the later stages of glucocorticoid receptor maturation. This group of TPR proteins includes the large immunophilins cyclophilin 40 (Cyp40), FKBP52 and FKBP51 and protein phosphatase 5 (PP5). [6]. It is becoming clear that, although the immunophilins bind to receptor complexes via hsp90, their roles are not interchangeable [7]. FKBP52 up-regulation results in an increase in the hormone binding affinity of the glucocorticoid receptor complex [7]. Conversely, high cellular levels of FKBP51 inhibit hormone binding to the receptor [8]. The immunosuppressive drug FK506 appears to modulate the glucocorticoid maturation pathway by inducing the replacement of FKBP51 with PP5 in the steroid receptor complex. This step is believed to relieve the FKBP51 inhibition of steroid hormone binding [7]. The identity of the TPR protein in the glucocorticoid receptor complex has also been shown to determine the subcellular location of the receptor, with FKBP52 complexes being found principally in the nucleus and FKBP51 complexes mainly in the cytoplasm [9]. Consequently it appears to be physiologically important for the maturation complex to be able to ensure that a specific TPR protein binds to the C-terminus of hsp90.
PP5 possesses little catalytic activity in its basal state. This is due to interactions between PP5’s N- and C-terminals limiting substrate access to the catalytic site [10, 11]. The autoinhibition can be removed by the addition of long chain fatty acids [12, 13], long chained acyl CoA esters, the 12 kDa C-terminal domain of hsp90 (C-90) [14] and full length hsp90 [11]. Although relatively high concentrations of the peptide are required, an 8 residue peptide composed of hsp-90’s C-terminal sequence (hsp90 C-8) is also capable of activating PP5 [11]. PP5 is believed to modulate the activity of DNA-PKcs, ATM kinase and ATR kinase, three kinases that act as checkpoint proteins in DNA repair [15–18]. It has also been demonstrated that PP5 protects the cell from apoptosis following oxidative stress [19]. Unfortunately, the role played by PP5 in glucocorticoid receptor maturation is unclear. The PP5 bound to the complex may be acting as a phosphatase, anchoring the receptor complex to cytosolic dynein [20] or merely acting as a protein capable of displacing the inhibitory FKBP51 protein from the maturation complex [7].
The crystal structures of many of the TPR proteins involved in glucocorticoid receptor maturation have been resolved. The X-ray data indicate that most of the TPR binding domains consist of 3 adjacent TPR motifs, with each motif normally possessing 2 anti-parallel α helices with a tight turn between helices. [4, 21–24]. Although PP5 TPR binds to hsp90 and Hop TPR1 binds to hsp70, their structures are almost superimposable [11]. Most of the TPR proteins involved in steroid receptor maturation bind to the steroid receptor complexes via the C-terminal domain of a heat shock protein [1]. Mutation studies indicate that the EEVD and MEEVD residues, found at the C-termini of hsp70 and hsp90 respectively, are essential for heat shock protein binding to the TPR domains [25–27]. The data also demonstrate that both the side chain and terminal carboxylic groups of hsp90’s C-terminal aspartate are essential for TPR binding, suggesting that the heat shock protein binds to the TPR domains via a two carboxylate clamp. The investigation of hsp90/Hop TPR2A interactions indicated that the adjacent valine is also essential for binding [27]. A second series of mutation studies were performed using hsp90 C-90. As expected, the studies indicated that the mutation of the terminal EEVD sequence to AAVA removed C-90’s ability to bind to PP5, FKBP52 and Hop [26]. These studies also indicated that the replacement of the aspartate and glutamate residues located in the −8 to −12 region of the C-terminal motif by alanine resulted in the marked diminishment of C-90’s ability to bind to all 3 TPR proteins. Mutation of C-90’s E651 and E653 residues resulted in diminished C-90 binding only to Hop [26]. Mutant studies have also indicated that PP5’s TPR domain contains 4 positively charged residues that play major roles in hsp90 binding [28]. Not surprisingly, the binding domains of other TPR proteins involved in glucocorticoid receptor maturation also possess 4 positively charged residues that correspond to those found in PP5 TPR [26]. The discrete roles played by the hsp90 binding TPR proteins in the steroid receptor maturation process suggest that these proteins possess unique secondary recognition sites that impart binding specificity. Both FKBP52 and FKBP51 possess residues near the C-terminal, a short distance from the TPR domain, that interact with hsp90 [29].
It appears that the heat shock proteins’ C-terminal domains play an essential role in hsp/TPR interactions. One of the aims of the current study is to use phage display to investigate the roles played by residues near the C-termini in these interactions. Unfortunately, hsp90’s terminal carboxylic group is essential for TPR binding [27]. This meant that it was not feasible to utilise many of the bacteriophages commonly used in phage display, such as M13, because they express random peptides with a free terminal amino, not carboxylic, group. Consequently, we utilized the bacteriophage T7, which expresses random peptides with a free carboxyl terminus on its viral coat [30], in the phage display experiments.
METHODS AND MATERIALS
Creation of a random T7 phage library
An oligonucleotide containing 12 NNK triplets and flanked by a 5' EcoRI site and a 3' HindIII site was synthesized. The complementary strand was created using the Klenow fragment (Promega) and then cleaved at the HindIII and EcoRI sites before dephosphorylation with alkaline phosphatase (Promega). Pre-cut T7 415-1b arms (Novagen) were phosphorylated using T4 polynucleotide kinase (Promega) and ligated to the duplex. The ligated DNA was then packaged using reagents from Novagen. After titering with the plaque assay, the library was amplified using the method of Castillo et al. [31].
Creation of a second generation T7 library
Phage#1 (Figure 1) was used as the parent for a second generation library using the 70:10:10:10 method [32]. At a given position, the method uses a mixture consisting of 70% of the nucleotide appearing in the parent and 10% of each of the other 3 nucleotides. The polynucleotide sequences were then annealed to the primer and the random T7 library synthesis method, described above, was followed.
