Abstract
Alcohol dependence (AD) is a heritable substance addiction with adverse physical and psychological consequences, representing a major health and economic burden on societies worldwide. Genes thus far implicated via linkage, candidate gene and genome-wide association studies (GWAS) account for only a small fraction of its overall risk, with effects varying across ethnic groups. Here we investigate the genetic architecture of alcoholism and report on the extent to which common, genome-wide SNPs collectively account for risk of AD in two US populations, African-Americans (AAs) and European-Americans (EAs). Analyzing GWAS data for two independent case-control sample sets, we compute polymarker scores that are significantly associated with alcoholism (P=1.64 × 10−3 and 2.08 × 10−4 for EAs and AAs, respectively), reflecting the small individual effects of thousands of variants derived from patterns of allelic architecture that are population-specific. Simulations show that disease models based on rare and uncommon causal variants (MAF<0.05) best fit the observed distribution of polymarker signals. When scoring bins were annotated for gene location and examined for constituent biological networks, gene enrichment is observed for several cellular processes and functions in both EA and AA populations, transcending their underlying allelic differences. Our results reveal key insights into the complex etiology of AD, raising the possibility of an important role for rare and uncommon variants, and identify polygenic mechanisms that encompass a spectrum of disease liability, with some, such as chloride transporters and glycine metabolism genes, displaying subtle, modifying effects that are likely to escape detection in most GWAS designs.
Keywords: alcohol dependence, GWAS, polymarker scores, synthetic association, rare variants, pathway analysis
Introduction
Alcohol dependence (AD) is a complex, highly heritable disorder characterized by compulsive, excessive consumption of alcohol, resulting in physical, psychological and social impairment (American Psychiatric Association 1994) that constitutes a significant health and economic burden in the US (Harwood 2000), with 4–5% of the population affected at any given time (Li et al. 2007). Family, twin and adoption studies have consistently shown a substantial genetic contribution to disease etiology (Goodwin et al. 1974; McGue 1999; Nurnberger et al. 2004), with heritability estimates ranging from 50–80% (Heath et al. 1997; Knopik et al. 2004). To date a number of genes have been implicated in alcoholism susceptibility via linkage analysis, candidate gene approaches and genome-wide association studies (GWAS), including the often replicated GABRA2 (Edenberg et al. 2004; Bierut et al. 2010) and ADH4 (Guindalini et al. 2005; Luo et al. 2004; Edenberg et al. 2006), among others (Wang et al. 2004; Xuei et al. 2006; Chen et al. 2009; Zlojutro et al. 2011; Bierut et al. 2012). However, these genetic loci collectively account for only a small fraction of the risk of AD, with effects varying across ethnic groups (Gelernter & Kranzler 2009).
This shortfall in explained genetic variance, popularly referred to as “missing heritability” (Maher 2008; Manolio et al. 2009), has been widely observed for other complex disease phenotypes, leading many to re-evaluate the validity of the common disease-common variant hypothesis and suggest a more central role for rare variants, epigenetics and/or genetic interactions in pathogenesis. New analytical approaches, however, have begun to bridge the heritability gap, indicating that much of the additive genetic variance of complex traits, such as human height (Yang et al. 2010), intelligence (Davies et al. 2011) and schizophrenia (Lee et al. 2012), are arguably captured by common GWAS markers.
In this paper we investigate the polygenic architecture of alcoholism by evaluating the extent to which common, genome-wide SNPs collectively capture the variation in susceptibility in two US populations, European-Americans (EAs) and African-Americans (AAs). To achieve this, we aggregated genotypic data from case-control samples into sets of quantitative scores, representing varying thresholds of GWAS P-values or particular classes of minor allele frequency (MAF), and tested their association to AD, as well as their fit to simulated disease models. In addition, we computed empirical, additive genetic relationships between case-control subjects with the available GWAS data and estimated from them the total variance in AD liability that is accounted by common SNPs via linear mixed models, as proposed by Yang and colleagues (2010). Lastly, in an effort to identify some of the specific genetic mechanisms that underlie the biology of AD, the designated scoring bins of putative risk alleles were annotated to gene locations and tested for gene enrichment for various biological ontologies and signaling pathways in EA and AA populations using a permuted approach.
Materials and methods
Population Samples
Routines for aggregating genome-wide SNP counts into composite scores (Fig. S1) were designed using GWAS data from case-control subjects representing European-American (n = 1,274) and African-American (n = 285) populations, as ascertained by the Collaborative Study on the Genetics of Alcoholism (COGA) (Edenberg et al. 2010), a national consortium designed to study the genetic predisposition to develop alcoholism and related phenotypes. Alcoholic probands were recruited from inpatient and outpatient treatment centers, whereas controls were selected from Health Maintenance Organizations (HMOs), drivers’ license records, and dental clinics, with the objective of obtaining representative samples of the communities at each recruitment site (Reich et al. 1998). All cases were diagnosed for DSM-IV alcohol dependence at each clinical assessment if assessed multiple times. To avoid pleiotropic genetic components that contribute to multiple substance abuse phenotypes, non-alcoholic controls did not meet diagnostic criteria for other illicit substance abuse or dependence (although cases could). Furthermore, controls were required to be 25 years or older and to have consumed alcohol at some point in their lives to ensure that their unaffected status was not due to lack of exposure to alcohol. These procedures were approved by the Institutional Review Boards of all COGA sites, and all participants gave informed consent.
Developed scoring routines were applied to independent GWAS data for EA (n = 1,573) and AA (n = 841) case-control subjects from the Study of Addiction: Genetics and Environment (SAGE) (Bierut et al. 2010). For this data set, AD cases and non-dependent controls were selected from three large, complementary studies: COGA, Family Study of Cocaine Dependence (FSCD), and Collaborative Genetic Study of Nicotine Dependence (COGEND). All COGA subjects were excluded to ensure independence between the discovery and target samples (although it should be noted that not all of the cases from the COGA case-control study were a part of SAGE). Cases (n = 958) were identified as having a lifetime history of AD using DSM-IV criteria. Control subjects (n = 1,456) were required to report a history of drinking and have no significant AD symptoms or any other substance dependencies. The Institutional Review Board at each contributing institution approved the protocols, and all subjects provided written informed consent for genetic studies.
