SUMMARY
Expression of the Down syndrome cell-adhesion molecule (Dscam) is increased in the brains of patients with several neurological disorders. Although Dscam is critically involved in many aspects of neuronal development, little is known about either the mechanism that regulates its expression or the functional consequences of dysregulated Dscam expression. Here, we show that Dscam expression levels serve as an instructive code for the size control of presynaptic arbor. Two convergent pathways, involving dual leucine zipper kinase (DLK) and fragile X mental retardation protein (FMRP), control Dscam expression through protein translation. Defects in this regulation of Dscam translation lead to exuberant presynaptic arbor growth in Drosophila somatosensory neurons. Our findings demonstrate a previously unknown aspect of Dscam function and provide insights into how dysregulated Dscam may contribute to the pathogenesis of neurological disorders.
INTRODUCTION
The Down syndrome cell-adhesion molecule (Dscam) is important for the development of neural circuits in both invertebrates and vertebrates (Fuerst et al., 2009; Fuerst et al., 2008; Millard and Zipursky, 2008; Schmucker and Chen, 2009; Yamagata and Sanes, 2008). In Drosophila, Dscam undergoes extensive alternative splicing to generate as many as 38,016 different isoforms (Schmucker et al., 2000). This diversity is critical for neurite self-recognition (Hattori et al., 2007; Hattori et al., 2008; Zipursky and Sanes, 2010). For example, loss of Dscam function results in a dramatic increase in intraneuronal dendritic crossings in the dendritic arborization (da) neurons (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007) and a failure in sister branch segregation of the axons of mushroom body neurons (Hattori et al., 2007; Wang et al., 2002).
In addition to self-recognition, Drosophila Dscam regulates synaptic target selection and axon guidance in several types of neurons (Chen et al., 2006; Hummel et al., 2003; Millard et al., 2010; Wang et al., 2002; Zhu et al., 2006). For instance, in mechanosensory neurons of the adult fly, Dscam mutants exhibit profound loss of axon terminal branches as a result of defective branch extension and target selection (Chen et al., 2006).
Despite the absence of the remarkable molecular diversity seen in insects, vertebrate Dscam is also essential for neurite self-avoidance and synaptic target selection (Blank et al., 2011; Fuerst et al., 2009; Fuerst et al., 2008; Yamagata and Sanes, 2008), suggesting that the functions of Dscam in neuron morphogenesis and circuit assembly are evolutionarily conserved (Zipursky and Sanes, 2010).
Little is known about how Dscam is regulated, but several observations suggest that its expression must be tightly controlled. Dscam expression is dynamically regulated in developing brains (Maynard and Stein, 2012; Saito et al., 2000). In mouse, Dscam protein levels peak at postnatal day 7 to 10 in the cerebral cortex, coinciding with a period of extensive axonal branching (Larsen and Callaway, 2006), and decreases after postnatal day10 (Maynard and Stein, 2012). Moreover, Dscam expression is elevated in several brain disorders, including Down Syndrome (DS) (Saito et al., 2000), intractable epilepsy (Shen et al., 2011), and bipolar disorder (Amano et al., 2008). These findings suggest that appropriate regulation of Dscam expression may be important for development, and that inappropriate or dysregulated Dscam expression may lead to developmental abnormalities and disease. However, the mechanisms that regulate Dscam expression and the function of such regulations are thus far unknown.
In the present study, we describe an important role for the regulation of Dscam expression in determining the size of the presynaptic arbor. We found that while isoform diversity of Dscam is critical for presynaptic arbor targeting, Dscam expression level determines the size of the presynaptic arbor. We further define novel regulatory mechanisms that control the size of the presynaptic arbor by regulating the translation of Dscam protein. These findings emphasize the importance of the regulation of Dscam expression during development, and the potential consequences of dysregulated Dscam expression in disease.
RESULTS
Dscam Instructs Presynaptic Arbor Growth
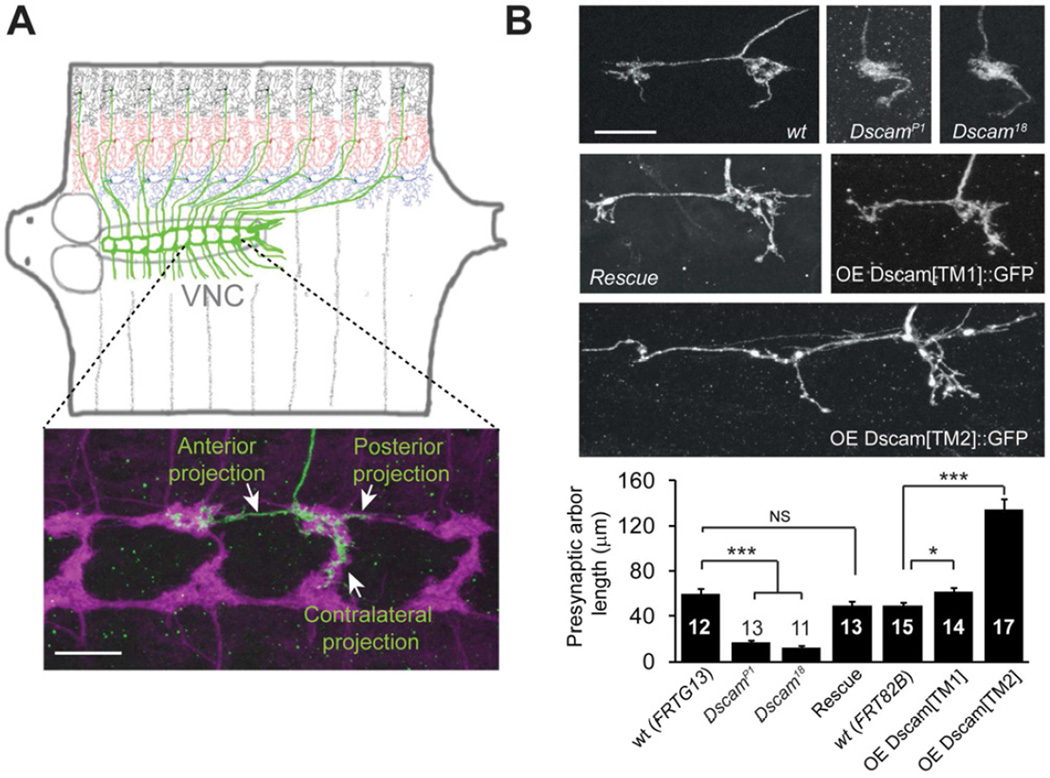
We studied the role of Dscam in presynaptic arbor development in Drosophila larval class IV dendritic arborization (C4da) neurons (Grueber et al., 2002), a system that was used to establish the function of Dscam in dendritic self-recognition (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). The cell bodies and dendrites of C4da neurons are located in the larval body wall where they sense nociceptive stimuli (Hwang et al., 2007; Kim et al., 2012; Xiang et al., 2010); the axons project to the ventral nerve cord (VNC) (Figure 1A, top). In the VNC, the axon terminal of each C4da neuron consists of anterior, posterior, and contralateral branches (Figure 1A, bottom, green). These axon terminals are presynaptic arbors, as shown by enrichment of the presynaptic marker synaptotagmin ∷ GFP (syt ∷ GFP) (Figure S1A). The presynaptic arbors of C4da neurons collectively form a ladder-like structure in the VNC (Figure 1A, bottom, magenta).
Figure 1. Dscam Instructs Presynaptic Arbor Growth.
(A) Top: Schematic representation of Drosophila larval C4da neurons. In each hemi-segment, Three C4da neurons, ddaC (black), v’ada (red), and vdaB (blue), elaborate dendrites on the body wall and send axons (green) to the VNC to form a ladder-like structure. Bottom: the presynaptic arbor of a single ddaC neuron (green). The flip-out technique was used to express the membrane GFP marker mCD8 ∷ GFP. The presynaptic terminals of C4da neurons, which express another membrane marker, mouse CD2 (magenta), collectively form a ladder-like structure. (B) Representative images and quantification of the presynaptic arbors of single C4da neurons that are wild-type (wt), null mutants of Dscam (DscamP1 or Dscam18), null mutants rescued by one copy of a transgene harboring the Dscam genomic DNA (Rescue), overexpressing the dendritic (OE Dscam[TM1] ∷ GFP) or overexpressing the axonal (OE Dscam[TM2] ∷ GFP) isoform. The MARCM technique was used in these experiments, and the arbors of single ddaC neurons are shown. Sample numbers are indicated in each bar. Scale bars: 10 mm. Also see Figure S1.
We investigated the requirement of Dscam in presynaptic arbor development by using the Mosaic Analysis with a Repressible Cell Marker (MARCM) (Lee and Luo, 1999). Single C4da neurons homozygous for Dscam null mutations, DscamP1 (Schmucker et al., 2000) or Dscam18 (Wang et al., 2002), exhibited markedly reduced presynaptic arbor growth (Figure 1B). These dramatic defects in presynaptic arbor growth were completely restored by the introduction of a transgene harboring Dscam genomic DNA (Figure 1B, Rescue), confirming that loss of Dscam function led to the observed defects.
