Abstract
Background
Therapies that inhibit androgen receptor (AR) are needed for treatment of castration-resistant prostate cancer (CRPC). The ErbB3 binding protein 1 (EBP1) reduces protein expression of both AR and its target genes in CRPC. Although EBP1 regulates AR in hormone-sensitive prostate cancer cells by both destabilizing AR mRNA and inhibiting protein translation, the mechanism of EBP1 down regulation of AR in CRPC is unknown.
Materials and Methods
Western blot and quantitative PCR analysis of cell lysates and polysomes were used to assess AR mRNA, protein expression and translation.
Results
In contrast to hormone- dependent cells, EBP1 did not change steady state levels of AR mRNA or AR mRNA stability in hormone refractory cells. EBP1 did slow protein translation of AR mRNA. The ErbB3/4 ligand heregulin further diminished AR translation in EBP1 - transfected cells, but not in control cells.
Conclusion
These studies suggest that one pathway of EBP1 down-regulation of AR levels may be lost in CRPC.
Keywords: Prostate cancer, EBP1, mRNA translation, androgen receptor
Prostate cancer begins as an androgen- dependent tumor that regresses in response to therapies that reduce testosterone concentration. Despite this treatment, the disease progresses to a uniformly fatal hormone-refractory or castration-resistant prostate cancer (CRPC). The androgen receptor (AR) regulates malignant progression in CRPC (1). Thus, new approaches to reduce AR protein expression and silence AR signaling are important for the therapy of CRPC.
ERBB family members and their ligands have been implicated in the development of castration resistance due to their stimulatory effects on AR function (2). Androgen- independent sublines of human prostate cancer xenografts express higher levels of ErbB2 than androgen-dependent sublines and ectopic expression of ErbB2 sensitizes cells to extremely low levels of androgen (3). ERBB2/3 kinase activity stabilizes AR protein levels via inhibition of ubiquitin-mediated degradation and increased recruitment of AR to AR-responsive promoters (2). ERBB3 has also recently emerged as an important factor in prostate cancer progression. Overexpression of ERBB3 is linked to a less favorable prognosis in prostate cancer (4). A high frequency of nuclear ERBB3 expression is associated with hormone-refractory prostate cancer (5) and metastasis (6).
Our laboratory has demonstrated that EBP1, an ERBB3 binding protein, is an AR co-repressor (7–9). EBP1 expression is significantly reduced in CRPC (10). Ectopic expression of EBP1 in the hormone-refractory LNCaP variant C81 cell line leads to reversion to an androgen-dependent phenotype and to lower expression of AR protein (10,11). EBP1 affects both transcription and post-transcriptional regulation of the AR gene via its ability to bind DNA (12), RNA (13) and protein (9). The ability of EBP1 to bind RNA led us to explore its role in post-transcriptional AR regulation. We previously demonstrated that the C-terminal domain of EBP1 bound to both a (UC)- rich region in the 3' UTR of AR mRNA that affects mRNA stability (14) and a 5' CAG rich sequence in the coding sequence, predicted to affect protein translation (15). EBP1 overexpression in hormone-dependent prostate cancer cells resulted in reduced AR mRNA steady state levels, mRNA stability and translation (13). However, EBP1 post-transcriptional regulation of AR was not examined in CRPC.
The purpose of this study was to examine whether ectopic expression of EBP1 in hormone refractory prostate cancer cells would lower AR protein expression via destabilizing AR mRNA and slowing protein translation as is found in hormone sensitive cells. In addition, we wished to determine if the ERBB3/4 ligand heregulin (HRG) could destabilize AR mRNA and slow protein translation in hormone refractory cells.
Materials and Methods
Cell culture and reagents
The generation of C81 cells stably transfected with EBP1 cDNA was previously described (10). HRGß1 and EGF were purchased from R&D Systems (Minneapolis, MN,USA) and Cell Signaling (Danvers, MA,USA), respectively.
Linear sucrose gradient fractionation
Logarithmically growing C81 cells and C81–EBP1 transfected cells were harvested and cytoplasmic extracts obtained in Triton-X 100 based polysome extraction buffer (13). Sucrose gradient fractionation was performed as described previously (13). Each gradient was fractionated into 1-ml aliquots using a gradient fractionator (Brandel, Gaithersburg, MD, USA) and monitored by optical density measurement (A254). Each fraction was diluted with an equal volume of water and RNA was isolated using Trizol (Invitrogen, Carlsbad, CA,USA). AR mRNA levels in individual fractions were measured using quantitative real-time PCR (RT-qPCR) as described below. For analysis of EBP1, EIF2α, phospho-EIF2α and actin protein expression, 15 μl of each fraction was denatured in 15 μl of 1×Laemmli buffer and analyzed by Western blotting as described below. Where indicated, both C81 vector control and EBP1- transfected cells were serum-starved for 24 hours and then treated with HRG (20 ng/ml) for 24 hours prior to analysis. Results shown represent one of three independent experiments.