Genome-Wide Genotyping
Genotyping was performed by the Center for Inherited Disease Research (CIDR) at John Hopkins University using the Illumina® Infinium II assay protocol (Gunderson et al. 2006) for hybridization to Illumina® HumanHap 1M BeadChips (Illumina, San Diego, CA), with a blind duplicate reproducibility of 99.97% and 99.98% for the COGA and SAGE samples, respectively. Details are reported by Bierut et al. (2010) and Edenberg et al. (2010). Protocols and GWAS data for the COGA (n = 1,003,800 SNPs) and SAGE (n = 1,040,106 SNPs) samples are available on the National Center for Biotechnology Information (NCBI) database dbGaP. For each sample set, subjects were assigned to EA and AA population groups via principal component (PC) analysis of the genotype data, corresponding to two major population clusters observable in PC space (Table 1; Fig. S2 and S3).
Table 1.
Descriptive statistics for COGA and SAGE data sets.
| European-American | African-American | |
|---|---|---|
| COGA | ||
| Sample Size | 1,274 | 457 |
| Cases (Controls) | 767 (507) | 329 (128) |
| Males (Females) | 676 (598) | 245 (212) |
| Mean Age | 41.17 yrs | 39.87 yrs |
| SAGEa | ||
| Sample Size | 1,573 | 841 |
| Cases (Controls) | 599 (974) | 359 (482) |
| Males (Females) | 616 (957) | 389 (452) |
| Mean Age | 35.71 yrs | 39.59 yrs |
COGA, Collaborative Study on Genetics of Alcoholism; SAGE, Study of Addiction: Genetics and Environment; yrs, years.
All COGA subjects were excluded to ensure independence between the two data sets.
Polymarker Scoring
COGA has conducted a series of analyses that evaluate the predictive utility of GWAS data for alcoholism and related phenotypes (Yan et al. 2011). Here, we have expanded the scope of this work by examining what this information tells us about the disorder’s underlying genetic architecture. Using a two-stage, risk prediction framework similar to the one employed by Purcell et al. (2009) to characterize the polygenic basis of schizophrenia, we aggregated variation across nominally associated GWAS loci into quantitative scores or “genomic profiles” and correlated these predictors with observed AD status in independent target samples from SAGE (Fig. S1).
For the design of the genome-wide scoring routines, autosomal GWAS data (n = 1,003,800) was pruned of SNPs in strong linkage disequilibrium (LD) with other markers (pairwise r2 threshold of 0.25, within a 200-SNP sliding window), ensuring that the scores computed in our target samples represent the aggregate effect of a large number of predominantly independent markers. The retained genotype data for EA (n = 193,979) and AA samples (n = 332,687) were further trimmed for minor allele frequency (MAF ≥ 0.05), call rate (≥ 0.98) and deviation from Hardy-Weinberg (HW) equilibrium (p ≥ 1×10−3), leaving 124,291 and 256,549 SNPs in the two respective population samples available for developing the scoring routines.
Genome-wide association tests were conducted with the program PLINK (Purcell et al. 2007), using the standard measured genotype method, with covariates age and sex (quantile-quantile plots are provided in Fig. S4). SNPs were then delineated into bins according to incremental thresholds of association test P-values, as well as MAF ranges, from which scores were defined as the total number of “risk” alleles carried by a given target sample, weighted by the log odds ratio (OR) for AD as estimated from the COGA data. Scores were calculated for the SAGE data, limited to SNPs with an allele frequency > 1%, in HW equilibrium (P ≥ 1×10−4), and with a minimum call rate of 98% (n = 948,658). To measure how well the SAGE target scores predict AD risk, logistic regression analyses of case-control status were performed to quantify the amount of variation accounted for by the scores, as determined by Nagelkerke’s pseudo-R2, representing the difference in R2 estimates for the null model, with terms for the intercept, age, sex and genotyping rate, and the alternative model that includes the polymarker scores.
Variance Component Analysis of AD Liability
Using the method proposed by Yang et al. (2010), the amount of variance in AD risk that is explained simultaneously by genome-wide SNPs was estimated by treating the effects of SNPs as statistically random. The model for this analysis is y = Σ wibi + e, where y is the phenotypic value, bi is the effect of the ith SNP, wi is a scaling factor equivalent to (xi − 2pi)/(2pi (1 − pi))1/2 with pi the allele frequency and xi the genotype indicator of the ith SNP (values of 0, 1 or 2), and e is a random environmental effect (Visscher et al. 2010). In matrix notation this is equivalent to y = g + e, where g = Wb is a vector of genetic values calculated from the SNP alleles each individual carries, with var(g) = WW′σb2 (WW′ is the matrix of genetic relationships between individuals). Using the software GCTA (Yang et al. 2011), we computed the genetic relationship matrix (GRM) for our LD-pruned genotype data, combining the COGA and SAGE samples for the EA (n = 2,763) and AA (n = 1,167) study populations, with the exclusion of individuals with estimated relatedness greater than 0.025 (i.e., corresponding to third cousins or closer). The GRMs were then fitted to the linear models for AD status, parameterized on an unobserved continuous liability scale via a probit transformation (Lee et al. 2011), using a restricted maximum likelihood (REML) approach, with the covariates age and sex. The estimates of AD variation explained by the GRMs were corrected for ascertainment bias using population-specific prevalence rates (0.038 and 0.036 for EAs and AAs, respectively) (Grant et al. 2004).