Conversely, we found that gain of Dscam function promoted presynaptic terminal growth. Alternative splicing of Dscam mRNA generates two transmembrane domain (TM) isoforms that differ in their subcellular distribution (Wang et al., 2004). The TM1 isoform is preferentially localized in dendrites, while the TM2 isoform is preferentially localized in the axon (Wang et al., 2004). Overexpression of a Dscam transgene containing TM2 caused abnormally long presynaptic arbors, resulting in a 2.7-fold increase in presynaptic terminal length (Figure 1B, OE Dscam[TM2] ∷ GFP). In contrast, overexpression of a Dscam transgene containing TM1 caused only a 24% increase in presynaptic growth (Figure 1B, OE Dscam[TM1] ∷ GFP). These results demonstrate that Dscam plays an instructive role in the presynaptic arbor growth of C4da neurons.
The Role of Dscam in Presynaptic Arbor Growth is Independent of the Ectodomain Diversity of Dscam
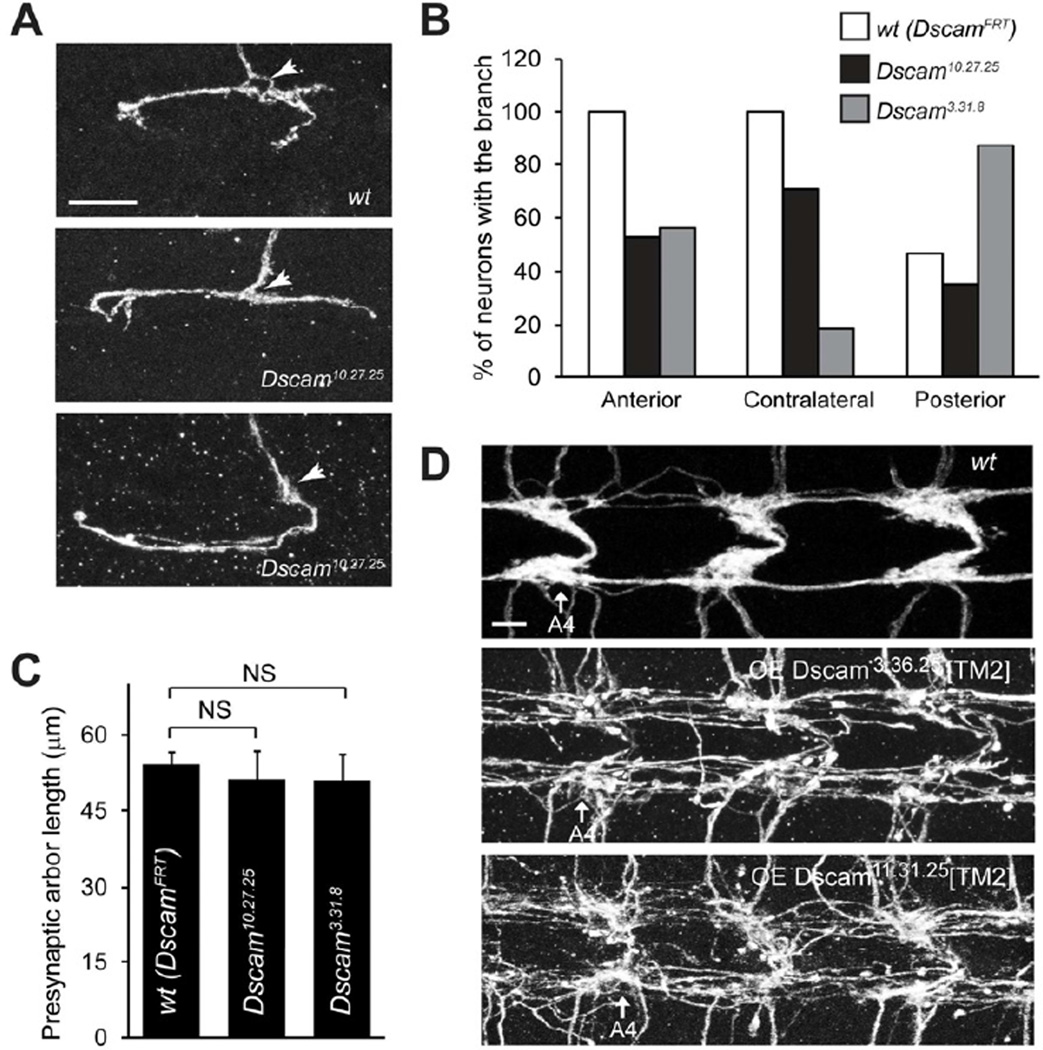
In Drosophila, Dscam mRNA undergoes extensive alternative splicing in ectodomain-encoding exons 4, 6, and 9, resulting 19,008 potential isoforms of the ectodomain (Schmucker et al., 2000). This ectodomain diversity is essential for Dscam’s known functions in neurite self-avoidance (Hattori et al., 2007; Hattori et al., 2008; Zipursky and Sanes, 2010) and axon targeting (Chen et al., 2006; Hattori et al., 2009). We wondered whether the reduced presynaptic arbor size in Dscam null mutant neurons is secondary to self-avoidance or targeting defects caused by loss of ectodomain diversity. To address this, we first used a Dscam allele with a 75% reduction in isoform diversity (Wang et al., 2004) to assess the effect of reduced diversity on presynaptic arbor development. Reducing Dscam diversity by 75% did not affect the development of presynaptic terminals in C4da neurons (Figure S2). Furthermore, we employed the intragenic MARCM technique to examine presynaptic arbor development of neurons expressing a single ectodomain isoform from the endogenous locus (Hattori et al., 2007). Importantly, Dscam expression levels in these mutants are comparable to those of wild-type (Hattori et al., 2007). C4da neurons expressing the single Dscam isoform containing exons 4.10, 6.27, and 9.25 (referred to as Dscam10.27.25) exhibited defective targeting of the synaptic terminals (Figure 2A and 2B). 47% of the Dscam10.27.25 ddaC neurons completely lost their anterior branches and 29.4% lost their contralateral branches, while 100% of wild-type control clones (referred as DscamFRT) had both branches (Figure 2A and 2B). Similar targeting defects were observed in C4da neurons homozygous of a second allele, Dscam3.31.8 (Figure 2B). Strikingly, the presynaptic arbor sizes of Dscam10.27.25 and Dscam3.31.8 neurons were indistinguishable from those of wild-type neurons (Figure 2C). These results strongly suggest that the ectodomain diversity is dispensable for Dscam-mediated control of presynaptic arbor size and that the reduced growth seen in Dscam mutant presynaptic arbors is not due to defective synaptic targeting.
Figure 2. Dscam Instruction of Presynaptic Arbor Growth is Independent of Ectodomain Diversity.
(A) Presynaptic arbors of a wild-type (DscamFRT) and two Dscam10.27.25 intragenic MARCM clones (ddaC). Arrows point to the entry points into the C4da neuropil. The middle panel shows a Dscam10.27.25 ddaC clone that lacks a contralateral branch but extends an unusually long posterior branch. The bottom panel shows a clone that lacks the anterior projection but forms an abnormally long contralateral projection. Scale bar: 10 mm. (B) Summary of the presynaptic arbor patterns of wt (DscamFRT), Dscam10.27.25, and Dscam3.31.8 intragenic MARCM clones. (C) Quantification of presynaptic arbor length of intragenic MARCM clones. Sample numbers: wt (DscamFRT), n = 15; Dscam10.27.25, n = 17; Dscam3.31.8, n = 16. (D) Presynaptic arbor overgrowth caused by expressing Dscam[TM2] transgenes are independent of ectodomain diversity. Two independent Dscam[TM2] transgenes containing different and randomly chosen ectodomains, Dscam3.36.25 and Dscam11.31.25 , were overexpressed in C4da neurons using the ppk-Gal4 driver. Presynaptic arbors of all C4da neurons were collectively visualized with ppk-CD4 ∷ tdTomato. C4da presynaptic arbors in abdominal segments 4 (A4) through 6 are shown. Scale bar: 5 µm. Also see Figure S2.
Consistently, overexpression of two independent Dscam[TM2] transgenes containing different and randomly chosen ectodomains: Dscam11.31.25 (Zhan et al., 2004) and Dscam3.36.25 (Wang et al., 2004), were both sufficient to induce exuberant presynaptic overgrowth (Figure 2D).
Collectively, these results demonstrate two separable functions of Dscam in the development of presynaptic terminals: an ectodomain diversity-dependent role in directing presynaptic terminal targeting, and an ectodomain diversity-independent role in controlling presynaptic arbor size.