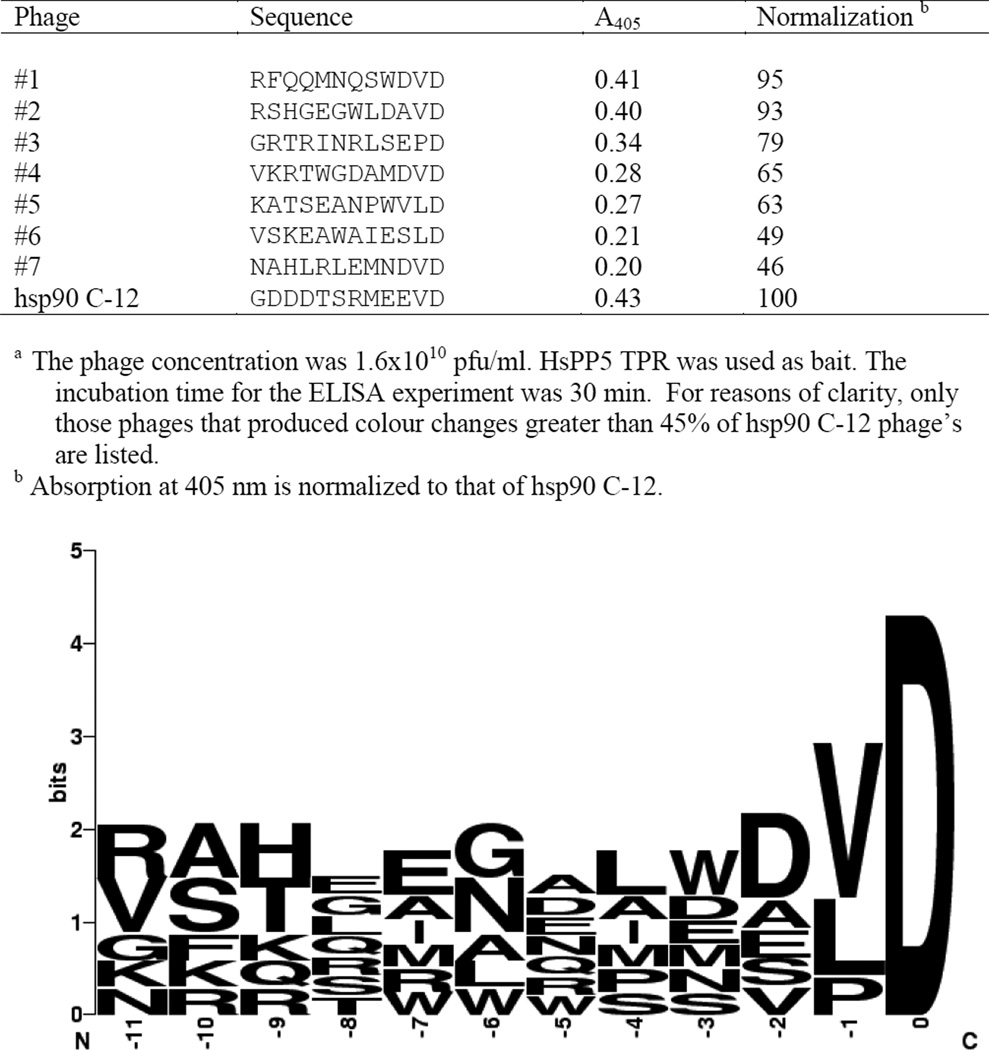
Figure 1.
The interaction of HsPP5 TPR with the VD phages using ELISA a.
A control phage was created by synthesising a duplex that encoded the GDDDTSRMEEVD sequence found at the C-terminal of hsp90 and ligating it with the pre-cut T7 415-1b arms (Novagen).
Biopanning
Wells of an Immobilizer Amino 96-well plate (NUNC) were coated with 1 µg of the target protein in 200 mM sodium hydrogen carbonate and the plate incubated overnight at 4°C. The wells were then extensively washed with phosphate buffered saline + 0.05% Tween 20 (PBST). A sample of a phage library was then added to the wells and the plate was incubated at room temperature for 2 h. The wells were washed thoroughly before log phase E. coli strain BL21 was applied to each well. The plate was then incubated at 37°C for 30 min before the E. coli was extracted and 0.1 ml of the infected bacteria added to 50 ml of a fresh sample of log-phase BL21 cells. The E. coli were incubated with shaking at 37°C until lysis of the 50 ml sample occurred. Meanwhile, the phage concentration of the bacterial sample removed from the plate was determined by plaque assay. Once the bacterial sample had lysed, it was centrifuged at 8000g to remove bacterial debris and the phage concentration of the clarified lysate was determined. The following day, the biopanning process was repeated, applying a sample of the partially purified phage, obtained during day 1, to a fresh batch of immobilized target protein. This procedure continued until the ratio of phage extracted from plate:phage loaded to plate remained constant. This process took approximately 5 rounds of biopanning.
Determination of phage concentration
Phage concentrations were determined using plaque assays [31]. Briefly, a mixture of melted agarose, E. coli and the desired dilution of a phage sample were applied to a warmed agar plate. The plate was then incubated until a bacterial lawn and T7 plaques appeared. The number of plaques on the plate, the phage sample dilution and the volume of phage sample applied to the plate were then used to calculate the phage concentration in the stock sample. In our hands the method worked within a very narrow temperature window. Agarose stocks that were too hot resulted in the underestimation of phage concentration due to T7 thermoliability. However, the use of cooler agarose samples resulted in the agarose/phage solution beginning to solidify before it could be evenly spread. We increased the temperature window by using low melting point agarose and then incubating overnight at room temperature to allow plaque formation.
Insert sequencing
A stab taken from a T7 plaque was added to 100 µl of 10 mM EDTA, pH 8.0. The sample was heated for 10 min at 65°C, allowed to cool and then clarified by centrifugation. The clarified sample was used as the template in a Hotmaster Taq (Eppendorf) PCR step using the following primers: 5'-GGCCTGTTCATGCACCGCTCTGCG-3' and 5'-CAGGTTGGAGTAAACATCAGGTAG-3'. The PCR product was then sequenced by Lone Star Labs, Inc.
Numbering of peptide sequences
The residues in a peptide sequence are identified by their distance from the C-terminal. For instance, the MEEVD sequence found at the C-terminus of hsp90 is numbered D 0, V-1, E-2, E-3 and M-4.
ELISA studies
1 µg per well of the target protein in 200 mM sodium hydrogen carbonate was loaded into wells of an Immobilizer Amino 96 well plate. After overnight incubation at 4°C, the plates were washed with PBST (used in all the wash steps). Phage samples were loaded into the wells and the plate was incubated at room temperature for 2 h. Unbound phage was removed from the wells before the plate was extensively washed. Mouse anti-T7 IgG (Novagen) was then added to each well and the plate incubated for 1 h at room temperature. The antibody was removed and the plate washed before rabbit anti-mouse IgG–alkaline phosphatase (Novagen) was added to each well. The plate was again incubated for 1 h at room temperature, the antibody removed and the wells washed. At this point alkaline phosphate substrate solution (Sigma) was added to each well and the plate stored in the dark at room temperature for a set time. The wells’ absorption at 405 nm was then determined.
The amino acid sequence data were collated and visualised using the WebLogo program [33]. Where appropriate, figures include the WebLogo analysis of the results located beneath the ELISA data. The size of a letter representing an amino acid is a function of the frequency at which the residue appears at a particular position in the incorporated table.