Simulation of Genome-Wide Scores for Different Disease Models
Using the program GCTA, case-control phenotypes for six disease architectures were simulated using real genotype data from the COGA and SAGE data sets, pruned of SNPs in strong LD, as described above. The phenotypes were generated from a simple additive genetic model yj = Σi xijbi + ej, where xij is the number of reference alleles for the ith causal variant of the jth individual, bj is the allelic effect of the ith causal variant, and ej is the residual effect generated from a normal distribution with mean 0 and variance of (xijbi)(1 − 1/h2). The six selected disease models differ with regards to the number of causal loci (100, 1,000 or 5,000) and their allele frequency profiles (MAF < 0.05 or MAF ≥ 0.05). For each of the population samples, a new AD status was assigned via a disease liability threshold, with the number of cases matching those in the original phenotype data. Causal loci were randomly selected from LD-pruned SNPs excluded from the initial two-stage, genome-wide scoring analysis, which have not been filtered for MAF and thus include rare variants (Fig. S1), with 100 replicates drawn for each disease model. The heritability of the disease phenotypes was set at a conservative 0.65, the median of estimates reported for AD in a pair of published studies (Heath et al. 1997; Knopik et al. 2004). Effect sizes were fixed for each model, making the variance accounted for by a causal locus proportional to the total number of loci in a given disease model and its respective minor allele frequency. With the program PLINK and the R statistical package (R Development Core Team 2011), genome-wide association tests, followed by the aforementioned two-stage, scoring routines, delineated according to MAF class, were conducted on the simulated disease phenotypes and the corresponding COGA or SAGE genotype data.
Gene Enrichment Analysis for Biological Ontologies
For the final analytical approach, the focus was shifted from the general, genetic architecture of AD to the detection of specific polygenic mechanisms giving rise to the disorder, as permuted gene enrichment analyses were conducted on the bins of potential risk alleles applied in the scoring calculations described above. For each population-specific bin, representing one of twenty GWAS P-value thresholds defined by increments of 0.05, alleles exhibiting contrasting directions of effect between the discovery and target samples (accounting for ∼50% of the markers) were assumed to be predominantly due to chance and removed from analysis to help control statistical noise. The remaining SNPs were then assigned to genes based on the UCSC hg18 gene coordinates, with the boundaries extended +/− 20 kb to include regions that may have cis-regulatory functions. The resulting gene lists were tested for enrichment for genes belonging to various biological ontologies (n = 507) and receptor signaling pathways (n = 227), as defined in the ResNet Mammalian v. 7.0 database curated by Ariadne Genomics (Bethesda, MD). Unlike the “GO” vocabulary from the Gene Ontology Consortium, the Ariadne ontologies are mostly based on narrowly defined cellular processes and molecular functions, thus limiting the redundancies between biological categories. Each ResNet ontology and pathway was limited to only member genes marked by genotyped SNPs in the LD-pruned GWAS data from COGA, with only those retaining 2 or more genes examined for enrichment (n = 651 and 639 ontologies/pathways for the AA and EA GWAS data sets, respectively). Gene enrichment was evaluated via Fisher’s exact tests using the R package, with permuted lists (n = 1,000) randomly assembled from genes marked by the LD-pruned GWAS data (totaling 16,740 and 14,777 for AAs and EAs, respectively), with each gene weighted for its SNP coverage. Empirical P-values represent the number of times the P-values from permuted Fisher’s exact tests are smaller than the value from the observed test.
Results
Application of Population-Matched Scoring Routines
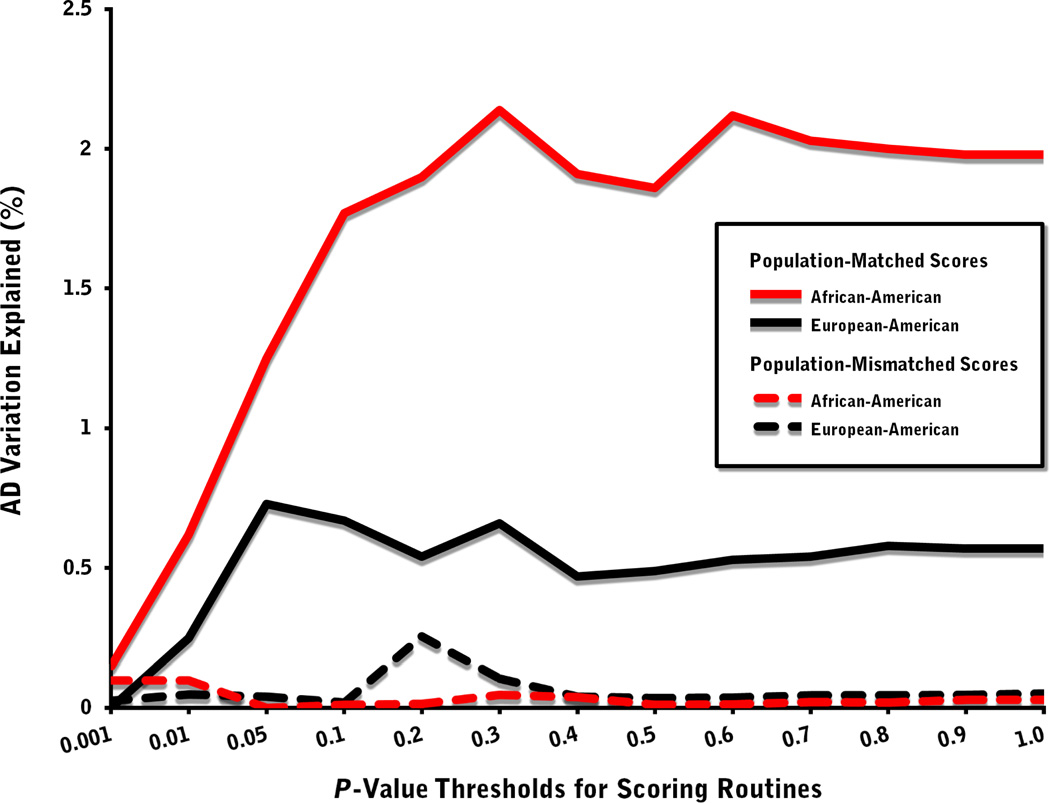
When target scores derived from associations in the COGA data set are used to predict case/control status for the matched population (i.e. EA or AA) in SAGE, the R2 estimates for both EAs and AAs are modest, but statistically significant (Fig. 1). Maximum values are observed for association P-value thresholds set at less than 0.05 (n = 6,790 risk alleles) for EA and 0.30 (n = 76,218 risk alleles) for AA target samples, accounting for 0.73% (P = 1.64 × 10−3) and 2.14% (P = 2.08 × 10−4) of the variation in AD status, respectively (Table S1); although both sets of R2 values begin to plateau at around the 0.05 or 0.10 thresholds. Given the heritability estimates of 50–80% for AD liability (Heath et al. 1997; Knopik et al. 2004), these results fall well short of the total additive genetic variation believed to underlie the illness. This discrepancy can be attributed in part to the statistical noise arising from the inclusion of non-associated markers, as well as the large number of small, individual estimates of AD effect, whose standard errors reduce the accuracy of the aggregate scores in predicting disease outcome despite their small sizes.