The DLK Signaling Pathway Controls Presynaptic Arbor Growth by Regulating Dscam Expression
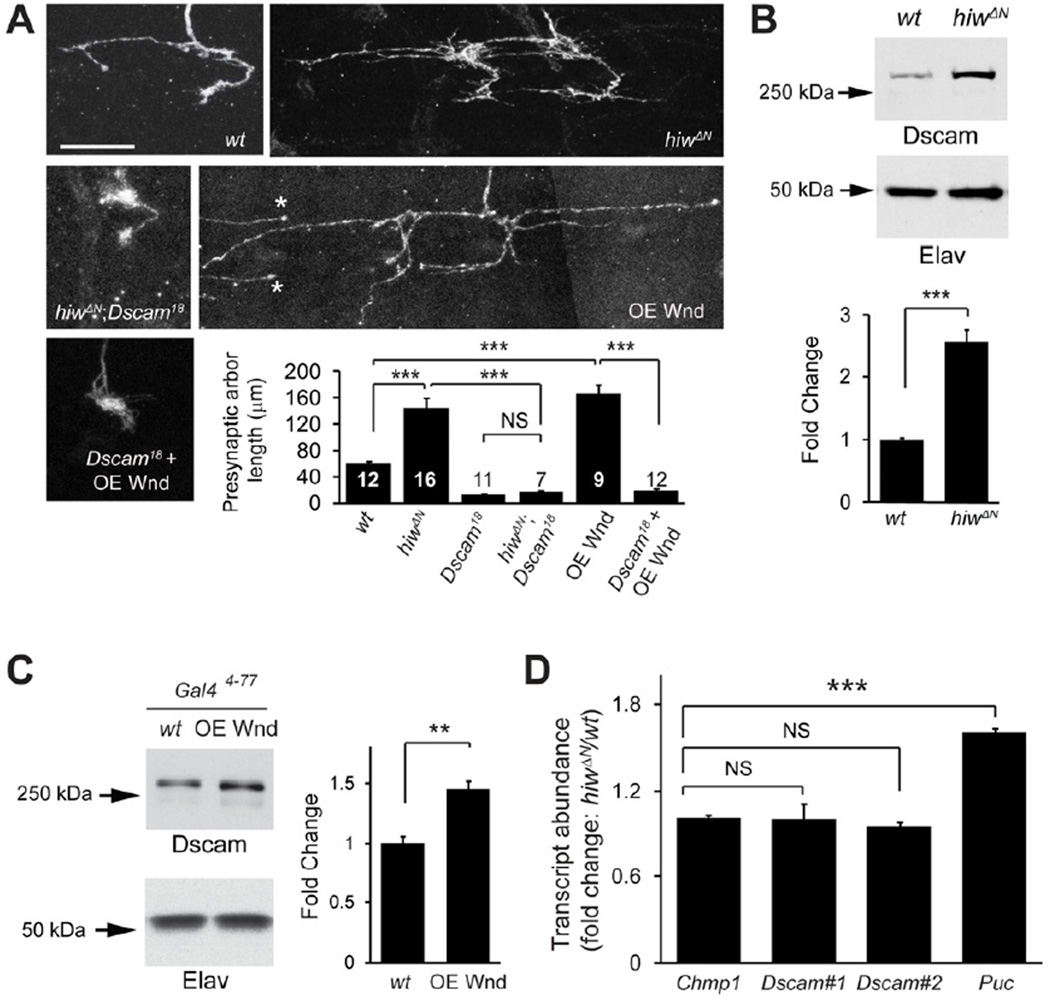
The instructive role of Dscam in presynaptic arbor growth led us to hypothesize that expression level of Dscam determines the size of the presynaptic arbor. To test this hypothesis, we sought to identify the molecular mechanisms that regulate Dscam expression. We screened a number of signaling pathways known to regulate synaptic and axonal growth and found that loss of highwire (hiw) caused dramatic presynaptic overgrowth and ectopic synapses (Figure 3A, and S1) in C4da neurons, which resembled the phenotype of Dscam[TM2]-overexpressing neurons (Figure 1B, and S1).
Figure 3. Hiw and Wnd Control Presynaptic Arbor Growth by Regulating Dscam Expression.
(A) Dscam is required for the overgrowth of presynaptic arbors in hiw mutant neurons and Wnd-overexpressing neurons. Shown are representative presynaptic arbors of single ddaC neurons generated by MARCM in wild-type (wt), hiwΔN hemizygote (hiwΔN), hiwΔN and Dscam18 double mutant (hiwΔN;Dscam18), Wnd overexpression (OE Wnd)(* marks two presynaptic terminals from adjacent neurons), and overexpressing Wnd in Dscam18 mutant (Dscam18 + OE Wnd). Scale bar: 10 µm. Quantification of presynaptic arbor length for each genotype is shown. Data for wild-type (FRTG13) and Dscam18 are the same as that shown Figure 1B. Sample numbers for each condition are shown in the bars. (B) Dscam expression is elevated in hiwΔN mutants. Top: Western blots from brains of wild-type (w1118) and hiwΔN mutant third instar larvae. Bottom: Quantification of Western blots (n = 5). The intensities of Dscam bands were normalized to those of the neuron-specific protein Elav and presented as fold change. (C) Dscam expression is elevated in neurons overexpressing Wnd. Top: Western blots of brain lysates from third instar larvae overexpressing Wnd under the control of Gal44–77 (OE Wnd). For consistency, only one copy of Gal44–77 was used as a wild-type control (wt). Bottom: Quantification of Western blots (n = 4). (D) hiwΔN does not affect the levels of Dscam transcripts. The relative transcript levels of Chmp1 (n = 8), Dscam (two independent sets of primers, #1 and #2, against the invariant exon 24 of Dscam mRNA) (n = 4 and 8, respectively), and Puckered (Puc) (n = 4) from wt and hiw mutant larval brains were measured by real-time PCR. Also see Figure S1 and S3.
Hiw encodes the Drosophila homolog of the evolutionarily conserved E3 ubiquitin ligase PAM/Hiw/RPM-1 (PHR) (Fulga and Van Vactor, 2008; Lewcock et al., 2007; Schaefer et al., 2000; Zhen et al., 2000). The PHR proteins downregulate the dual leucine zipper kinase (DLK) to restrict synaptic growth (Collins et al., 2006; Lewcock et al., 2007; Nakata et al., 2005). Consistently, we found that this signaling module, consisting of Hiw and the Drosophila DLK, Wallenda (Wnd), operates in C4da neurons to regulate presynaptic arbor size (Figure S3).
To determine whether the Drosophila DLK pathway and Dscam genetically interact to control presynaptic arbor growth, we did epistasis analysis by generating Dscam null mutant (Dscam18) MARCM clones in either a hiw mutant (hiwΔN) background or in C4da neurons overexpressing Wnd (OE Wnd). Both hiw mutant and Wnd-overexpressing C4da neurons exhibited dramatically overgrown presynaptic arbors (Figure 3A). Notably, such overgrowth was completely abolished in both conditions in Dscam mutant clones. The presynaptic arbors of hiw and Dscam (hiwΔN;Dscam18) double-mutant clones, and Dscam clones with Wnd-overexpression (Dscam18 + OE Wnd) were morphologically indistinguishable from those of Dscam MARCM clones (Figure 3A), suggesting that Dscam is essential for presynaptic arbor regulation by the Hiw-Wnd pathway.
This epistasis also raised the possibility that the Hiw-Wnd pathway regulates Dscam expression to control presynaptic arbor size. We examined Dscam protein levels in the brains of hiw mutant larvae by Western analysis. Compared to wild-type, Dscam protein levels were increased by 2.5-fold in hiw mutant brains (Figure 3B). Consistently, overexpressing Wnd in a subset of neurons significantly increased Dscam expression in larval brains (Figure 3C). Taken together, these results suggest that the Drosophila DLK pathway controls presynaptic arbor growth by regulating Dscam expression. They also underscore the importance of regulating Dscam expression for proper presynaptic arbor size.
The DLK Pathway Regulates Dscam Expression through the 3'UTR of Dscam
We next asked how the DLK pathway regulates Dscam expression. The DLK pathway has been shown to regulate axon growth and regeneration through transcription or mRNA stability (Collins et al., 2006; Watkins et al., 2013; Yan et al., 2009). We therefore tested whether the Hiw-Wnd pathway regulates Dscam mRNA levels with quantitative real-time PCR on wild-type and hiw larval brains. Using two independent primer sets against the invariant exon 24 of Dscam mRNA, we did not detect any significant difference in Dscam transcript amounts (Figure 3D). As a positive control, hiw mutations caused an increase in the transcripts of Puckered, which is known to be up-regulated by loss of hiw in motoneurons (Xiong et al., 2010). Moreover, the Hiw-Wnd pathway does not regulate Dscam promoter activity, because the expression of a Dscam[TM2] ∷ GFP transgene, under the control of the Dscam promoter, was not significantly different between wild-type and hiw mutant brains (Figure S3C).