Construction of human TPR proteins and TPR domains
PCR methodology was used to engineer restriction sites at the 5' and 3' terminals of cDNA constructs encoding the desired proteins. The cDNAs were ligated into the TOPO 2.1 plasmid (Invitrogen), transformed into TOP10 E. coli (Invitrogen) and the plasmids sequenced to confirm the absence of point mutations. The cDNA inserts were ligated into the pET15b plasmid for bacterial expression.
In addition to full-length proteins, constructs containing the TPR binding domains were designed. HsHop TPR1 (amino acids 1–118), HsHop TPR2A (223–352), HsPP5 TPR (1–181) and HsFKBP52 TPR (255–457) were isolated as NdeI-XhoI fragments. NdeIBamHI fragments were produced encoding HsHop TPR2B (353–477), HsFKBP51 TPR (253–459) and HsCyp40 TPR (154–370).
The cDNA fragments were then ligated into pET15b that had been digested with the appropriate restriction enzymes. The plasmids were then transformed into BL21 (DE3) cells for protein expression and purification.
Cloning of rat PP5 TPR mutants
The series of rat PP5 TPR mutants in pCMV6 created by Russell et al. [28] were used as templates. Mutant RnPP5 TPR constructs in pCMV6 were transformed into DM1 cells, the plasmids extracted and digested with XhoI and StuI. The fragments containing the mutated regions of the TPR constructs were then ligated into wild-type RnPP5 TPR in pET15b. The DNA sequences were confirmed before the plasmids were transformed into BL21 (DE3) cells.
Protein purification
Protein expression in log phase BL21 (DE3) cells containing the pET15b RnPP5 TPR constructs was induced with 1.0 mM IPTG for 5 h at 30°C. Cells were harvested by centrifugation and stored overnight at −20°C. The remaining steps were performed at 4°C. The cell pellets were taken up in 20 mM Tris, 5 mM imidazole, 500 mM NaCl, pH 8.0, containing 0.5 mg/ml lysozyme, EDTA-free protease inhibitor cocktail (Roche) and 20 mM benzamidine (Sigma). Samples were stirred for 30 min before the cells were lysed by sonication and the cell debris removed by centrifugation at 18,000g for 1 h. The supernatants were loaded onto 5 ml Hi Trap chelation columns (GE Health Care) pre-loaded with NiCl2 and pre-equilibrated with 5 mM imidazole in 20 mM Tris, 500 mM NaCl pH 8.0. The His-tagged proteins were eluted with a gradient of 35–500 mM imidazole in 20 mM Tris, 500 mM NaCl, pH 8.0. The eluted RnPP5 TPR fractions were combined and dialyzed against 50 mM Hepes, 100 mM NaCl pH 7.4. A similar method was used for the human TPR proteins and TPR constructs.
RESULTS
The T7 random peptide library methodology was used to create a 1.4 ×1012 pfu library containing 4.4 × 108 unique plaques. The library was then used to biopan HsPP5 TPR. Surprisingly, a phage encoding MEEVD, hsp90’s C-terminal sequence, was not detected. Figure 1 indicates that the MEEVD sequence can be retrieved from the sequence data. Unfortunately, the motif is spread amongst 3 separate phages. The data indicate that, amongst this family of phages, the C-terminal D was the only residue to be absolutely conserved. It is noticeable that within the region −11 to −5 there was very little evidence of residue selection during the biopanning process. However, selection becomes more apparent near the C-terminus with the majority of the isolated phages possessing hydrophobic residue at positions −4 and −2. The table incorporated into Figure 1 indicates that phages #1 and #2 produced approximately the same colour change in ELISA experiments as the hsp90 C-12 control phage. This suggests that phages #1 and #2 form complexes with HsPP5 TPR that are almost as stable as the hsp90 C-12/HsPP5 TPR complex. It is noteworthy that both phages possess a V at position −1, suggesting that valine is the optimal residue at this position.
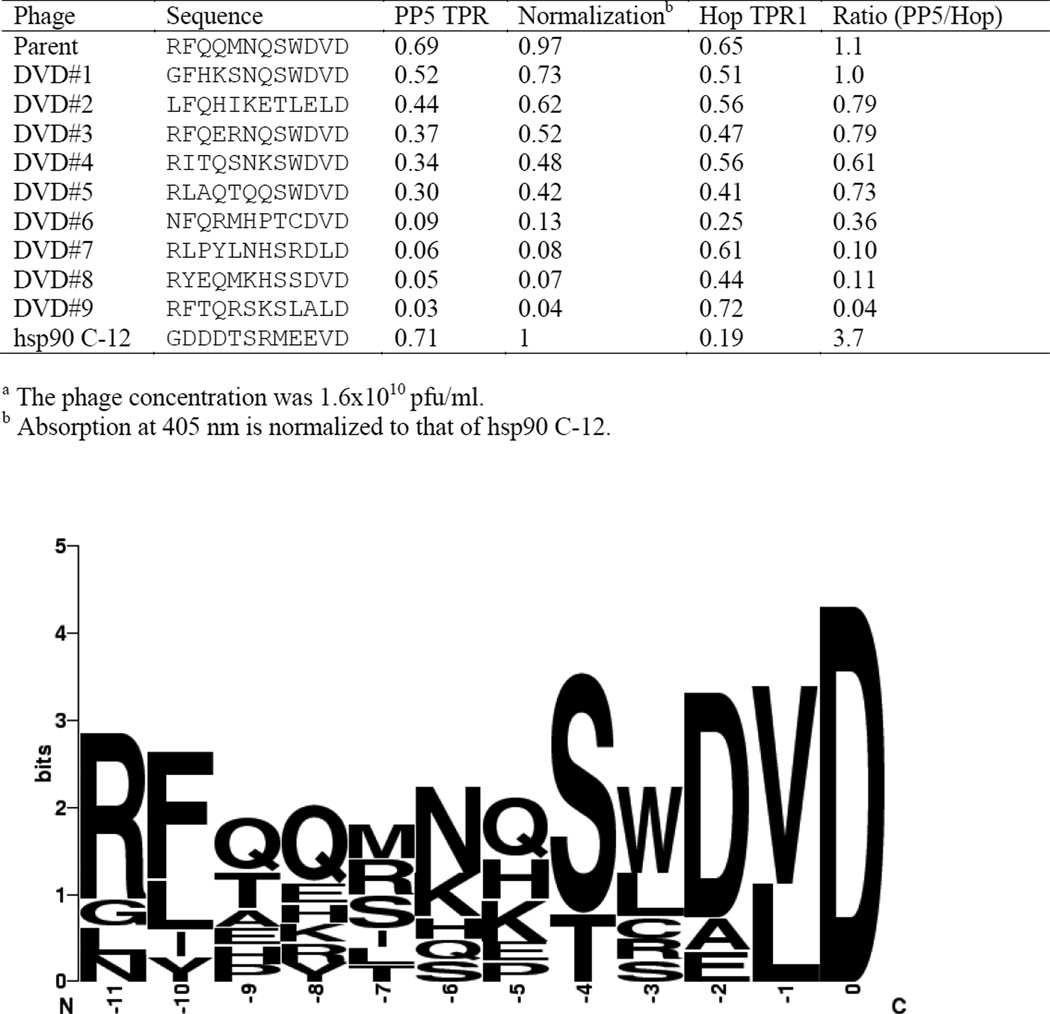
The peptide sequence displayed on phage #1 (Figure 1) was used as a template for a second-generation library created by the 70:10:10:10 method. The library contains 8.8×106 unique sequences that were amplified to form a 1.3 ×1012 pfu library and was used to biopan HsPP5 TPR (Figure 2). The isolated phages were sequenced and their ability to bind to HsPP5 TPR assayed. Not unexpectedly, sequencing revealed that many of the tight binding phages express peptides with a C-terminus very similar to the parental sequence. The data indicate that again all the isolated peptides possessed a C-terminal D, a hydrophobic residue at −1 and predominately possess a negatively charged residue at position −2. However, phages isolated during biopanning with the second generation phage library possessed some sequence homology away from the C-terminus. R and S were the most common residues at positions −11, −10 respectively and only S or T occurred at −4. The majority of phages in the region between −9 and −5 possessed residues that are either positively charged or contained an amide side group. While the parental phage possessed several residues that act as H-bond donors, none of residues in this region are charged. This finding was unexpected as hsp90 possesses aspartate residues in positions −10 to −8. Surprisingly, all of the HsPP5 TPR binding phages obtained from the 70:10:10:10 biopanning appeared to show a relatively high affinity for the hsp70 specific Hop TPR1 domain. Indeed many of these phages appeared to possess a much higher affinity for Hop TPR1 than for the hsp90 specific HsPP5 TPR domain.