Figure 1. Variance in alcohol dependence (AD) explained by genome-wide scores for African-American and European-American subjects.
Polymarker scoring routines based on AD status were designed for thirteen GWAS significance thresholds (plotted against the x axis) using COGA data and applied to SAGE target samples. The y axis represents Nagelkerke’s pseudo R2, the amount of variation in AD accounted by the SAGE scores, computed for routines that are both population-matched and mismatched across the COGA and SAGE data sets.
Variance Component Analysis
To obtain a more accurate estimate of AD variance explained by genome-wide markers, we conducted variance component analysis using the method proposed by the Yang et al. (2010). Based on this approach, we estimate from our LD-pruned GWAS data that 37.8% (s.e. = 10.4%) and 35.1% (s.e. = 27.8%) of the variation in AD risk is captured by common SNPs in EAs and AAs, respectively (Table S2). Although the heritability of AD is not fully recovered in these results, at least for the larger heritability estimates when one considers the substantial standard errors, it is reasonable that any unaccounted, additive genetic variation could be “hidden” from our statistical purview due to causal variants not being in strong LD with the GWAS markers, with the most probable candidates being those with small MAFs (Purcell et al. 2009; Visscher et al. 2012).
Application of Population-Mismatched Scoring Routines
Despite having nearly equivalent levels of AD risk variation captured by common genetic markers, EAs and AAs appear to have distinctly different allelic architectures. Genome-wide scores generated from routines that are mismatched for population (i.e., EA COGA discovery sample and AA SAGE target sample, or vice versa) do not predict AD risk (Fig. 1), with R2 values generally less than 0.1% and βs displaying opposite directions of effect (Table S3). This stands in sharp contrast to the genome-wide scoring results reported by Purcell et al. (2009) for a larger sample of schizophrenia subjects, in which AA cases were found to carry significantly more European-derived risk alleles than AA controls (P = 0.008; R2 = 0.4%). Though the aggregate differences in allele frequencies and LD patterns between EAs and AAs are expected to lead to attenuated associations, our findings suggest a larger degree of allelic heterogeneity may exist between these two populations for the genetic liability of AD than for schizophrenia and perhaps other psychiatric disorders.
Scoring Delineated by Association P-Value and MAF Class
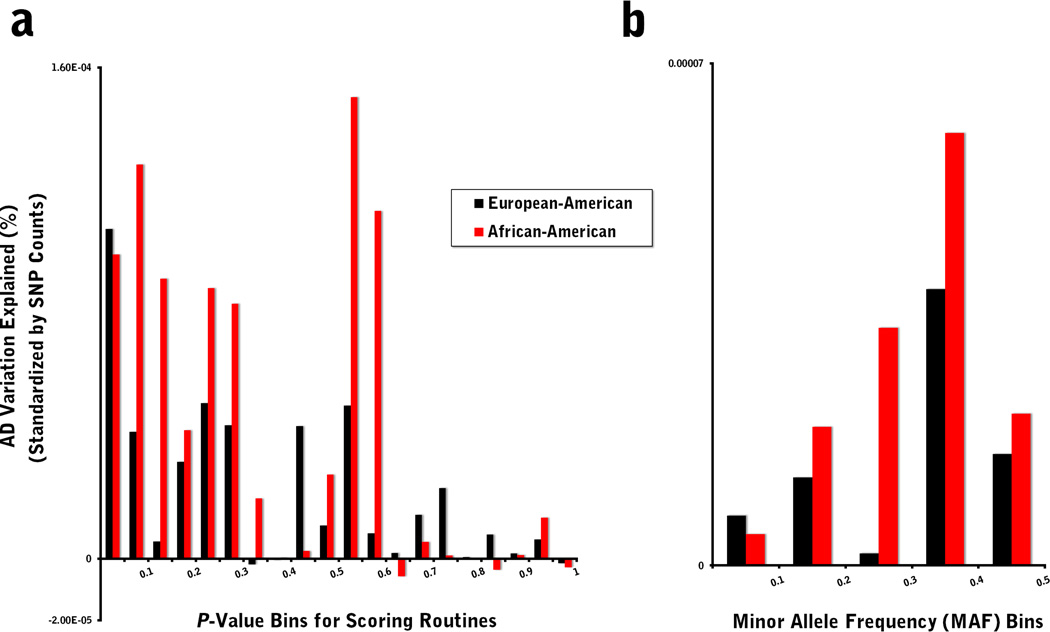
To further dissect the allelic architecture of alcoholism in our two study populations, we re-ran the scoring routines on non-overlapping bins of risk alleles, based either on GWAS P-values or classes of minor allele frequency. For the target samples, we observed significant R2 values for scores representing weakly associated risk alleles, including ones for significance thresholds as permissive as 0.50 ≤ P < 0.55 (OR: 1.05–1.15; R2 = 0.30%; P = 0.027) and 0.55 ≤ P < 0.60 (OR: 1.07–1.26; R2 = 1.42%; P = 0.0012) for EAs and AAs, respectively (Fig. 2a; Table S4). When broken down by frequency, a skew in the R2 distribution towards more common markers is evident (Fig. 2b; Table S5), with a peak at 0.3 ≤ MAF < 0.4 for both population samples (EA: R2 = 0.57%, P = 0.0047; AA: R2 = 2.13%, P = 0.00013), suggesting an important role for highly common variants in the liability of AD if one assumes a robust LD relationship between score alleles and the unknown causal loci.