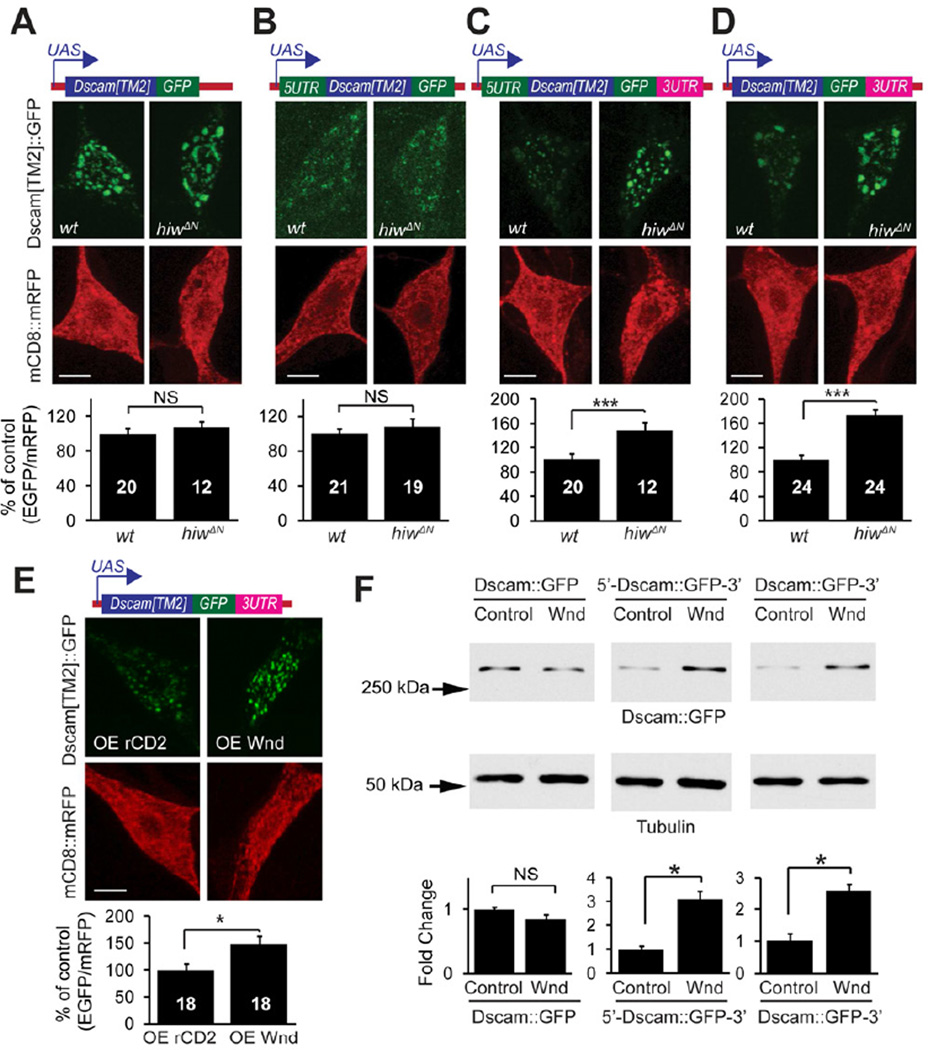
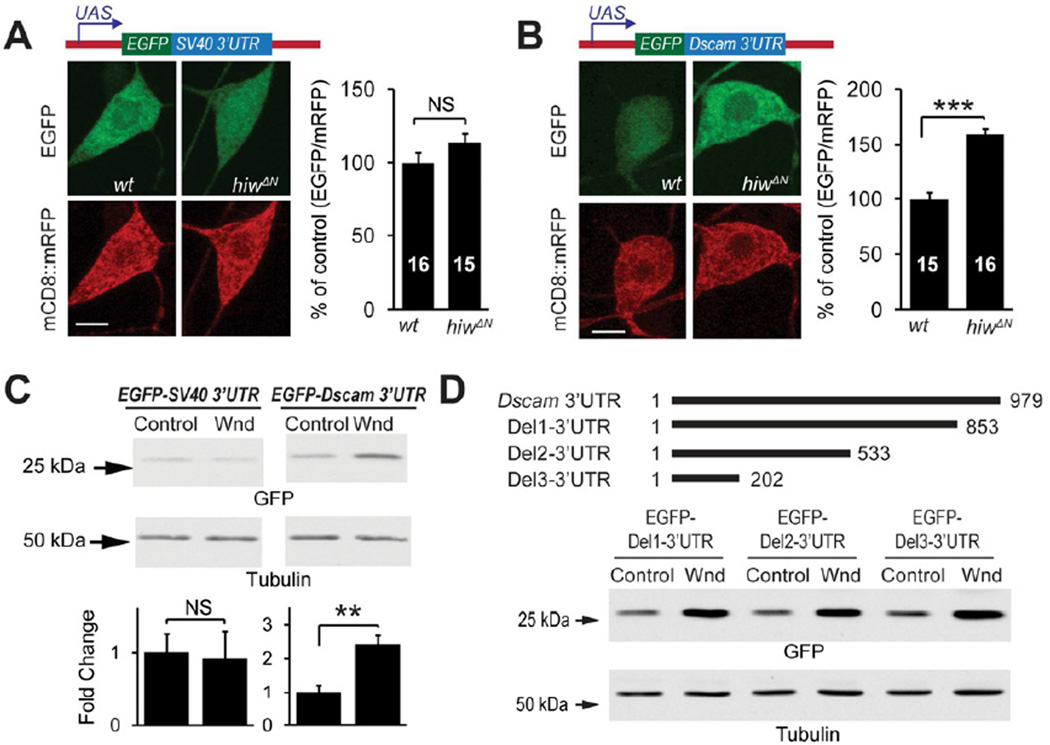
These results suggest that Hiw-Wnd pathway regulates Dscam expression through a previously unknown mechanism, possibly at the level of protein translation. The untranslated regions (UTRs) of mRNAs are key components of protein translational control (Wilkie et al., 2003). In order to determine the requirement of the UTRs in Dscam expressional control, we generated Dscam transgenes fused to GFP with or without Dscam 5’ and/or 3’ UTRs (Figure 4). The expression of a Dscam transgene lacking both UTRs (Dscam ∷ GFP) was not affected by hiw mutations (Figure 4A). Similarly, expression of a transgene with only the 5’ UTR(5'-Dscam ∷ GFP) was also unaffected by hiw function (Figure 4B). In contrast, the expression levels of a transgene with both the 5’ and 3’ UTRs (5'-Dscam ∷ GFP-3') and those of the transgene with only the 3’UTR (Dscam ∷ GFP-3') were significantly elevated in hiw mutant neurons (Figure 4C and 4D). Consistently, overexpressing Wnd enhanced the expression of the Dscam transgene with only 3’UTR in C4da neurons (Figure 4E) as well as Drosophila Schneider 2 (S2) cells in culture (Figure 4F). These results denote that Hiw-Wnd pathway controls Dscam expression through the 3'UTR of Dscam mRNA.
Figure 4. Hiw and Wnd Regulate Dscam Expression through Dscam 3'UTR.
(A–D) The Dscam 3’UTR is required by hiw to regulate Dscam expression in C4da neurons. Dscam[TM2] ∷ GFP transgenes with or without UTRs (schematically shown at the top) were co-expressed with mCD8 ∷ mRFP in wild-type (wt) or hiwΔN C4da neurons using the C4da driver Gal44–77. Quantification of the immunofluorescence intensities is shown at the bottom. Sample numbers are shown in the bars. (E) Wnd promotes Dscam expression through the Dscam 3’UTR in C4da neurons. Dscam[TM2] ∷ GFP transgene containing 3’UTR, was co-expressed with either Wnd (OE Wnd) or the membrane protein rCD2 (OE CD2) as a control, together with mCD8 ∷ mRFP, by the C4da driver Gal44–77. In A–E, Dscam[TM2] ∷ GFP immunofluorescence in ddaC cell bodies were normalized to that of mCD8 ∷ mRFP and presented as % of controls. Scale bars: 5 mm. (F) Wnd promotes Dscam expression through the Dscam 3’UTR in cultured S2 cells. Dscam constructs were transfected into S2 cells along with either an empty vector (control) or a Wnd-expression construct (Wnd). Dscam ∷ GFP expression was examined using Western blotting with an anti-GFP antibody. The intensities of Dscam ∷ GFP bands were normalized to those of tubulin and presented as fold change for statistical analysis (n = 4)
Next, we tested whether the Dscam 3'UTR is sufficient for translational control by the Hiw-Wnd pathway. We generated reporter transgenes by fusing EGFP cDNA with either the 3’UTR of Dscam mRNA or that of SV40 as a control (Figure 5A and B). Hiw mutations specifically enhanced the expression of the Dscam 3'UTR reporter in C4da neurons (Figure 5A and 5B). Consistently, expression of Wnd in cultured S2 cells markedly increased expression of the Dscam 3’UTR reporter (Figure 5C). We further found that the first 202 nucleotides of Dscam 3'UTR are sufficient for the Wnd-regulation (Figure 5D). Taken together, these results suggest that the Dscam 3'UTR is necessary and sufficient for translational regulation by the Drosophila DLK pathway.
Figure 5. The 3'UTR ofDscammRNA is Sufficient for the Regulation by Hiw-Wnd Pathway.
(A and B) Dscam 3’UTR is sufficient to enhance expression in hiw loss-of-function neurons in vivo. EGFP reporter transgenes containing either SV40 3’UTR (A) or Dscam 3’UTR (B) were expressed in wild-type (wt) or hiwΔN C4da neurons. EGFP immunofluorescence in the cell bodies of ddaC was normalized to that of mCD8 ∷ mRFP, and presented as % of control. Sample numbers are shown in the bars. Scale bar: 5 µm. (C) Western blots of lysates from S2 cells expressing the EGFP reporters SV40 3’UTR (EGFP-SV40 3’UTR) or Dscam 3’UTR (EGFP-Dscam 3’UTR), along with a Wnd-expression construct (Wnd) or an empty vector (control). EGFP expression levels were normalized to tubulin levels and are presented as fold change (bottom panel, n = 4). (D) Mapping of the regions of the Dscam 3’UTR required for Wnd-mediated regulation. EGFP reporter constructs containing serial deletions in the Dscam 3’UTR (schematics shown at the top) were transfected into S2 cells along with Wnd-expression construct (Wnd) or the empty vector (control). EGFP expression levels were normalized to tubulin levels and presented as fold change (bottom panel, n = 4).