RNA isolation and PCR analysis
RNA was prepared from C81 vector control or EBP1 transfected cells untreated or treated with HRG using Trizol reagent (10) or from sucrose gradient fractions as described above. RNA was DNAse-treated and converted into cDNA using the AMV reverse-transcription system (Promega, Madison, WI, USA) in the presence of random hexamers (Invitrogen). The cDNA was used for conventional PCR or RT-qPCR for detection of AR mRNA with specific gene primers as previously described (13). A MYIQ real-time PCR detection system and SYBR green PCR mix (Bio-Rad, Richmond, CA, USA) were used to carry out the real-time PCR. The relative quantitation of targeted genes was determined by the comparative ΔΔCt (threshold) method using actin as an internal control (13). All data were analyzed from three independent experiments and statistical significance was validated by Student's t-test.
Western blotting analysis
Western blot analysis of C81 vector control and EBP1-transfected cells treated with or without HRG (20 ng/ml) was performed as described previously (16). The EBP1 antibody was from Millipore (Billerica, MA, USA), the monoclonal anti-β-actin and anti-GADPH antibodies were from Sigma (St. Louis, MO, USA), and the monoclonal antibody against AR was from Santa Cruz (Santa Cruz, CA,USA). Polyclonal antibodies against EIF2α, phospho-EIF2α (Ser51), p44/42 MAPK, phospho-p44/42 MAPK (Thr202/Tyr204), SAPK/JNK and phospho-SAPK/JNK (Thr183/Tyr185) were from Cell Signaling.
Statistical analysis
RT-qPCR assays were performed in triplicate and repeated at least three times and Western blotting assays were repeated three times. All data presented represent one individual experiment. Where appropriate, means comparison were made using a two-tailed Student's t- test with alpha equal to 0.05.
Results
EBP1 reduces AR protein levels without changing AR mRNA levels in CRPC
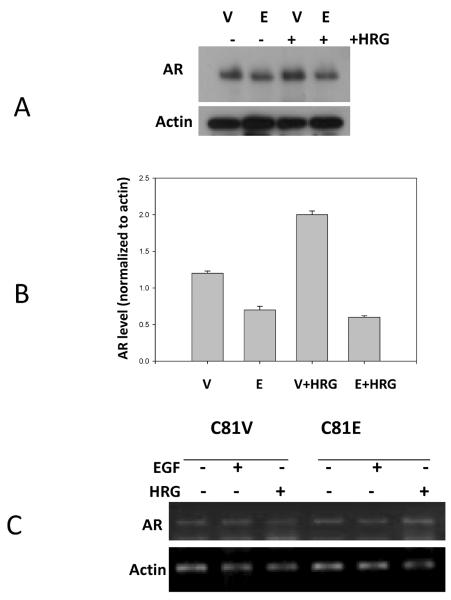
Previous work from our laboratory showed that ectopic expression of EBP1 reduced the levels of AR protein in C81 hormone refractory cells (10). However, microarray data indicated that the steady-state level of AR mRNA in EBP1 transfectants was not changed (10). In contrast, we previously found that EBP1 reduced the steady state level of AR mRNA in LNCaP hormone-responsive cells and that HRG enhanced this decrease (13). We therefore tested the effects of EBP1 overexpression and HRG treatment on steady state-levels of AR mRNA and protein in hormone refractory C81 vector and EBP1 transfectants. We found, as previously shown (10), that AR protein levels decreased in EBP1 transfectants as compared with vector controls (Figure 1A and B). HRG increased the levels of AR protein in C81 vector control cells and slightly decreased AR expression levels in EBP1 transfectants (Figure 1A and B). In keeping with previous data (10), we found that mRNA levels of AR were approximately equal in C81 vector control and EBP1-transfected cells. Neither EGF nor HRG affected AR mRNA levels in either control or EBP1-transfected cells (Figure 1C). Neither ectopic expression of EBP1 nor HRG treatment affected AR mRNA decay (data not shown), in keeping with data on AR mRNA expression levels.