Figure 2. Scoring analysis stratified by non-overlapping bins of score risk alleles based on (a) GWAS P-values and (b) minor allele frequencies.
Variance explained was standardized by SNP counts for the respective bins.
Simulation of Genome-Wide Scores
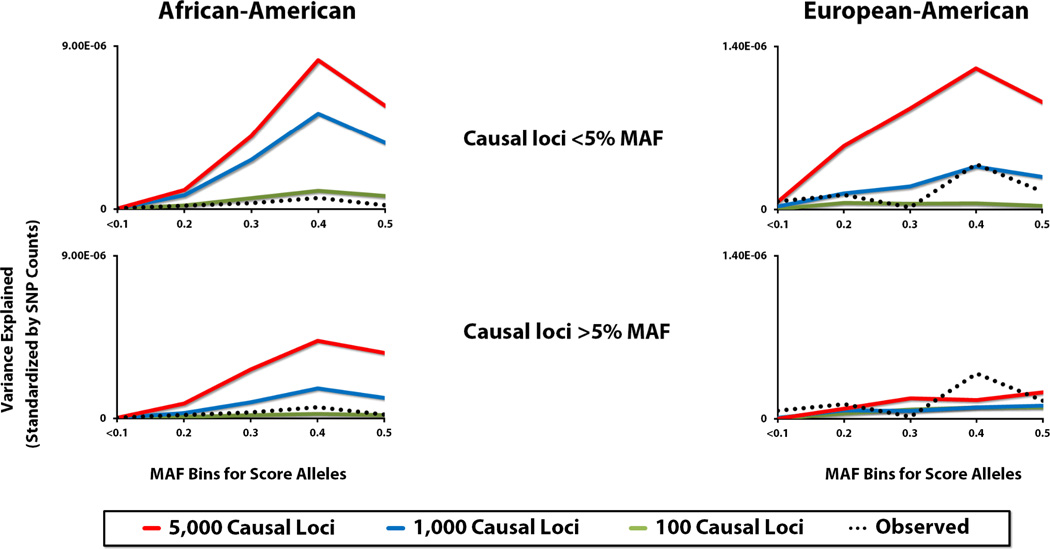
To explore whether or not this is indeed the case, we simulated a series of disease models and conducted the same two-stage, genome-wide scoring delineated by MAF class (Fig. 3). Surprisingly, the strongest R2 signals in both populations are for simulated diseases arising entirely from rare and uncommon risk alleles, with modes overlapping the observed peak at the 0.3 ≤ MAF < 0.4. For AAs the observed R2 values fall slightly below those generated for the model based on 100 causal loci (with a maximum of 0.022 variance explained by any individual variant; goodness of fit R2 = 0.78, P = 0.046), whereas the best fitting model for EAs is for 1,000 causal loci (maximum variance explained of 0.0037; R2 = 0.49, P = 0.19). For disease models representing the other part of the frequency spectrum (i.e., common alleles), the fit to the observed results is poor for EAs (R2 = 0.07, P = 0.68 for 5,000 causal loci), with the genome-wide scores explaining substantially less of the variation in the disease phenotypes. For AAs the signals are more concordant; however they also are noticeably attenuated relative to those obtained for rare/uncommon risk alleles, with the model based on 1,000 causal loci fitting best to the observed R2 values (R2 = 0.79, P = 0.044). In addition to these six models, we also tested mixed models representing rare and common causal loci drawn randomly from the MAF spectrum. As one would expect, the simulated R2 profiles are intermediate to those reported for the models discussed above (Fig. S5), with the ones based on 100 and 5,000 causal loci fitting best to the observed results for AAs (R2 = 0.80, P = 0.04) and EAs (R2 = 0.38, P = 0.27), respectively.
Figure 3. Variance explained by genome-wide scoring routines for observed and simulated disease phenotypes according to MAF class.
The variances explained, derived from MAF bins comprised of different score alleles, are presented for six disease models for each study population. The models represent either 100, 1,000 or 5,000 causal variants, which were randomly drawn from SNP data excluded from the original design of scoring routines, limited to either rare/uncommon markers (<5% MAF) or common markers (>5% MAF). Each model was replicated 100 times. Disease heritability was set at 0.65, with causal effect sizes fixed for all loci. Observed R2 results for AD are given as black, dotted lines.