FMRP Suppresses Dscam Expression to Restrict Presynaptic Arbor Growth
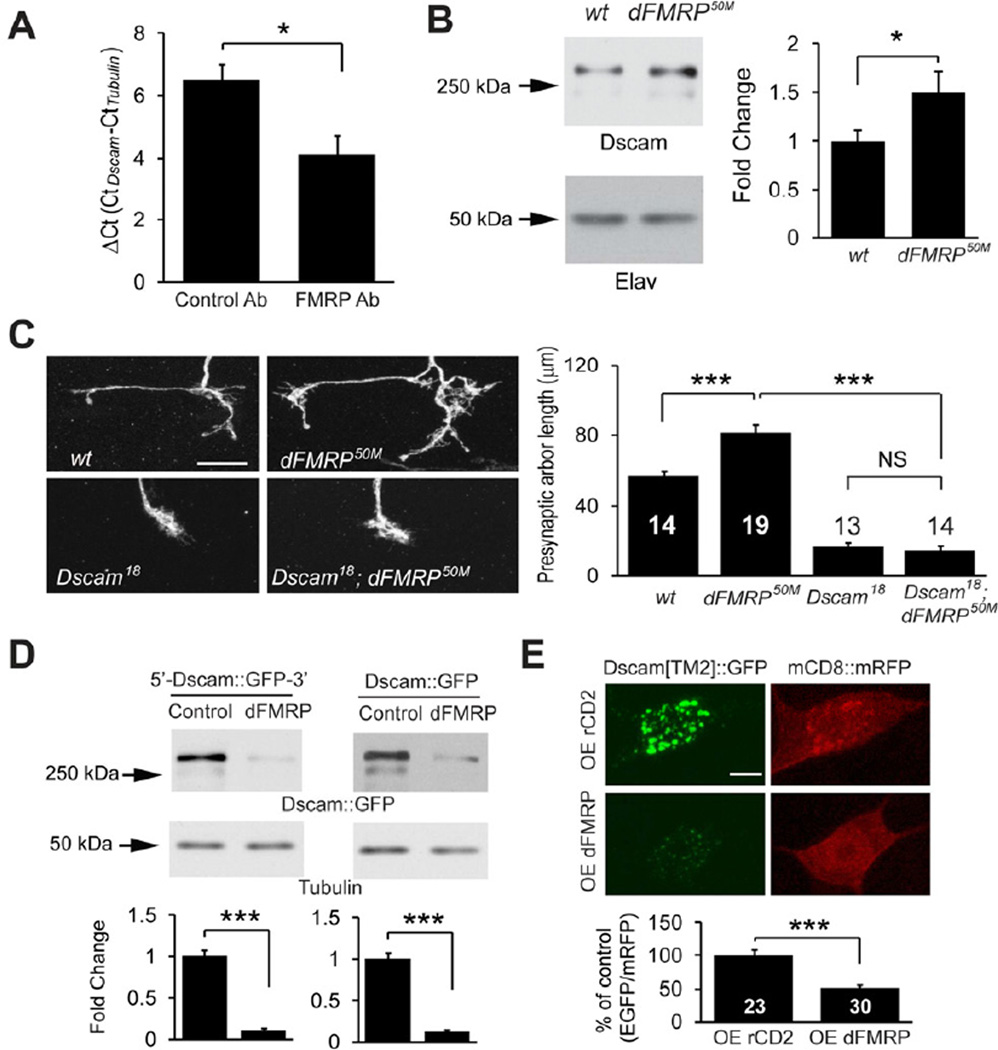
The RNA-binding protein fragile X mental retardation protein (FMRP) is involved in the post-transcriptional regulation of a number of target mRNAs (Santoro et al., 2012). FMRP has been reported to bind to Dscam mRNA in mammalian neurons (Brown et al., 2001; Darnell et al., 2011), but the functional relevance of this binding is unknown. We wondered whether FMRP might also regulate Dscam protein translation. We tested the association between Drosophila FMRP (dFMRP) and Dscam mRNA in larval brain lysates by RNA-immunoprecipitation. Compared to a control antibody, anti-dFMRP antibody pulled down more Dscam mRNA as assessed by real-time PCR (Figure 6A). The difference in cycle number (DCt) between dFMRP-and control immunoprecipitates translates into a 5.8-fold more association of Dscam mRNA to dFMRP immunoprecipitates, suggesting that dFMRP binds to Dscam mRNA in Drosophila. We then examined whether FMRP regulates Dscam expression. Western blot analysis of larval brain lysates showed that dFMRP null mutations led to a 49% increase in Dscam protein levels (Figure 6B), which is consistent with the role of FMRP as a translational repressor (Laggerbauer et al., 2001). Furthermore, in keeping with a previous study of the Drosophila neuromuscular junction (NMJ) (Zhang et al., 2001), dFMRP mutations in C4da neurons caused mild but significant overgrowth of presynaptic terminals that was completely abolished by Dscam null mutations (Figure 6C and 6D). Taken together, these results suggest that dFMRP regulates Dscam expression to restrain presynaptic arbor growth.
Figure 6. FMRP Suppresses Dscam Expression to Restrict Presynaptic Arbor Growth.
(A) dFMRP associates with Dscam mRNA in vivo. RNA-immunoprecipitation, followed by reverse transcription and real-time PCR, was done using larval brain lysates (n = 3). The difference in DCt between control and dFMRP-immunoprecipitates reflects a 5.8-fold binding of Dscam mRNA to FMRP. (B) Western analysis of Dscam expression in the brains of wild-type (w1118) and dFMRP50M third instar larvae. The intensities of Dscam bands were normalized to those of the neuron-specific protein Elav and presented as fold change (n = 13). (C) Dscam is required by dFMRP to restrict presynaptic arbor growth. Images show the presynaptic arbors of ddaC MARCM clones of wt, dFMRP50M, sDscam18, and Dscam18/dFMRP50M double mutant (Dscam18; dFMRP50M) neurons. Scale bar: 10 µm. Right: Quantification of presynaptic arbor length for each condition. Sample numbers are shown in or above the bars. (D) dFMRP regulates Dscam expression through the coding region of Dscam in S2 cells. Shown are Western blots of lysates of cultured S2 cells expressing Dscam[TM2] ∷ GFP with Dscam 5’ and 3’UTR (5’-Dscam ∷ GFP-3’) or with SV40 3’UTR (Dscam ∷ GFP) in the presence of a dFMRP-expression construct (dFMRP) or the empty vector (Control). Dscam[TM2] ∷ GFP levels were normalized to tubulin levels and presented as fold change (n = 4). (E) dFMRP suppresses Dscam expression through the Dscam coding region in vivo. Dscam[TM2] ∷ GFP and mCD8 ∷ RFP were expressed in C4da neurons using Gal44–77, along with either rCD2 (control) or dFMRP (OE dFMRP). Dscam[TM2] ∷ GFP levels in ddaC cell bodies were normalized to mCD8 ∷ mRFP levels and presented as % of control (right panel). Also see Figure S4.
FMRP Suppresses Dscam Expression Through the Coding Region
While Wnd expression greatly enhanced the expression levels of the EGFP reporter containing Dscam 3'UTR in S2 cells (Figure 5C), dFMRP overexpression did not change the expression levels of the same reporter (Figure S4A), suggesting that the regulation by dFMRP is independent of Hiw-Wnd pathway.
Recent studies have uncovered that FMRP acts on the coding regions of some mRNAs to control translation (Ascano et al., 2012; Darnell et al., 2011). We thus tested the involvement of Dscam coding region in the regulation by FMRP. Overexpressing dFMRP in S2 cells strongly inhibited the expression of both Dscam transgenes either with or without UTRs (Figure 6D), suggesting that dFMRP suppresses Dscam translation via Dscam coding region. Similarly, dFMRP overexpression in C4da neurons reduced the expression of a Dscam[TM2] ∷ GFP transgene that does not contain Dscam UTRs (Figure 6E).
Consistent with the change in expression, dFMRP overexpression reduced presynaptic arbor overgrowth caused by Dscam[TM2] ∷ GFP overexpression (Figure S4B). Moreover, dFMRP mutations increased presynaptic arbor sizes in C4da neurons overexpressing Dscam (with both 5’ and 3’UTRs) (43.0 ± 15.3 % increase) proportionally to those without Dscam overexpression (38.2 ± 7.1 % increase) (Figure S4C–E). Consistent with the notion that dFMRP suppresses Dscam translation by acting on the coding region, dFMRP null mutations led to similar percentage of increase in presynaptic arbors between neurons expressing Dscam transgene with Dscam UTRs, and those without Dscam UTRs (Figure S4C–E). Taken together, these results demonstrate that FMRP regulates Dscam expression through the coding region.