Figure 2.
The specificity of the second generation library using human TPR domainsa.
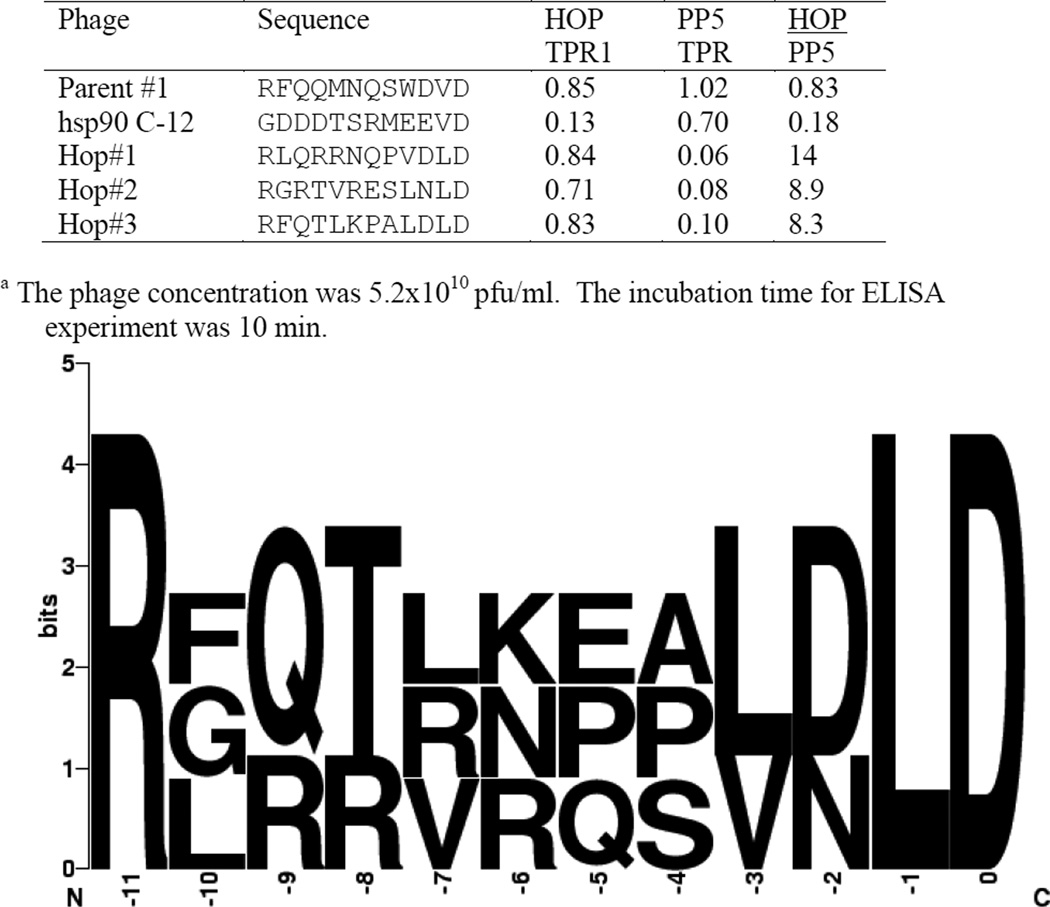
We investigated this finding by using the second-generation library to biopan the hsp70 specific HsHop TPR1 domain (Figure 3). The phages obtained from this biopanning experiment were surprisingly similar to those obtained when HsPP5 TPR was used as bait. Figure 3 demonstrates that while the position 0 D was the only residue to be completely conserved, the residues located in position −1 are either V or L, position −2 is mainly D and position −3 is largely hydrophobic. It is noteworthy that the residues in positions −9 to −5 again contain mainly positively charged or amide side groups. It is apparent that the phages obtained in this biopanning experiment have a higher affinity for HsHop1 TPR than for HsPP5 TPR. The five peptide sequences that demonstrated the highest binding specificity possessed a LD C-terminal sequence. Three phages showing a high degree of specificity were further quantitatively examined (Figure 4) using the hsp90 C-12 phage and the parental phage (#1) as controls. The figure confirms that all three phages act as hsp70 analogues. Interestingly, the three LD phages appear to have approximately the same affinity for HsHop TPR1 as the parental phage. Previous studies have suggested that hsp90’s ability to bind to Hop TPR2A (hsp90 specific) and not to Hop TPR1 (hsp70 specific) is due to the presence of a methionine at position −4 [4, 27]. Consequently, the ability of an HsPP5 TPR binding phage with a −4 methionine, isolated during the initial biopanning experiment, to bind to the HsHop TPR domains and to HsPP5 TPR was investigated (Table 1). Contrary to expectations not only could this phage form a stable complex with HsHop TPR1, but it appeared to have a low affinity for HsHop TPR2A.
Figure 3.
The binding specificity of HsTPR1 T7 phagesa.