Figure 1. Differential effects of HRG on AR expression in C81 vector and EBP1 transfectants.
A: C81 vector (V) and EBP1-transfected (E) cells were serum-starved overnight and treated with HRGß1 (20 ng/ml) for 24 hours. Cell lysates were analyzed by Western blotting for expression of AR and actin as indicated. A representative Western blot of three independent experiments is shown. B: Densitometric analysis of the blot in A. C: Total RNA was extracted from vector control and EBP1 transfected C81 cells treated with HRG (20 ng/ml, 24 hours) or EGF (50 ng/ml, 24 hours) to detect steady-state levels of AR and actin mRNA by RT-PCR. A representative gel of three independent experiments is shown.
EBP1 affects AR mRNA association with polysomes
As ectopic expression of EBP1 reduced AR protein but not mRNA levels, we examined the effects of EBP1 overexpression and HRG treatment on AR protein translation.
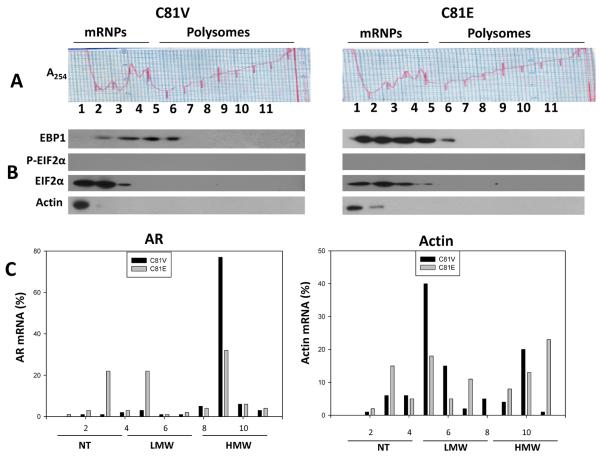
Equal amounts of cytoplasmic extracts of logarithmically growing C81 vector and C81 EBP1-transfected cells were fractionated by sucrose gradient centrifugation. Absorbance readings at A254 indicated the distribution of monosomes and polysomes. The pattern was comparable in C81 vector control (Figure 2A, left panel) and C81 EBP1 transfectants (Figure 2A, right panel), indicating that overexpression of EBP1 did not trigger significant changes in general protein translation. Western blot analysis of the different fractions of the gradient demonstrated the association of EBP1 with 40S, 60S and 80S ribosomes. EBP1 co-purified with elongation-initiation factor 2α (EIF2α), a component of the translation initiation complex, as has been previously reported in HeLa cells (17). EIF2α was not phosphorylated in either C81 vector or EBP1- transfected cells (Figure 2B).
Figure 2. Overexpression of EBP1 reduces AR mRNA translation.
A: Cytoplasmic extracts of C81 vector (left) and C81 EBP1 transfected cells (right) growing in logarithmic culture conditions were fractioned by centrifugation on 10–50 w/v sucrose gradients. Eleven fractions were collected and absorbance at 254-nm was recorded. B: Western blot analysis was used to measure the levels of EBP1, p-EIF2α, EIF2α, and actin in each fraction from C81 vector control (left panel) and C81 EBP1-transfected (right panel) cells. C: The levels of AR and actin mRNAs in each gradient fraction were measured by RT-qPCR and plotted as a percentage of the total AR (left panel) or actin mRNA levels (right panel) in that sample. The translational activity associated with each fraction is indicated as NT (not translated), LMW (low molecular weight polysomes, moderately translated) and HMW (high-molecular-weight polysomes, actively translated) as previously described (13). Data represent one of three independent experiments showing similar results.
The relative abundance of AR mRNA in each polysome fraction was next used to measure the degree of engagement of AR mRNA with the translational apparatus. The percentage of AR mRNA in actively translating polysomes declined markedly following stable transfection of exogenous EBP1 cDNA (Figure 2C, left panel), in keeping with the observation that AR protein levels are reduced in EBP1-transfected cells as compared to controls. The polysome profile of a control house-keeping transcript (actin mRNA) was not significantly altered in C81E cells as compared to C81V cells (Figure 2C, right panel). This finding suggests that ectopic expression of EBP1 reduces AR protein translation.