The DLK Pathway and FMRP Converge on Translational Control of Dscam to Regulate Presynaptic Arbor Growth
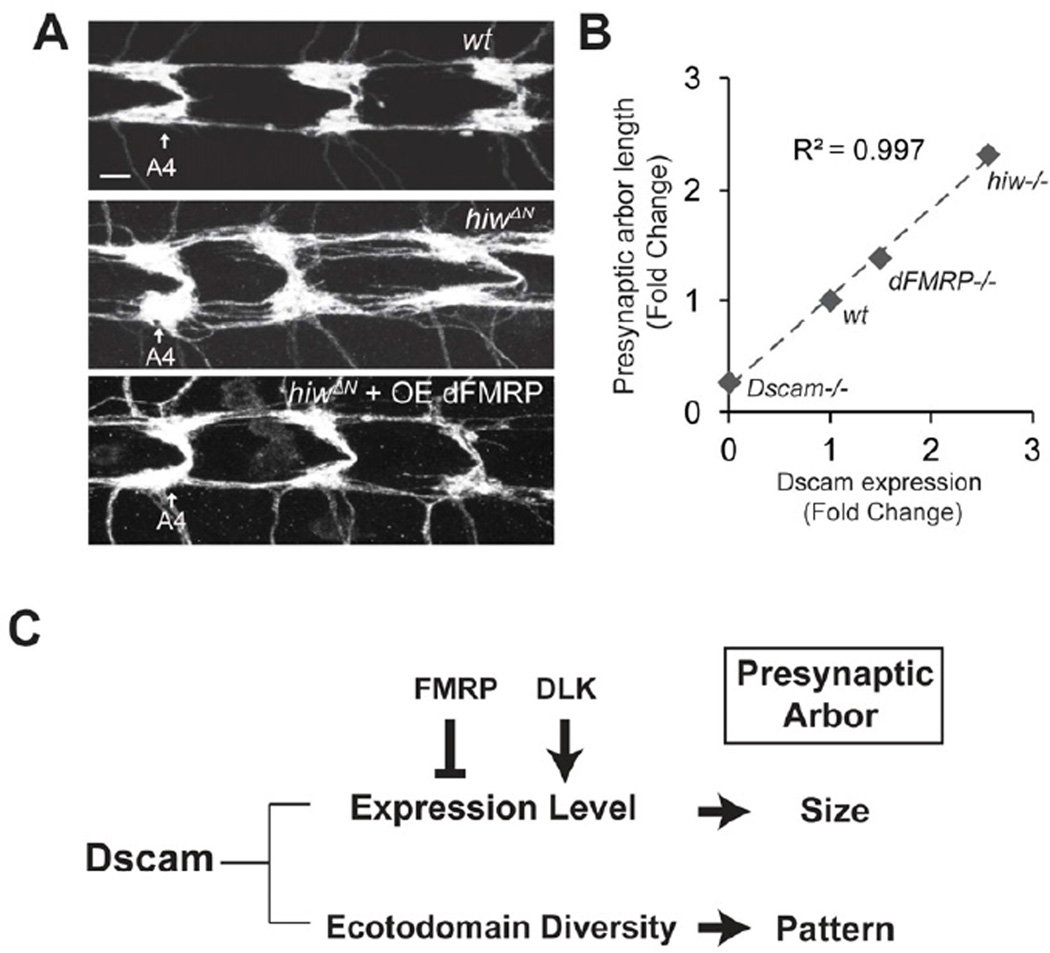
Although both the DLK pathway and FMRP regulate Dscam translation, they exert their influences on different parts of Dscam mRNA. The Dscam 3’UTR was sufficient to mediate regulation by Wnd (Figure 5C), but not by dFMRP (Figure S4A). Moreover, the Dscam coding region does not respond to the regulation by the Hiw-Wnd pathway (Figure 4A), but it mediates the suppression by dFMRP (Figure 6D–E). Thus, these two regulatory mechanisms appear to operate in parallel (Figure 7C). If these two pathways converge on Dscam expression to direct presynaptic arbor growth, the suppression of Dscam function by dFMRP would counteract the enhanced Dscam function in hiw mutants. Indeed, overexpressing dFMRP significantly suppressed the presynaptic arbor overgrowth caused by either hiw mutations (Figure 7A) or Wnd overexpression (Figure S5).
Figure 7. Dscam Expression Levels Correlate with Presynaptic Arbor Sizes.
(A) Hiw and FMRP pathways converge at the level of Dscam translational control to regulate presynaptic arbor growth. dFMRP was expressed with ppk, Gal4 either in wild-type (wt) or hiw mutant (hiwΔN + OE dFMRP) C4da neurons. C4da presynaptic arbors were visualized with mCD8 ∷ GFP. C4da presynaptic arbors in abdominal segments 4 (A4) through 6 are shown. Scale bar: 5 µm. (B) Dscam expression levels correlate with presynaptic arbor length. The relative presynaptic arbor sizes of single ddaC neurons from different genetic backgrounds were normalized to corresponding wild-type controls and denoted as fold change. Dscam expression levels from Western analyses were normalized to corresponding wild-type controls and presented as fold change. The presynaptic arbor size of Dscam18 was used for Dscam−/−. Dscam18is a protein-null allele (Soba et al., 2007). R2 value was derived from linear regression. (C) The present study suggests a model that explains how presynaptic arbor patterning and size control may be differentially controlled by a shared molecule, Dscam. The Dscam ectodomain diversity determines the pattern of presynaptic terminals, whereas its expression level instructs presynaptic terminal size. The DLK pathway and FMRP regulate Dscam expression levels to control presynaptic arbor size. Also see Figure S5.
Having established the importance of Dscam expression regulation for presynaptic arbor growth, we sought to determine the degree of correlation between presynaptic arbor sizes and Dscam protein levels. We plotted relative Dscam expression levels, as assayed by Western analysis (Figure 3B, and Figure 6B), against relative presynaptic arbor sizes of single C4da neurons (Figure 1B, 3A, and 6C) in different genetic backgrounds. The statistical analysis showed a striking linear correlation, with a coefficient of determination (R2) of 0.997 between Dscam levels and presynaptic arbor sizes (Figure 7B). This not only suggests that Dscam expression levels are tightly controlled for precise presynaptic arbor growth, but also emphasizes the function of Dscam expression levels in determining presynaptic arbor sizes.
DISCUSSION
In this study, we found that in addition to the ectodomain diversity, the expression level of Dscam serves as a code for neuronal development. We identified two regulatory mechanisms, one involving the kinase DLK and another involving the RNA-binding protein FMRP, which control Dscam expression at the level of protein translation. Defects in either of these regulatory pathways lead to aberrant growth of presynaptic arbors. The importance of this regulation is underscored by the strong correlation between the expression levels of Dscam and the sizes of presynaptic arbors.
An Instructive Role of Dscam in Presynaptic Arbor Growth
After reaching their target regions, axons branch and extend to form presynaptic arbors. A presynaptic arbor of a given neuron type typically develops a specific pattern and size, which is critical for establishing appropriate number of synaptic connections with specific targets. How the patterning mechanism relates to the ultimate size that each presynaptic arbor assumes is unknown. Here, we propose that both the patterning and size control of presynaptic terminals can be instructed by a common regulator, such as Dscam. The isoform diversity of Dscam determines the pattern of presynaptic terminals, whereas the expression levels of Dscam instruct the sizes of these terminals (Figure 7C).
Is the function of Dscam in presynaptic arbor size control a consequence of its dendritic functions? Several lines of evidence argue against this possibility. First, while expressing the axon-enriched TM2 isoforms caused dramatic increase of presynaptic arbor growth, expressing the dendrite-enriched TM1 isoforms led to only a minimal increase in presynaptic arbor growth (Figure 1B), suggesting that axonal Dscam regulates presynaptic growth. Second, overexpressing TM2 isoforms did not elicit any significant change in dendrite growth but caused dramatic increase in presynaptic arbor growth (Figure S1B–C), demonstrating that the axonal function of Dscam is separable from its dendritic functions. Third, whereas Dscam ectodomain diversity is required for dendritic self-avoidance, it is dispensable for presynaptic arbor growth (Figure 2). Therefore, the instructive role of Dscam levels in presynaptic arbor growth is independent of the dendritic functions of Dscam.
How might Dscam instruct presynaptic arbor growth? Dscam is a type I transmembrane protein with a cytoplasmic domain that is heavily tyrosine-phosphorylated (Schmucker et al., 2000). The cytoplasmic domain of Dscam interacts with the signaling molecule Pak1 (Li and Guan, 2004; Schmucker and Chen, 2009), which is important for the guidance of embryonic Bolwig's nerve (Schmucker et al., 2000). However, we observed no defect in C4da presynaptic arbor growth in either loss-of-function or gain-of-function of Pak1 (data not shown), indicating that Dscam does not act through Pak1 to instruct presynaptic arbor growth in C4da neurons. It remains to be determined how expression levels of Dscam instruct intracellular signaling and organelles to control the sizes of presynaptic arbors.
A Novel Regulatory Mechanism by the DLK Signaling Pathway
Given the strong correlation between Dscam expression level and presynaptic arbor size (Figure 7B), Dscam expression seems to be tightly controlled to ensure proper neural connectivity. Here we provide evidence for the translational control of Dscam by the DLK pathway. In Drosophila and C. elegans, respectively, Hiw orthologs regulate the turnover of Wnd and DLK1 (Collins et al., 2006; Nakata et al., 2005). Studies in C. elegans, Drosophila and mammals have demonstrated that DLK regulates axon growth and regeneration through either transcription programs or mRNA stabilization (Collins et al., 2006; Nakata et al., 2005; Watkins et al., 2013). However, our findings indicate that the regulation of Dscam expression by the DLK pathway does not occur through transcription or mRNA stability (Figure 3D). We thus propose that DLK has the novel function of enhancing protein translation through the 3’UTR of target mRNAs. How might Wnd enhance Dscam translation? Wnd, as a kinase, is likely to require downstream effecter(s) to regulate mRNA translation. It has been reported that Dscam mRNAs are translated in the dendrites of hippocampal neurons in culture, possibly through CPEB1 (Alves-Sampaio et al., 2010). In the future, it will be interesting to test if Wnd acts on CPEB1 to regulate Dscam translation.
Relevance to Neurological Disorders
Our findings on the function of Dscam in presynaptic arbor growth are relevant to neurological disorders not only because Dscam expression is elevated in several of these disorders, but also because growth of presynaptic arbors is involved in epilepsy and axon regeneration (Cavazos et al., 1991; Houser et al., 1990; Marco and DeFelipe, 1997; Sutula et al., 1988). Dscam protein level is elevated in intractable epilepsy (Shen et al., 2011), which involves aberrant mossy fiber sprouting (Sutula et al., 1989). Of note, increased occurrence of epileptic seizures is often associated with DS (Musumeci et al., 1999; Stafstrom, 1993) as well as fragile X syndrome (FXS) (Musumeci et al., 1999; Stafstrom, 1993), which is caused by loss of FMRP function (Verkerk et al., 1991). Our study suggests that elevated Dscam levels may contribute to the pathogenesis of these disorders by causing excessive presynaptic arbor growth. It also establishes a functional link between Dscam and FMRP, raising the intriguing possibility that Dscam might be a mechanistic link between DS and FXS, the two most prevalent genetic causes of mental retardation.