Figure 4.
The specificity of T7 phage obtained by bio-panning HsHop TPR1 with the second generation library monitored at 405 nma.
Table 1.
Binding specificity of T7 phage with Met at −4 position monitored at 405 nm a.
| Phage | Sequence | Hop TPR1 |
Hop TPR2A |
Hop TPR2B |
PP5 TPR |
|---|---|---|---|---|---|
| #1 | RFQQMNQSWDVD | 0.43 | 0.27 | 0.40 | 0.83 |
| #7 | NAHLRLEMNDVD | 0.32 | 0.04 | 0.27 | 0.56 |
| hsp90 C-12 | GDDDTSRMEEVD | 0.01 | 0.52 | 0.24 | 0.75 |
The phage concentration was 2.0 × 1010 pfu/ml. The incubation time in the ELISA experiment was 30 min.
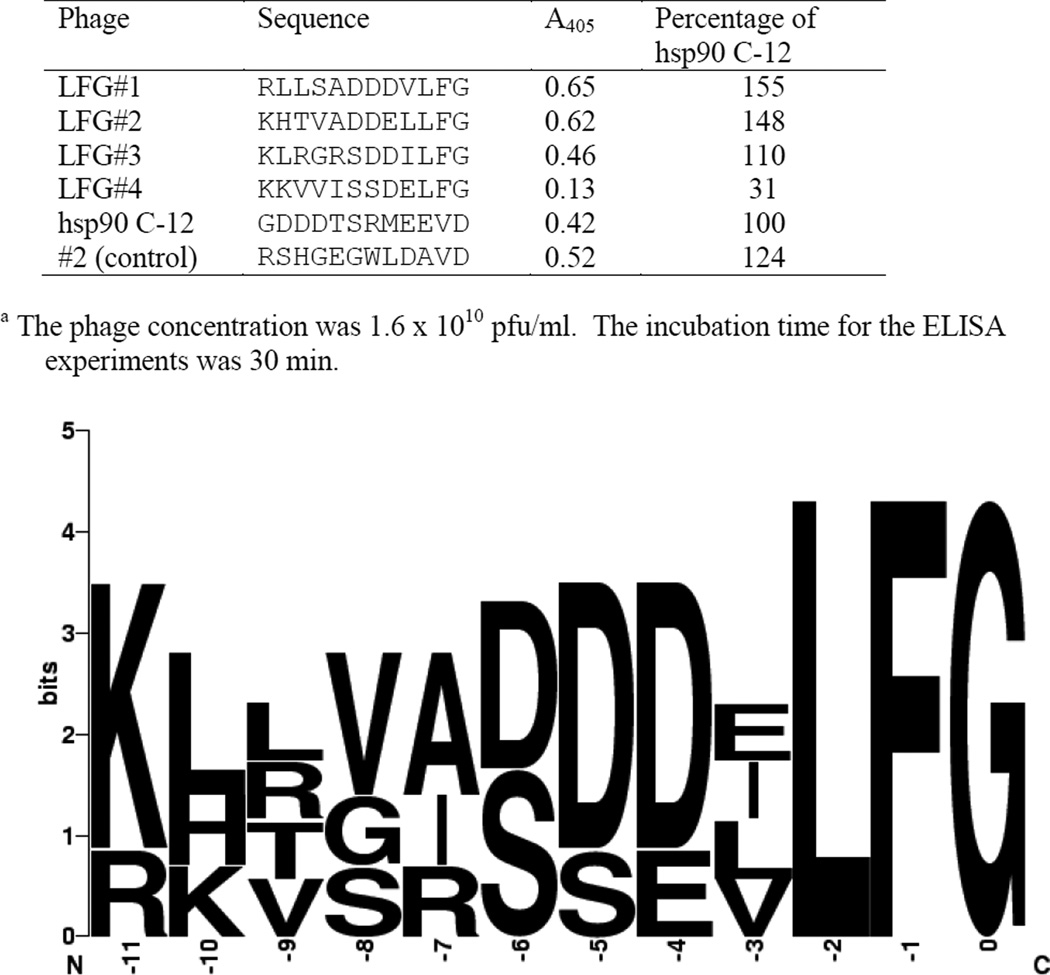
The initial biopanning experiment also produced a large number of copies of phages that possess a LFG C-terminal sequence (Figure 5). These sequences appear to have 3 regions. The residues 0 to −2 are uncharged, the residues located at −4 to −6 possess either a negatively charged or uncharged residues and there is a high concentration of positively charged residues at positions −11 and −10. Subsequent ELISA experiments (Figure 5) indicate that three of the phages appear to form a more stable complex with HsPP5 TPR than the hsp90 C-12 phage. The LFG phages exhibit a much greater degree of homology than the VD phages suggesting that more residues in the LFG sequence are involved in interactions with HsPP5 TPR than with the VD sequences. The binding specificities of VD phages #1 and #2 (Figure 1) and LFG phage #1 (Figure 5) were then investigated using a broader range of TPR proteins involved in steroid receptor maturation. The TPR proteins and their TPR domains were cloned, expressed and purified. (A full length Cyp40 construct was not used in this study due to proteolysis problems encountered during purification.)
Figure 5.
Binding of LFG phages to HsPP5 TPRa.
Table 2 demonstrates that the hsp90 C-12 phage formed a stable complex with HsPP5 TPR and HsHop TPR2A but was unable to form a stable complex with HsHop TPR1. This finding is consistent with published data [4]. However, the data also indicated that the hsp90 C-12 phage was able to form a stable complex with HsHop TPR2B. This was surprising as a Hop mutant containing Hop TPR2B, but not Hop TPR1 or 2A, was unable to pull down hsp90 [34]. Table 2 indicates that the two VD phages and the hsp90 C-12 control phage produced similar colour changes in the ELISA experiments when full-length HsPP5 or full length Hop was used as bait. Phage #2 possessed some of the expected specificity, as it appeared to exhibit a higher affinity for HsPP5 TPR and HsHop TPR2A&2B domains than the hsp70 binding HsHop TPR1 construct. Unfortunately, phage #1 produced much higher ELISA signals when the hsp70 specific HsHop TPR1 domain was used as bait than the hsp90 binding HsHop TPR2A&B domains. The LFG phage was the one new construct to show significant binding specificity during the experiment. This phage produced large increases in A405 in wells containing HsPP5 TPR. Replacing the HsPP5 TPR with HsHop TPR2A or 2B halved the colour change and the phage did not appear to be able to form a stable complex with HsHop TPR 1. Table 2 also indicates that large increases in A405 were not observed in wells containing full-length HsPP5. This is probably a function of the dissociation constant for hsp90/PP5 TPR interactions being significantly lower than for hsp90/full-length PP5 interactions [11]. Surprisingly, the table demonstrates that large colour changes were not observed in the ELISA experiments when any of the large-immunophilin TPR constructs were used as bait.