Gene Enrichment Analysis
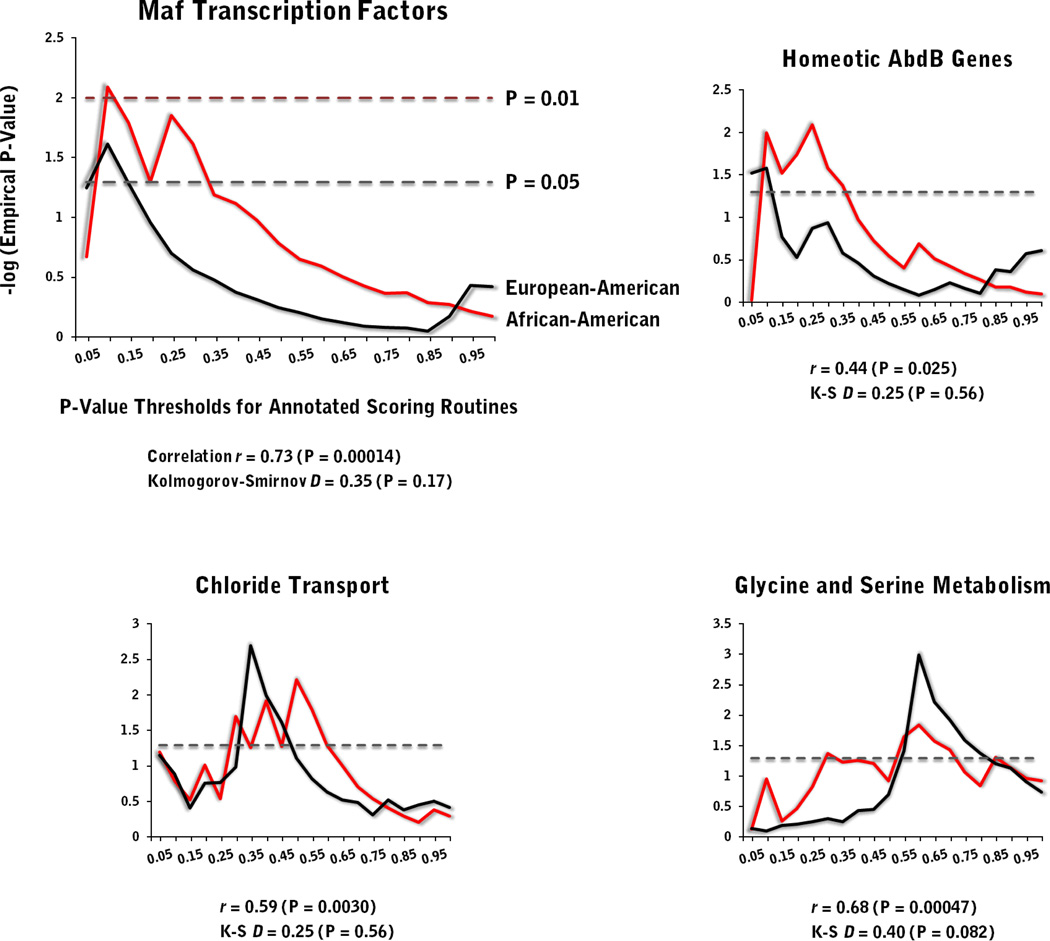
To identify potentially causative biological mechanisms for AD, we examined our scoring bins, ones defined by cumulative GWAS P-value thresholds, for discernible ontological patterns, including those comprised of alleles with small, statistically non-significant effects on disease risk. The permuted Fisher’s exact tests show that about 90% of the examined ontologies exhibit no significant evidence of gene enrichment (empirical P ≤ 0.05) for any of the twenty P-value thresholds for either population (Table S6), with the percentages slightly higher for the signaling pathways. Of the biological relationships that do show significant enrichment, approximately half are for single thresholds, with only a limited number displaying significance across three or more of the tested levels (n = 15 and 19 for EAs and AAs). From this latter group, the following four ontologies show evidence of significant enrichment in both population samples (in parentheses are the sizes of the ontologies after being matched against the gene coverage of population-specific GWAS data, along with the top empirical P-values observed for the various EA and AA gene lists, respectively): Maf transcription factors (n = 6 genes; P-values = 0.024 and 0.008); homeotic (Hox) AbdB genes (n = 16 genes; P = 0.026 and 0.008); chloride transport (n = 62 and 66 genes; P = 0.002 and 0.006); and glycine and serine metabolism (n = 27 and 33 genes; P = 0.001 and 0.014).
Discussion
Through the aggregation of genome-wide, genotypic data, we present molecular evidence for a substantial polygenic component to the risk of alcoholism. Although accounting for only a modest amount of variation in AD risk (R2 values less than 3%; Fig. 1), our polymarker scores are nonetheless significantly associated to AD in both EA and AA target samples, even for putative risk alleles with GWAS P-values as lax as 0.55 ≤ p < 0.60 (Fig. 2a), underscoring the statistical power issues faced by genome-wide studies of similarly complex, polygenic traits. When populations were mismatched between the discovery and target samples for the scoring routines, the resulting scores became poor predictors of alcoholism, suggesting that the genetic liabilities stem from patterns of allelic architecture that are predominantly population-specific, a finding that is consistent with the various novel genetic associations and linkage signals reported in ethnic studies (Gelernter & Kranzler 2009).
For a more accurate estimate of the proportion of AD variation captured by GWAS markers, we conducted variance component analysis via mixed linear modeling, with allelic effects treated as statistically random. Using this approach (Yang et al. 2010), we determined that around one-third of the phenotypic variation is collectively accounted for by common SNPs in our EA and AA samples. Thus, if recent estimates of AD heritability are reliable, this result still leaves much of the additive genetic variation to be explained, with a potentially important role for rare causal variants. One example that is particularly instructive is rs1229984, a functional variant in ADH1B known to modify the conversion of alcohol to acetaldehyde, with a low frequency in non-Asian populations (∼1–3%) and, as a result, is poorly tagged by genotyped markers in current GWAS arrays. However, when this coding variant was directly genotyped in the COGA sample, a genome-wide significant association with AD was revealed, with a strong protective effect (Bierut et al. 2012).
To explore the relative contributions of common versus rare causal variants to the genetic liability of AD, we simulated a series of disease models and conducted the same two-stage, genome-wide scoring for EA and AA samples, with routines delineated by MAF class. What we find is that the best fitting models, overall, are those based entirely on rare causal variants (Fig. 3). Although these simulations examined only a limited number of possible disease architectures and therefore do not preclude the possibility of thousands or tens of thousands of common loci solely contributing to AD risk, especially for heritabilities larger than the one tested in our models (65%), it does indicate that polymarker scoring based on GWAS data for complex phenotypes can detect the small, collective effects of rare and uncommon genetic determinants and that there could be as few as one hundred of them. This contrasts with the conclusion reached by Purcell et al. (2009) in their models that simulated both disease status and genotype data, asserting that rare variants could not alone account for R2 signals generated from genome-wide, polymarker scoring of psychiatric disorders such as schizophrenia. This discrepancy between the studies may stem from design differences, as our simulations are based on real genotype data, which could have produced divergent features in the respective LD structures, or perhaps be a reflection of fundamental differences in the genetic architectures of these two psychiatric disorders.