Recent studies have shown that axon injury activates the DLK pathway, which is essential for subsequent axon regeneration (Hammarlund et al., 2009; Shin et al., 2012; Watkins et al., 2013; Xiong et al., 2010; Yan et al., 2009). In light of the present study, it will be interesting to determine whether the DLK pathway requires Dscam to instruct axon regeneration.
In summary, this study demonstrates that Dscam expression levels, regulated by the DLK pathway and FMRP, determine presynaptic arbor size. It further shows the functional significance of dysregulated Dscam expression in neuronal development and provides a model for studying the pathogenesis of neurological disorders with dysregulated Dscam expression.
EXPERIMENTAL PROCEDURES
Fly Strains
hiwΔN ,UAS-Hiw ∷ GFP (Wu et al., 2005); wnd1, wnd3, and UAS-Wnd (Collins et al., 2006); DscamP1 (Schmucker et al., 2000); Dscam18 (Wang et al., 2002); UAS-Dscam[TM2] ∷ GFP (3.36.25), UAS-Dscam[TM1] ∷ GFP (3.36.25), and DscamP-Dscam[TM2] ∷ GFP (3.36.25)(Wang et al., 2004); UAS-Dscam[TM2] (11.31.25) (Zhan et al., 2004); Dscam10.27.25, Dscam3.31.8 and DscamFRT (Hattori et al., 2007); dFMRP50M , UAS-dFMRP (Zhang et al., 2001); ppk-Gal4 (Kuo et al., 2005); ppk-CD4 ∷ tdTomato (Han et al., 2011); and UAS-Syt ∷ eGFP (Zhang et al., 2002).
DNA Constructs for Generating Transgenic Flies and S2 Cell Transfection
cDNA constructs of EGFP expression reporters and dFMRP were subcloned into the pUAST vector. Dscam cDNA containing variable exons 4.3–6.36–9.25−17.2 (Wang et al., 2004) were used to generate Dscam[TM2] ∷ GFP constructs with or without the 5' and/or 3'UTR of Dscam mRNA in the pUASTattB vector. Using standard methods (Bateman et al., 2006), UAS-Dscam[TM2] ∷ GFP (3.36.25) transgenic lines were generated using PhiC31 integrase-mediated site-specific insertion at the attP40 landing site. As such, there is no position-effect on the transcription of these transgenes. The UAS-EGFP construct containing the Dscam 3’UTR was used to generate serial deletion constructs of the Dscam 3'UTR for mapping the required sequence for Wnd regulation. The genomic Dscam transgene used for rescue experiments and the wnd cDNA construct were, respectively, generous gifts from Dr. Tzumin Lee (Howard Hughes Medical Institute) and Dr. Catherine Collins (University of Michigan).
Labeling of the Presynaptic Arbors with Genetic Mosaic Techniques
The single C4da presynaptic arbors in Figure 1A were labeled by the flip-out technique with CD2 flanked by two FRT sequences sandwiched between UAS and mCD8 ∷ GFP. Excision of CD2 was achieved by heat-shock-induced flippase expression. The resulting C4da clones expressed mCD8 ∷ GFP; the rest of the C4da neurons expressed CD2. A modified flip-out technique with an excisable GAL80 (Gordon and Scott, 2009) was used to express the membrane marker mCD8 ∷ mRFP and the presynaptic marker synaptotagmin ∷ GFP under the control of ppk promoter in Figure S1A.
The MARCM technique (Lee and Luo, 1999) was used to generate and label homozygous Dscam18, DscamP1, and dFMRP50m C4da neurons, and to overexpress Dscam[TM2] ∷ GFP and Wnd. MARCM clones were induced as previously described (Ye et al., 2011). The same MARCM technique was also used to label presynaptic arbors of single ddaC neurons in hiwΔN hemizygous third-instar larvae.
To generate single C4da neurons expressing a single isoform of the ectodomain (Dscam10.27.25, Dscam3.31.8 ), we applied the intragenic MARCM technique (Hattori et al., 2007), A wild-type Dscam allele containing an FRT at the same genomic location as DscamSingle was used as control (DscamFRT).
Immunostaining and Imaging
Third instar larvae were immunostained as described (Ye et al., 2011). The primary antibodies used were mouse anti-GFP (Invitrogen) and rabbit anti-RFP (Rockland). Confocal imaging was done with a Leica SP5 confocal system equipped with 63x immersion oil lenses. To minimize the variation in presynaptic arbor sizes among C4da neurons in different body segments, only neurons in abdominal segments 4, 5, and 6 were imaged. Images were collected with z-stacks of 0.3-µm step size. The resulting 3D images were projected into 2D images using a maximum projection method. To ensure that fluorescence intensities reflected protein levels, image acquisition was adjusted to minimum signal saturation. The same imaging setting was applied throughout the imaging process. After image transformation into 2D images, the mean fluorescence intensity of the region of interest was measured with NIH ImageJ software.
Quantification of Presynaptic Arbor Size
The Neurolucida software was used to trace and measure the length between an axon’s entry point into the C4da neuropil and the axon endings. Branches shorter than 5 µm were excluded from analysis.
Western Blots
To analyze reporter expression in cultured cells, S2 cells maintained in Schneider’s medium with 10% fetal bovine serum were transfected with Lipofectamine 2000™ (Invitrogen). A construct containing the tubulin promoter fused to the cDNA of GAL4 was co-transfected with pUAST constructs. Two days after transfection, cells were harvested by centrifugation, homogenized in SDS sample buffer, separated by SDS-PAGE, and analyzed by Western blot.
To analyze Dscam protein levels in vivo, brains were removed from wandering third instar larvae and homogenized in SDS sample buffer. Homogenates of equal numbers of brains (3–5) from control and experimental groups were separated by SDS-PAGE and analyzed by Western blot. The primary antibodies used were mouse monoclonal anti-Dscam (Shi et al., 2007), rabbit polyclonal anti-GFP (Invitrogen), mouse monoclonal anti-tubulin (Sigma), mouse monoclonal anti-dFMRP (Developmental Studies Hybridoma Bank), rat monoclonal anti-Elav (Developmental Studies Hybridoma Bank), and mouse monoclonal anti-βGal (Developmental Studies Hybridoma Bank).
RNA-Immunoprecipitation
Larval brains (~150) were dissected from wandering third instars in PBS and washed two times with PBS. Crude homogenates were generated by homogenizing brains in lysis buffer (50 mM Tris/HCl, pH 7.5; 150 mM KCl; 1 mM EDTA; 0.5% TX100) in the presence of RNase inhibitor and protease inhibitor and centrifuged for 30 min at 20,000 x g, 4°C. The equal amount of supernatant was incubated for 1hr at 4°C with Dynabeads Protein-G (Life Technologies) precoupled with same amount of monoclonal anti-dFMRP antibody 5B6 (Developmental Studies Hybridoma Bank) or normal mouse IgG as a negative control. Beads were washed 5 times with lysis buffer, supplemented with 10 mg glycogen (Invitrogen) and 10 pg of firefly luciferase mRNA (Promega), then processed for RNA extraction.
Real-Time PCR
Total RNA was extracted from brains of third instar larvae, using a standard Trizol protocol (Invitrogen). First-strand cDNA was synthesized with Invitrogen SuperScript III First-Strand Synthesis SuperMix (Invitrogen). cDNA from 10 ng RNA was used for each real-time PCR reaction (15 µl), using the Absolute QPCR SYBR Green mix (Thermo Scientific) with Applied Biosystems 7300. After the cycle number at the threshold level of log-based fluorescence (Ct) had been collected for each sample, ΔCt for each test gene was calculated by subtracting the Ct number of the reference gene (elav) from that of the test gene (Cttest-Ctelav) (Yuan et al., 2006). This normalizes transcript levels of test genes to elav. Our extensive tests showed that hiw mutations do not alter elav or Chmp1 transcript levels. The ΔCt of each test gene was statistically compared between wild-type and hiw, and then converted to fold change. The Mann-Whitney test was used to determine the statistical significance of changes in different transcripts.
For RNA-immunoprecipitation, ΔCt for Dscam mRNA was calculated by subtracting the Ct number of the reference mRNA (α-tubulin) from that of Dscam mRNA. We used α-tubulin mRNA as the reference because mammalian α-tubulin mRNA does not bind to FMRP (Darnell et al., 2011). Three independent RNA-immunoprecipitation experiments were done and the values of ΔCt were compared between control antibody and anti-dFMRP antibody by using two-tailed paired Student t-test.
Primer sets used were: elav, 5’-CTGCCAAAGACGATGACC-3’ and 5’-TAAAG CCTACTCCTTTCGTC-3’; Chmp1, 5’-AAAGGCCAAGAAGGCGATTC-3’ and 5’-GGGCAC TCATCCTGAGGTAGTT-3’; Puckered, 5’-AAAGTCCCAATGAGAGCC-3’ and 5’-CGTGCA TCTTCGATAAAGTC-3’; Dscam #1, 5’-CTTACGATTGTGCTCATTACTC-3’ and 5’-CAGTT TCGATTTGTTCTGTTGG-3’; Dscam #2, 5’-ATCGAAACTGTTCAATGCAC-3’ and 5’-CTT GAGTGTATCTGTGTTTCGG-3’; firefly luciferase, 5'- CTCACTGAGACTACATCAGC-3' and 5'- TCCAGATCCACAACCT TCGC-3'; α-tubulin, 5'-GCCAATTAGGCGATTGAGATTC-3' and 5'-AGCACTCGGACTGTGCGTTT-3'.