Table 2.
The binding specificity of HsPP5 TPR binding T7 phages monitored at 405 nma.
| Phage Sequence |
#1 RFQQMNQSWDVD |
#2 RSHGEGWLDAVD |
hsp90 C-12 GDDDTSRMEEVD |
LFG#1 RLLSADDDVLFG |
|---|---|---|---|---|
| Protein | ||||
| FL PP5 | 0.10 | 0.17 | 0.08 | 0.02 |
| PP5 TPR | 0.88 | 0.60 | 0.58 | 0.48 |
| FL Hop | 0.74 | 0.56 | 0.66 | 0.00 |
| Hop TPR 1 (70)b | 0.75 | 0.30 | 0.00 | 0.00 |
| Hop TPR 2A | 0.26 | 0.82 | 0.65 | 0.20 |
| Hop TPR 2B | 0.22 | 0.58 | 0.58 | 0.22 |
| FL FKBP51 | 0.00 | 0.00 | 0.00 | 0.00 |
| FKBP51 TPR | 0.05 | 0.09 | 0.09 | 0.01 |
| FL FKBP52 | 0.00 | 0.00 | 0.00 | 0.00 |
| FKBP52 TPR | 0.02 | 0.05 | 0.02 | 0.03 |
| Cyp40 TPR | 0.04 | 0.12 | 0.03 | 0.01 |
The phage concentration was 1.7 × 1010 pfu/ml. The incubation time for the ELISA experiments was 30 min.
Hop TPR1 is hsp70 specific, the other proteins and constructs are capable of binding to hsp90.
Since phage #1 and the LFG phage #1 display significantly different binding specificities than the hsp90 C-12 control phage, the possibility exists that the newly isolated phages bind to different regions of PP5 TPR than the control phage. A range of RnPP5 TPR mutants, originally described by Russell et al. [28], was subcloned into a bacterial expression vector and the purified recombinant proteins used to test this hypothesis (Table 3). The data indicate that the hsp90 C-12 phage requires K32, R74, K97 and R101 to produce the maximal colour change in the ELISA experiments. This finding is consistent with previous investigations of hsp90 binding to RnPP5 TPR [28]. The data also indicate that, of the residues mutated in this study, only K97 is required for binding to phage #1. This suggests that phage #1 and the hsp90 C-12 phage do not bind to identical regions of the TPR domain. However, none of the mutated PP5 TPR residues appear to be essential for the binding of either of the two LFG phages. This suggests that the LFG and hsp90 C-12 peptide sequences may be interacting with different regions of the RnPP5 TPR construct.
Table 3.
Identification of RnPP5 TPR residues required for optimal T7 phage binding a.
| Phage Sequence |
#1 RFQQMNQSWDVD |
hsp90 C-12 GDDDTSRMEEVD |
LFG#1 RLLSADDDVLFG |
LFG#2 KHTVADDELLFG |
|---|---|---|---|---|
| Wild Type | 1.14 | 1.26 | 0.95 | 0.89 |
| E29A | 0.91 | 1.06 | 0.77 | 0.75 |
| K32A | 0.96 | 0.09 | 0.74 | 0.76 |
| K40A | 1.01 | 0.95 | 0.85 | 0.90 |
| E56A | 0.96 | 1.08 | 0.78 | 0.77 |
| I63A | 0.84 | 0.83 | 0.79 | 0.83 |
| R74A | 0.75 | 0.24 | 0.69 | 0.75 |
| E76A | 0.98 | 1.12 | 0.89 | 0.93 |
| Y80A | 1.08 | 1.09 | 0.76 | 0.73 |
| K97A | 0.15 | 0.02 | 0.60 | 0.62 |
| R101A | 0.91 | 0.00 | 0.53 | 0.54 |
| R113A | 0.84 | 0.72 | 0.66 | 0.74 |
| R117A | 1.00 | 1.06 | 0.80 | 0.78 |
The phage concentration was 1.5 × 1010 pfu/ml. The incubation time for the ELISA experiments was 30 min.
DISCUSSION
This investigation utilized a pair of complex phage libraries. Consequently, it was slightly surprising that the initial HsPP5 TPR biopanning experiment produced a series of phages with a degree of homology being observed only in the 2 residues at the C terminus. This strongly suggests that the −1and 0 residues were the only ones to play important roles in the binding of this family of phages to HsPP5 TPR. However, this finding is in broad agreement with previous studies that used physical chemical techniques to study TPR/hsp90 interactions. Brinker et al.’s [27] first mutation study of heat shock protein binding to Hop TPR1 and TPR2A indicated that hsp90’s VD C-terminal motif was essential for Hop TPR2A binding and consequently did not include positions 0 and −1 in their more detailed studies. An investigation of hsp90 C-5/PP5 TPR interactions using NMR techniques [35] produced similar results. The data indicated that the terminal aspartate is bound to the TPR domain at all times and that the −1 valine interacts with an adjacent hydrophobic pocket within the TPR domain. The investigation found that, although the glutamate residues in the C-terminal MEEVD sequence may interact with the TPR domain via transient salt bridges, these residues do not form stable contacts with PP5. This has led Cliff et al. [35] to postulate that the tight binding observed between PP5 TPR and hsp90 C-5 is largely the result of interactions between hsp90’s terminal aspartate and the TPR domain. Brinker et al. [27] also demonstrated that amongst the residues in the TSRMEE sequence, located at positions −2 to −7 of the C-terminus of hsp90, only the −3 glutamate was required for optimal binding to Hop TPR2A. Their data indicate that the presence of negatively charged residues in positions −2 and −3 in hsp90 C-8 enhanced binding to Hop TPR2A. However, with the exception of position −3, large uncharged residues, such as tryptophan, in positions −2 to −7 were also capable of enhancing hsp90 C-8 binding to Hop TPR2A. Our initial HsPP5 TPR biopanning experiment produced a significant number of phages expressing peptide sequences containing the terminal VD motif and with predominantly the combination of uncharged and negatively charged residues in positions −2 to −7 predicted by Brinker et al. [27].