The exact contributions of rare and common genetic variants to the underpinnings of AD remain unknown, but consistent with both the neutral and selection theories of genetic variation, our results, principally those for the EA sample, point to a strong likelihood for a concentration of weak causal variants from the low end of the MAF spectrum that can lurk beneath stringent genome-wide significance boundaries (Heath et al. 2011). Moreover, these findings support the theoretical possibility of “synthetic association”, a phenomenon described and coined by Dickson et al. (2010), in which the aggregate risk effects of extended genomic blocks of rare variants can create genome-wide significant associations with weakly tagged, common SNP markers, complicating the interpretation of GWAS results as it relates to the localization of causal variants. Despite other simulation studies and empirical evidence that lend support to this genetic mechanism of association, including the well-known instance involving the NOD2 locus and Crohn’s disease (Anderson et al. 2011), several recent papers have disputed the prevalence of synthetic association for complex phenotypes, drawing upon the paucity of replicable linkage signals that should be amenable to similar rare variant effects (Orozco et al. 2010; Anderson et al. 2011), as well as the modality of GWAS marker signals towards higher frequencies (Wray et al. 2011) and the observance of trans-ethnic associations (Waters et al. 2010). Although our findings indicate the plausibility of recapitulating rare variant effects through polymarker scores derived from common GWAS markers (e.g., 0.3 ≤ MAF < 0.4), this should not be interpreted as support of a rare variant-only model for the genetic architecture of alcoholism, as mixed models also exhibit robust fits (Fig. S5). The simulations conducted here represent only a cursory exploration of potential disease models, and thus does not discount other neutral evolutionary models for common genetic variation, especially given the positive relationship between risk allele frequency and disease variance explained (Visscher et al. 2012).
Lastly, this study delved beyond the allelic architecture of alcoholism, searching for wider biological patterns among alleles of varying association strengths by means of permuted gene enrichment analysis. Of the ontologies and signaling pathways that show significant enrichment in our data set, four are particularly compelling, as they represent broad signals (i.e., significance across three or more GWAS P-value thresholds) and are shared by both EAs and AAs: a) Maf transcription factors, which regulate cell differentiation and potentially brain segmentation (Cordes & Barsh 1994; Sadl et al. 2003); b) Hox AbdB genes, a family of transcription factors involved in embryogenesis and axial patterning; c) chloride transport, which plays a crucial role in synaptic inhibition through the activity of GABA and glycine neurotransmitters; and d) glycine and serine metabolism, for which glycine is an important inhibitory neurotransmitter. When their empirical P-values from the enrichment analyses are plotted, they reveal remarkably similar trends between the two populations (Fig. 4), with overlapping peaks, significant correlations (r ranging from 0.73 to 0.44), and non-significant Kolmogorov-Smirnov (K–S) distances between the P-value distributions. All of this, as well as substantial sharing between annotated gene lists at peak enrichment thresholds (Table S7), suggest commonalities in the genetic mechanisms responsible for AD liability that transcend population differences in the underlying allelic architectures. For Maf transcription factors and AbdB genes, the strongest signals for enrichment occur at small GWAS P-values (< 0.10), indicating large to modest effects on AD risk by genes belonging to these particular groups, whereas chloride transport (which includes GABA receptors that have been often implicated in AD) and glycine/serine metabolism reveal peaks at markedly higher thresholds (< 0.60), pointing to more subtle effects that are likely to escape detection in most single marker association tests. These enrichment differences may represent molecular signatures of a hierarchical etiology, in which the effects of the Maf and AbdB transcription factors on developmental and pathophysiological pathways related to AD are more proximate to the disease endpoint than chloride transport and glycine-related neurochemical systems. (Gaino & Fishell 2002; Pandey 2004; Yamauchi 2005; Lee & Messing 2008; Aguirre et al. 2010; Moonat et al. 2010; Kaun et al. 2011)
Figure 4. Empirical P-values of four, top-ranking biological ontologies based on permuted (1,000X) Fisher’s exact tests of gene enrichment in European- and African-American samples.
Allele bins, delineated by genome-wide association P-values at cumulative increments of 0.05, were annotated for gene location using UCSC hg18 coordinates.
From the other ontologies and pathways tested for enrichment in this study, some also exhibit similar trends in their empirical P-value distributions between the two study populations, of which several appear to be potentially meaningful to AD and neuronal function, including NOTCH → EP300 signaling (Gaino & Fishell 2002; Aguirre et al. 2010; Kaun et al. 2011), organic anion transport (Moonat et al. 2010), and calcium-dependent protein kinases (Yamauchi 2005; Lee & Messing 2008) (Fig. S6; Table S8). However it should be noted that many of the significant enrichment signals are indeed population-specific (Fig. S7 and S8), hinting that some important differences in the genetic etiology of alcoholism may exist between EAs and AAs.
In conclusion, we report that a significant proportion of variance in AD risk can be explained by common SNPs of small effect in an aggregate manner, with allelic architectures that are specific to EA and AA populations. Although these findings would appear to support the widely held common disease-common variant hypothesis, our simulation models show that the modest effects of rare and uncommon susceptibility loci can be captured in genome-wide association signals for complex disease phenotypes, at least in aggregate. How big of a role rare variation actually has, if any, in the genetic liability of alcoholism is unknown, however there is growing evidence that it can have important effects on psychiatric disorders, including results from studies of copy number variants (CNVs) (Stone et al. 2008; Sanders et al. 2011), as well as early findings from exome sequencing efforts that reveal an abundance of rare genetic variation, much of which is functional (Keinan & Clark 2012; Kiezun et al. 2012; Tennessen et al. 2012). In addition, our GWAS data sets have implicated a number of biologically relevant pathways and mechanisms in both study populations, including various transcription factors known to affect brain development, as well as genes involved in inhibitory neurotransmission. The latter plays a key role in the brain’s reward system and has been previously linked to externalizing psychopathologies (e.g., antisocial personality disorder, childhood conduct disorder) that share a genetic predisposition with substance abuse disorders (Dick et al. 2006), thus providing compelling targets for future research on alcoholism, as well as population-specific pathways.