HRG attenuates AR mRNA translation only in EBP1 transfected cells
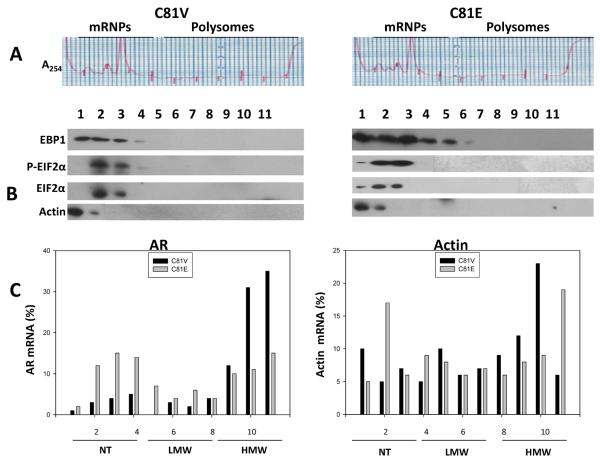
To investigate if HRG can regulate the association of EBP1 with polysomes, C81 vector and EBP1- transfected cell lines were treated with HRG and processed as described above. HRG treatment did not change the polysomal distribution of EBP1, EIF2α or actin in either cell line as compared to untreated cells. HRG treatment induced the phosphorylation of EIF2α in both C81 vector control and EBP1- transfected cells. However, the phosphorylation of EIF2α was more marked in the EBP1 transfectants than in vector controls (Figure 3B).
Figure 3. HRG attenuates AR mRNA translation only in EBP1 transfected cells.
C81 vector and EBP- transfected cells were serum-starved overnight and then treated with HRG (20 ng/ml) for 24 h. Cell lysates were processed as described in the Materials and Methods. A: Absorbance profiles at 254-nm. B: Western blot analysis of polysome fractions. C: The levels of AR and actin mRNAs in each gradient fraction were measured by RT-qPCR and plotted as a percentage of the total AR (left panel) or actin mRNA levels (right panel) in that sample. Data represent one of three independent experiments showing similar results.
AR mRNA continued to be associated with actively translated polysomes in C81 vector control cells in the presence of HRG (Figure 3C). In contrast, the percentage of AR mRNA associated with the non-translated fractions was about 50% greater than in untreated EBP1 transfected cells as determined using area under the curve software (PRISM, Irvine, CA, USA) (Figures 2C and 3C, left panels). The association of actin mRNA with polysomes was not significantly reduced by HRG treatment in either vector or EBP1 transfected cells (Figure 3C, right panel).
Effects of ectopic expression of EBP1 on EIF2α, Mitogen-activated protein kinase (MAPK) and Jun-N-terminal kinase (JNK) phosphorylation after HRG treatment
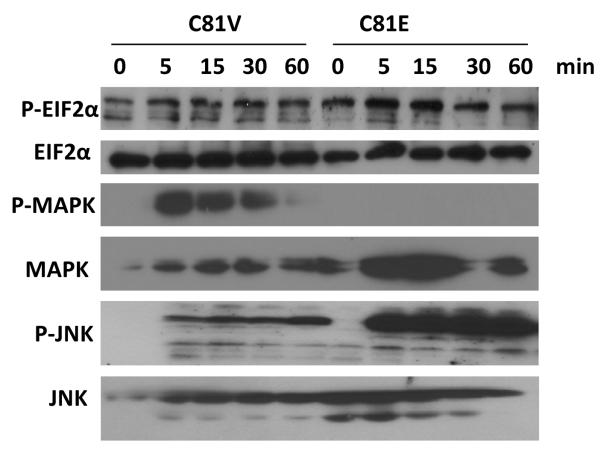
As EIF2α phosphorylation levels were higher in polysome fractions of EBP1 transfectants after HRG stimulation, we examined the activation status of HRG-regulated kinases that may affect EIF2α phosphorylation. A recent study demonstrated that inhibition of MAPK phosphorylation is associated with decreased stability of mitogen-activated kinase phosphatase 7 (MKP-7), a JNK phosphatase. The decreased activity of this phosphatase results in increased activation of JNK. The activation of JNK results in phosphorylation and subsequent inhibition of protein-phosphatase 1(PP1), an EIF2α-directed phosphatase. The inhibition of PP1 activity leads finally to increased phosphorylation of EIF2α (18). We therefore examined the ability of HRG to induce phosphorylation of MAPK and JNK in C81 vector control and EBP1 transfectants. We showed, using total cell lysates, that HRG stimulation resulted in greater EIF2α phosphorylation in EBP1 transfectants as compared to vector controls. Ectopic expression of EBP1 resulted in inhibition of HRG-stimulated phosphorylation of MAPK. In contrast, JNK phosphorylation was increased in response to HRG in EBP1- overexpressing cells as compared to vector controls (Figure 4).