The second-generation library used a VD phage selected for its ability to bind to HsPP5 TPR as a template. However, it proved to be far easier to isolate phages from this library that acted as hsp70 analogues rather than as hsp90 derivatives (Figures 2 and 3). This may be due to Hop TPR1 possessing a greater affinity for peptides containing a position −2 aspartate than the hsp90-binding TPR domains [27]. It appears likely that hsp70-specific Hop TPR1 and the hsp90-specific Hop TPR2A domains have evolved through duplication of a common ancestor [36]. Consequently, it is not that surprising that the hsp70 specific HopTPR1 domain was able to bind to some of the hsp90 analogues. The observation that phage #7, a VD phage with a −4 methionine, produced a significant colour change in ELISA experiments when Hop TPR1 but not when Hop TPR2A was used as bait (Table 1) was most unexpected. However, the presence of the −2 aspartate may explain the phage’s affinity for Hop TPR1, although its inability to produce a significant colour change when Hop TPR2A was used as bait remains a mystery. Figures 3 and 4 indicate that the replacement of the −1 valine with leucine produces peptides that are able to bind strongly to HsHop TPR1 but not to HsPP5 TPR. This might have been discovered by Brinker et al. [27], but they did not mutate the terminal VD sequence during their more detailed competition screening experiment. This serendipitous finding will enable a range of peptides to be designed that will bind specifically to hsp70 binding proteins. However, the binding specificity appears to be a little more complicated. Phage DVD #2, which contains the LD motif, binds strongly to both HsPP5 TPR and HsHop TPR1 (Figure 2). It may be significant that DVD #2’s C-terminal motif is ELD rather than the more common DVD.
The possibility that the high affinity binding observed when the LFG phages are added to HsPP5 TPR is merely a biopanning artefact cannot be discounted. p-Nitrophenol phosphate assays indicated that the C-12 peptide found on the coat of LFG phage #1 was unable to activate HsPP5 (data not shown). However, the isolation of a family of LFG phages with closely related sequences makes this possibility a lot less likely. Additionally, Table 2 indicates that the LFG phage #1 produces a large colour change in ELISA experiments only when HsPP5 TPR is used as bait. This strongly suggests that the phage is binding specifically to this construct. It may also be significant that while the LFG phage #1 and hsp90 C-12 peptide sequences have very different C-terminal residues, both sequences have a grouping of three negatively charged residues a short distance away from the C-terminus. Yang et al.’s molecular modelling [11] predicts that the residues at the extreme C-terminus of hsp90 interact with PP5 TPR’s binding groove. A significant portion of the outer surface of this TPR domain is positively charged [21] and it is possible that the DDD grouping found within both the hsp90 C-12 phage and the LFG phage#1 is interacting with this surface [11]. This theory is supported by our previous mutation studies that suggested that the DDD grouping played a major role in hsp90 binding to PP5 [26]. Scheufler et al. [4] reported that the GDDD sequence, located in positions −8 to −11 of hsp90, was responsible for lowering the Kd for Hop TPR2A/hsp90 C-terminal domain interactions from 33 µM (for C-8) to 8 µM (for C-12). Figures 1&2 indicate that there are many peptide sequences that are capable of acting as hsp90 analogues. It was initially surprising that the C terminal MEEVD sequence was not obtained during the two biopanning experiments that used HsPP5 TPR as bait. It may be significant that Figure 2 indicates that the hsp90 analogues isolated in the second biopanning experiment were also able to bind relatively strongly to the hsp70 specific HsHop TPR 1 domain. The possibility exists that although many peptide sequences are capable of binding to PP5 TPR, the peptide sequence GDDDTSRMEEVD, located at the C-terminus of hsp90, has evolved because it is capable of binding to the hsp90 dependant binding domains while possessing a low affinity for hsp70 specific domains. It appears highly likely that regions of heat shock proteins a little distance away from the C-terminus play an important role in the formation of stable heat shock protein/TPR complexes [37]. Previous studies have indicated that sites located in both the C-terminal and middle domains of hsp90 interact with Hop [38]. Mutation of hsp90’s E651 and D653 residues greatly reduced the stability of the C-90/Hop complex [26]. This makes the E651-D653 region an obvious candidate for the secondary site of either Hop TPR2A or 2B binding. The data of Chen et al. [39] support this theory. They produced a series of hsp90 mutants and measured their ability to bind to Hop, Cyp40, FKBP52 and FKBP51. They found that the replacement of the terminal EEVD sequence with AAVD greatly diminished the ability of hsp90 to bind to all 4 TPR proteins. Interestingly, they also reported that mutating residues some distance away from the C-terminus could specifically diminish the stability of some of the TPR/hsp90 complexes. Although the existence of a secondary binding site within the PP5 TPR domain has not been confirmed, the possibility that the LFG peptides are interacting with a secondary site remains tantalizing.
Very weak binding was observed during ELISA studies when the large-immunophilin TPR constructs were used as bait. This was probably due to the peptide sequences encoded on the phage coat being too short to form a stable complex with these constructs. Previous binding experiments utilized hsp90 sequences much larger than the 12 residue inserts used in this study. The size difference did not appear to be important when using the HsHop TPR domains or HsPP5 TPR as bait because these constructs appear to bind tightly to the C-terminus of hsp90. However, hsp90 appears to interact with both the immunophilins’ TPR motifs and to a region of their C-terminal sequence a short distance away from their TPR domains [29, 40]. Additionally, there is evidence that FKBP52 binds to a site near hsp90’s N-terminus [41].
Our experimental data suggest the presence of very limited TPR binding specificity amongst the hsp90 C-12 and VD phages. It seems highly likely that the C-terminus of hsp90 acts as a universal anchor, enabling Hop TPR2A&B and PP5 TPR domains to bind to this region with similar affinities. This indicates that hsp90/TPR protein binding specificity appears to be essential for steroid receptor maturation, is due to interactions at weaker secondary binding sites. This model also suggests that the major difference between the complexes formed by hsp90 with non-immunophilin and immunophilin TPR domains is the relative importance of the secondary binding sites in stabilising the complexes.
ACKNOWLEDGMENTS
The authors thank Dr. Brian K. Kay of the Argonne National Laboratory for his guidance in the production of the random phage library. This work was supported by the National Institutes of Health Grant DK 55877 (to M.C.).