Supplementary Material
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, and L. Bierut, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Texas Biomedical Research Institute (L. Almasy); and Virginia Commonwealth University (D. Dick). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). SOLAR is supported by R01 MH59490 from the National Institutes of Mental Health. Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Family-based genotyping was performed using the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by the Indiana Genomics Initiative of Indiana University (INGEN®); INGEN is supported in part by The Lilly Endowment, Inc. AA receives support from ABMRF/Foundation for Alcohol Research. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
L.J. Bierut, J.P. Rice and A.M. Goate are inventors on the patent ‘Markers for Addiction’ (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction.
Footnotes
No other authors reported any conflicts of interest to disclose.
References
- Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–327. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. fourth edition. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Anderson CA, Soranzo N, Zeggini E, Barrett JC. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E Gene Environment Association Studies Consortium. A genome-wide association study of alcohol dependence. PNAS. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr., Kuperman S, O’Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neruopsychiatr Genet. 2009;150B:359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, McGhee K, Lopez L, Gow AJ, Corley J, Redmond P, Fox HC, Haggarty P, Whalley LJ, McNeill G, Goddard ME, Espeseth T, Lundervold AJ, Reinvang I, Pickles A, Steen VM, Ollier W, Porteous DJ, Horan M, Starr JM, Pendleton N, Visscher PM, Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Kuperman S, Hesselbrock V, Schuckit M, Almasy L, Tischfield J, Porjesz B, Begleiter H, Nurnberger J, Jr., Xuei X, Edenberg HJ, Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet. 2006;36:577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr., O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit MA, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Ann Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiatry. 1974;31:164–169. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, Chang W, Bullis D, Musmacker J, King C, Lebruska LL, Barker D, Oliphant A, Kuhn KM, Shen R. Whole-genome genotyping. Methods Enzymol. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- Harwood H. Updating estimates of the economic costs of alcohol abuse in the United States: Estimates, update methods, and data. Report prepared by The Lewin Group for the National Institute on Alcohol Abuse and Alcoholism, 2000. Based on estimates, analyses, and data reported in Harwood, H.; Fountain, D.; and Livermore, G. The Economic Costs of Alcohol and Drug Abuse in the United States 1992. Report prepared for the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Department of Health and Human Services. Bethesda, MD: National Institutes of Health; 2000. [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14:612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, Hultman CM, Lichtenstein P, Magnusson P, Lehner T, Shugart YY, Price AL, de Bakker PI, Purcell SM, Sunyaev SR. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, Statham D, Whitfield JB, Martin NG. Genetic effects on alcohol dependence risk: Re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- Lee AM, Messing RO. Protein kinases and addiction. Ann NY Acad Sci. 2008;1141:22–57. doi: 10.1196/annals.1441.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, PGC-SCZ ISC,MGS, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Hewit tBG, Grant BF. The Alcohol Dependence Syndrome, 30 years later: a commentary. Addiction. 2007;102:1522–1530. doi: 10.1111/j.1360-0443.2007.01911.x. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M. Phenotyping alcoholism. Alcohol Clin Exp Res. 1999;23:757–758. doi: 10.1111/j.1530-0277.1999.tb04180.x. [DOI] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67:73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Wiegand R, Bucholz KK, O’Connor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, Bierut L, Hinrichs AL, Kuperman S, Hesselbrock V, Porjesz B. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61:1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010;19:R137–R144. doi: 10.1093/hmg/ddq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. International Schizophrenia Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr., Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Sadl VS, Sing A, Mar L, Jin F, Cordes SP. Analysis of hindbrain patterning defects caused by the Kreisler(enu) mutation reveals multiple roles of Kreisler in hindbrain segmentation. Dev Dyn. 2003;227:134–142. doi: 10.1002/dvdy.10279. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL, O’Donovan MC, Gurling H, Kirov GK, Blackwood DHR, Corvin A. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. International Schizophrenia Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. NHLBI Exome Sequencing Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Yang J, Goddard ME. A commentary on “Common SNPs explain a large proportion of the heritability for human height” by Yang et al. (2010) Twin Res Hum Genet. 2010;13:517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Goddard ME, Derks EM, Wray NR. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Mol Psychiatry. 2012;17:474–485. doi: 10.1038/mp.2011.65. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI, Jr., Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Reich T, Goate AM, Bierut LJ. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Waters KM, Stram DO, Hassanein MT, Le Marchand L, Wilkens LR, Maskarinec G, Monroe KR, Kolonel LN, Altshuler D, Henderson BE, Haiman CA. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e10001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Purcell SM, Visscher PM. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol. 2011;9:e1000579. doi: 10.1371/journal.pbio.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr., Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opiod system with alcohol dependence. Mol Psychiatry. 2006;11:1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Yamauchi T. Neuronal Ca2+/calmodulin-dependent protein kinase II - discovery, progress in a quarter of a century, and perspective: implication for learning and memory. Biol Pharm Bull. 2005;28:1342–1354. doi: 10.1248/bpb.28.1342. [DOI] [PubMed] [Google Scholar]
- Yan J, Aliev F, Kendler KS, Webb BT, Schuckit MA, Nurnberger JI, Jr., Edenberg HJ, Kramer JR, Agrawal A, Goate AM, Tischfield JA, Dick DM. Using genetic information from genome wide association studies in risk prediction for alcohol dependence in two samples; Washington, DC. XIXth World Congress on Psychiatric Genetics; September 10–14.2011. [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Jr., Rice JP, Schuckit MA, Foroud T, Edenberg HJ, Porjesz B, Almasy L. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropyschiatr Genet. 2011;156B:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.