Statistical Analysis
A two-tailed unpaired Student t-test was used for presynaptic arbor size analysis and Western blot analysis unless otherwise noted. The Mann-Whitney test was used for real-time PCR experiments. P-values smaller than 0.05 were considered statistically significant. All p values are indicated as: *: p < 0.05, **: p < 0.01, and ***: p < 0.001. Data are presented as mean ± SEM.
Supplementary Material
HIGHLIGHTS.
Dscam instructs presynaptic arbor growth, independent of ectodomain diversity.
Presynaptic arbor sizes strongly correlate with Dscam expression levels.
DLK activates Dscam translation to promote presynaptic arbor growth.
FMRP suppresses Dscam translation to restrict presynaptic arbor growth.
ACKNOWLEDGMENTS
We thank Drs. Tzumin Lee, Larry Zipursky, Catherine Collins, Chunlai Wu, Kendal Broadie, Chun Han, Liqun Luo, and Yuh Nung Jan generously for sharing reagents, Drs. Ting Han and John Kim for their help on the RNA-IP experiments, the members of Dr. Jiandie Lin’s lab for helping us to set up the real-time PCR experiments. We also thank Dr. Catherine Collins, Dr. Tzumin Lee, Dr. Hisashi Umemori, and Gabriella Sterne for critical comments on earlier versions of the manuscript. This work was supported by grants from NIH (R00MH080599 and R01MH091186), the Whitehall Foundation, and the Pew Scholars Program in the Biological Sciences to B.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alves-Sampaio A, Troca-Marín JA, Montesinos ML. NMDA-mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down’s syndrome. J. Neurosci. 2010;30:13537–13548. doi: 10.1523/JNEUROSCI.3457-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano K, Yamada K, Iwayama Y, Detera-Wadleigh SD, Hattori E, Toyota T, Tokunaga K, Yoshikawa T, Yamakawa K. Association study between the Down syndrome cell adhesion molecule (DSCAM) gene and bipolar disorder. Psychiatr. Genet. 2008;18:1–10. doi: 10.1097/YPG.0b013e3281ac238e. [DOI] [PubMed] [Google Scholar]
- Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank M, Fuerst PG, Stevens B, Nouri N, Kirkby L, Warrier D, Barres BA, Feller MB, Huberman AD, Burgess RW, et al. The Down syndrome critical region regulates retinogeniculate refinement. J.Neurosci. 2011;31:5764–5776. doi: 10.1523/JNEUROSCI.6015-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray Identification of FMRP-Associated Brain mRNAs and Altered mRNA Translational Profiles in Fragile X Syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cavazos JE, Golarai G, Sutula TP. Mossy fiber synaptic reorganization induced by kindling: time course of development, progression, and permanence. J. Neurosci. 1991;11:2795–2803. doi: 10.1523/JNEUROSCI.11-09-02795.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BE, Kondo M, Garnier A, Watson FL, P¸ettmann-Holgado R, Lamar DR, Schmucker D. The Molecular Diversity of Dscam Is Functionally Required for Neuronal Wiring Specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire Restrains Synaptic Growth by Attenuating a MAP Kinase Signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Darnell Jennifer C, Van Driesche Sarah J, Zhang C, Hung Ka Ying S, Mele A, Fraser Claire E, Stone Elizabath F, Chen C, Fak John J, Chi Sung W, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64:484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451:470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Van Vactor D. Synapses and growth cones on two sides of a highwire. Neuron. 2008;57:339–344. doi: 10.1016/j.neuron.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor Control in a Drosophila Taste Circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. U S A. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Chen Y, Matthews BJ, Salwinski L, Sabatti C, Grueber WB, Zipursky SL. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461:644–648. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL. Dscam-mediated cell recognition regulates neural circuit formation. Annu. Rev. Cell Dev.Biol. 2008;24:597–620. doi: 10.1146/annurev.cellbio.24.110707.175250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser CR, Miyashiro JE, Swartz BE, Walsh GO, Rich JR, Delgado-Escueta AV. Altered patterns of dynorphin immunoreactivity suggest mossy fiber reorganization in human hippocampal epilepsy. J.Neurosci. 1990;10:267–282. doi: 10.1523/JNEUROSCI.10-01-00267.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Bäumer P, Kondo M, Uemura T, Schmucker D. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal Targeting of Olfactory Receptor Neurons in Drosophila Is Controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. U S A. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel E-M, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum.Mol.Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Larsen DD, Callaway EM. Development of layer-specific axonal arborizations in mouse primary somatosensory cortex. J. Comp. Neurol. 2006;494:398–414. doi: 10.1002/cne.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Li W, Guan KL. The Down syndrome cell adhesion molecule (DSCAM) interacts with and activates Pak. J. Biol. J.Biol Chem. 2004;279:32824–32831. doi: 10.1074/jbc.M401878200. [DOI] [PubMed] [Google Scholar]
- Marco P, DeFelipe J. Altered synaptic circuitry in the human temporal neocortex removed from epileptic patientsExp. Exp Brain Res. 1997;114:1–10. doi: 10.1007/pl00005608. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite Self-Avoidance Is Controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Maynard KR, Stein E. DSCAM Contributes to Dendrite Arborization and Spine Formation in the Developing Cerebral Cortex. J. Neurosci. 2012;32:16637–16650. doi: 10.1523/JNEUROSCI.2811-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA. Drosophila Dscam Proteins Regulate Postsynaptic Specificity at Multiple-Contact Synapses. Neuron. 2010;67:761–768. doi: 10.1016/j.neuron.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SS, Zipursky SL. Dscam-mediated repulsion controls tiling and self-avoidance. Curr. Opin. Neurobiol. 2008;18:84–89. doi: 10.1016/j.conb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci SA, Hagerman RJ, Ferri R, Bosco P, Dalla Bernardina B, Tassinari CA, De Sarro GB, Elia M. Epilepsy and EEG findings in males with fragile X syndrome. Epilepsia. 1999;40:1092–1099. doi: 10.1111/j.1528-1157.1999.tb00824.x. [DOI] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y. Regulation of a DLK-1 and p38 MAP Kinase Pathway by the Ubiquitin Ligase RPM-1 Is Required for Presynaptic Development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Saito Y, Oka A, Mizuguchi M, Motonaga K, Mori Y, Becker LE, Arima K, Miyauchi J, Takashima S. The developmental and aging changes of Down's syndrome cell adhesion molecule expression in normal and Down's syndrome brains. Acta Neuropathol. 2000;100:654–664. doi: 10.1007/s004010000230. [DOI] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspectivec. Annu. Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23:147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Shen L, Xiao Z, Pan Y, Fang M, Li C, Chen D, Wang L, Xi Z, Xiao F, Wang X. Altered expression of Dscam in temporal lobe tissue from human and experimental animals. Synapse. 2011;65:975–982. doi: 10.1002/syn.20924. [DOI] [PubMed] [Google Scholar]
- Shi L, Yu HH, Yang JS, Lee T. Specific Drosophila Dscam juxtamembrane variants control dendritic elaboration and axonal arborization. J.Neurosci. 2007;27:6723–6728. doi: 10.1523/JNEUROSCI.1517-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A. Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74:1015–1022. doi: 10.1016/j.neuron.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang S-J, Yu H-H, Lee T, Jan LY, Jan Y-N. Drosophila Sensory Neurons Require Dscam for Dendritic Self-Avoidance and Proper Dendritic Field Organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom CE. Epilepsy in Down syndrome: clinical aspects and possible mechanisms. Am. J. Ment. Retard. 1993;98(Suppl):12–26. [PubMed] [Google Scholar]
- Sutula T, Cascino G, Cavazos J, Parada I, Ramirez L. Mossy fiber synaptic reorganization in the epileptic human temporal lobe. Ann. Neurol. 1989;26:321–330. doi: 10.1002/ana.410260303. [DOI] [PubMed] [Google Scholar]
- Sutula T, He XX, Cavazos J, Scott G. Synaptic reorganization in the hippocampus induced by abnormal functional activity. Science. 1988;239:1147–1150. doi: 10.1126/science.2449733. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee C-HJ, Lee T. Drosophila Dscam Is Required for Divergent Segregation of Sister Branches and Suppresses Ectopic Bifurcation of Axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Watkins TA, Wang B, Huntwork-Rodriguez S, Yang J, Jiang Z, Eastham-Anderson J, Modrusan Z, Kaminker JS, Tessier-Lavigne M, Lewcock JW. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc. Natl. Acad. Sci. U S A. 2013;110:4039–4044. doi: 10.1073/pnas.1211074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Wu C, Wairkar YP, Collins CA, DiAntonio A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J. Neurosci. 2005;25:9557–9566. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, Collins CA. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J.Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 Kinase Promotes mRNA Stability and Local Translation in C. elegans Synapses and Axon Regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Kim JH, Yang L, McLachlan I, Younger S, Jan LY, Jan YN. Differential regulation of dendritic and axonal development by the novel Kruppel-like factor Dar1. J. Neurosci. 2011;31:3309–3319. doi: 10.1523/JNEUROSCI.6307-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN., Jr. Statistical analysis of real-time PCR data. BMC bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: synaptotagmin-GFP. Genesis. 2002;34:142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.