Abbreviations
- TPR
tetratricopeptide repeat
- HsPP5
human protein phosphatase 5
- RnPP5
rat protein phosphatase 5
- Cyp40
cyclophilin 40
- hsp90
heat shock protein 90
- hsp70
heat shock protein 70
- FL
full length
- C-90
the 12 kDa C-terminal domain of hsp90
- hsp90 C-12
the 12 residues located at the C-terminus of hsp90
- pfu
plaque forming units
REFERENCES
- 1.Pratt WB, Toft DO. Regulation of signalling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 2.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 5.Carrigan PE, Sikkink LA, Smith DF, Ramirez-Alvarado M. Domain:domain interactions within Hop, the Hsp70/Hsp90 organizing protein, are required for protein stability and structure. Protein Sci. 2006;15:522–532. doi: 10.1110/ps.051810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratt WB, Galigniana MD, Harrell JM, DeFranco DB. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell. Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Davies TH, Ning YM, Sanchez ER. Differential control of glucocorticoid receptor hormone-binding function by tetratricopeptide repeat (TPR) proteins and the immunosuppressive ligand FK506. Biochemistry. 2005;44:2030–2038. doi: 10.1021/bi048503v. [DOI] [PubMed] [Google Scholar]
- 8.Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee A, Periyasamy S, Wolf IM, Hinds TD, Yong W, Shou W, Sanchez ER. Control of glucocorticoid and progesterone receptor subcellular localization by the ligand-binding domain is mediated by distinct interactions with tetratricopeptide repeat proteins. Biochemistry. 2008;47:10471–10480. doi: 10.1021/bi8011862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair C, Borchers C, Parker C, Tomer K, Charbonneau H, Rossie S. The tetratricopeptide repeat domain and a C-terminal region control the activity of Ser/Thr protein phosphatase 5. J. Biol. Chem. 1999;274:23666–23672. doi: 10.1074/jbc.274.33.23666. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Roe SM, Cliff MJ, Williams MA, Ladbury JE, Cohen PTW, Barford D. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 2005;24:1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MX, Cohen PTW. Activation of protein phosphatase 5 by limited proteolysis or the binding of fatty acids to the TPR domain. FEBS Lett. 1997;400:136–140. doi: 10.1016/s0014-5793(96)01427-5. [DOI] [PubMed] [Google Scholar]
- 13.Skinner J, Sinclair C, Romeo C, Armstrong D, Charbonneau H, Rossie S. Purification of a fatty acid-stimulated protein serine/threonine phosphatase from bovine brain and its identification as a homolog of protein phosphatase 5. J. Biol. Chem. 1997;272:22464–22471. doi: 10.1074/jbc.272.36.22464. [DOI] [PubMed] [Google Scholar]
- 14.Ramsey AJ, Chinkers M. Identification of potential physiological activators of protein phosphatase 5. Biochemistry. 2002;41:5625–5632. doi: 10.1021/bi016090h. [DOI] [PubMed] [Google Scholar]
- 15.Wechsler T, Chen BP, Harper R, Morotomi-Yano K, Huang BC, Meek K, Cleaver JE, Chen DJ, Wabl M. DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1247–1252. doi: 10.1073/pnas.0307765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughan CK, Mollapour M, Smith JR, Truman A, Hu B, Good VM, Panaretou B, Neckers L, Clarke PA, Workman P, Piper PW, Prodromou C, Pearl LH. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Molecular Cell. 2008;31:886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA damage-induced ATM activation. Genes Dev. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Bao S, Furumai R, Kucera KS, Ali A, Dean NM, Wang XF. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol. Cell. Biol. 2005;25:9910–9919. doi: 10.1128/MCB.25.22.9910-9919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita K, Saitoh M, Tobiume K, Matsuura H, Enomoto S, Nishitoh H, Ichijo H. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. EMBO J. 2001;20:6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galigniana MD, Harrell JM, Murphy PJ, Chinkers M, Radanyi C, Renoir JM, Zhang M, Pratt WB. Binding of hsp90-associated immunophilins to cytoplasmic dynein: direct binding and in vivo evidence that the peptidylprolyl isomerase domain is a dynein interaction domain. Biochemistry. 2002;41:13602–13610. doi: 10.1021/bi020399z. [DOI] [PubMed] [Google Scholar]
- 21.Das AK, Cohen PTW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J. 1998;15:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431–438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- 23.Sinars CR, Cheung-Flynn J, Rimerman RA, Scammell JG, Smith DF, Clardy J. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:868–873. doi: 10.1073/pnas.0231020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, Rao Z. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8348–8353. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey AJ, Russell LC, Whitt SR, Chinkers M. Overlapping sites of tetratricopeptide repeat protein binding and chaperone activity in heat shock protein 90. J. Biol. Chem. 2000;275:17857–17862. doi: 10.1074/jbc.M001625200. [DOI] [PubMed] [Google Scholar]
- 27.Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70·Hop·Hsp90 complexes. J. Biol. Chem. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 28.Russell LC, Whitt SR, Chen M-S, Chinkers M. Identification of conserved residues required for the binding of a tetratricopeptide repeat domain to heat shock protein 90. J. Biol. Chem. 1999;274:20060–20063. doi: 10.1074/jbc.274.29.20060. [DOI] [PubMed] [Google Scholar]
- 29.Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J. Biol. Chem. 2003;278:17388–17394. doi: 10.1074/jbc.M300955200. [DOI] [PubMed] [Google Scholar]
- 30.Dai M, Temirov J, Pesavento E, Kiss C, Velappan N, Pavlik P, Werner JH, Bradbury ARM. Using T7 phage display to select GFP-based binder. Protein Eng. Des. Sel. 2008;21:413–424. doi: 10.1093/protein/gzn016. [DOI] [PubMed] [Google Scholar]
- 31.Castillo J, Goodson B, Winter J. T7 displayed peptides as targets for selecting peptide specific scFvs from M13 scFv display libraries. J. Immunol. Methods. 2001;257:117–122. doi: 10.1016/s0022-1759(01)00454-9. [DOI] [PubMed] [Google Scholar]
- 32.Martens CL, Cwirla SE, Lee RY, Whitehorn E, Chen EY, Bakker A, Martin EL, Wagstrom C, Gopalan P, Smith CW, Tate E, Koller KJ, Schatz PJ, Dower WJ, Barrett RW. Peptides which bind to E-selectin and block neutrophil adhesion. J. Biol. Chem. 1995;270:21129–21136. doi: 10.1074/jbc.270.36.21129. [DOI] [PubMed] [Google Scholar]
- 33.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator, Genome Research, 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Prapapanich V, Rimerman RA, Honore B, Smith DF. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol. Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 35.Cliff MJ, Harris R, Barford D, Landbury JE, Williams MA. Conformational diversity in the TPR Domain-Mediated Interaction of Protein Phosphatase 5 with Hsp90. Structure. 2006;14:415–426. doi: 10.1016/j.str.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Torres JH, Papandreou N, Chumilier J. Cell Stress Chaperones. 2009;14:281–289. doi: 10.1007/s12192-008-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travers AA, Fares MA. Functional co-evolutionary networks of the hsp70-hop-hsp90 system revealed through computational analyses. Mol. Biol. Evol. 2007;24:1032–1044. doi: 10.1093/molbev/msm022. [DOI] [PubMed] [Google Scholar]
- 38.Onuoha SC, Coulstock ET, Grossmann JG, Jackson SE. Stuctural studies on the co-chaperone Hop and its complexes with hsp90. J. Mol. Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J. Biol. Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 41.Chadli A, Bruinsma ES, Stensgard B, Toft D. Analysis of hsp90 cochaperone interactions reveals a novel mechanism for TPR protein recognition. Biochemistry. 2008;47:2850–2857. doi: 10.1021/bi7023332. [DOI] [PubMed] [Google Scholar]