Figure 4. Effects of ectopic expression of EBP1 on EIF2α, MAPK and JNK phosphorylation after HRG treatment.
C81 vector control and EBP1-transfected cells were serum-starved overnight and then incubated with or without heregulin (20 ng/ml) for the times indicated. Whole-cell extracts were prepared and resolved as described in the Materials and Methods. Levels of phosphorylated and total EIF2α, MAPK, and JNK were determined by Western blotting.
Discussion
EBP1 is an AR mRNA binding protein that reduces AR mRNA levels in hormone-dependent prostate cancer cells by promoting AR mRNA decay and inhibiting protein translation (13). In this study, we examined if ectopic expression of EBP1 could also down-regulate AR in hormone-refractory cells. We found, in contrast to what is observed in hormone-dependent cells, that EBP1 inhibited AR translation but had no effect on steady-state levels of AR mRNA or mRNA stability in hormone-refractory cells. We suggest that the inability of EBP1 to destabilize AR mRNA may play a role in the development of CRPC.
The reasons for the failure of EBP1 to destabilize AR mRNA and reduce steady-state mRNA levels in hormone-refractory cells are unknown. EBP1 is likely to act as part of a complex to destabilize AR mRNA and key co-regulatory RNA-binding proteins such as HuR and poly-C binding proteins 1 and 2 (CP1 and CP2) (14) may be missing in hormone refractory cells. It is also possible that the 3'UTR regulatory element in AR mRNA, important to its stability, is mutated in CRPC.
However, as in hormone-dependent LNCaP cells, overexpression of EBP1 in hormone-refractory C81 cells resulted in a shift of AR mRNA towards translationally inactive ribosomes. This finding suggests that EBP1 might inhibit translation initiation of AR mRNA. EBP1 cosediments with 40S, 60S and 80S ribosomes in CRPC cells, consistent with previous reports that EBP1 is part of ribonucleoprotein protein translation complexes in HeLa cells (17,19). Furthermore, our previous finding that EBP1 binds a CAG trinucleotide repeat at the 5'-end of AR exon 1, predicted to form a stable stem-loop structure and regulate AR translation, supports this view (13). The fact that EBP1 binds viral internal ribosome entry sites supports its potential role in protein translation (20, 21).
We also found that in contrast to hormone-dependent cells, HRG increased AR protein levels in control C81 cells, but slightly reduced AR protein in EBP1 transfectants. Similarly, Cai et al. (22) found that HRG failed to affect AR levels in CWR-R1 hormone-refractory cells. These findings suggest that the inability of HRG to reduce AR protein levels may be a hallmark of the hormone-refractory phenotype. HRG was able to attenuate AR mRNA translation in EBP1- overexpressing cells, but not in vector controls that express EBP1 at extremely low levels (10). Similarly, we observed that knock-out of EBP1 in hormone-dependent LNCaP cells results in the inability of HRG to attenuate AR mRNA translation (13). Our findings suggest that HRG affects AR translation via an EBP1-mediated pathway. Another example of ERBB ligand regulation of AR mRNA translation is the HB-EGF-induced decrease of AR mRNA translation (23). The HB-EGF effect is mediated by the RNA-binding protein, hnRNP-K which, as in our study, binds sequences within the AR coding region that leads to reduced translation rates (24).
Consistent with the reduced translation rate, we found that phosphorylation and thus inactivation of EIF2α in response to HRG was enhanced in EBP1- transfected versus control cells. The mechanism of the HRG enhancement of EIF2α phosphorylation is not known. However, our results indicated that EBP1 overexpression inhibited HRG-induced MAPK phosphorylation and increased activation of JNK. It is known that JNK phosphorylates and inactivates the EIF2α phosphatase PP1, resulting in enhanced phosphorylation of EIF2α (25). Thus, the increased activation of JNK in EBP1 transfectants after HRG stimulation may have led to enhanced EIF2α phosphorylation.
In summary, our study suggests that EBP1 can inhibit AR translation in a model of CRPC, although its ability to destabilize AR mRNA is lost. The loss of a major mode of AR regulation in CRPC may contribute to malignant progression.
Acknowledgements
This work was supported by Maryland Technology Corporation (June 6, 2006) and Department of Defense grant W81XWH-07-0267 (YZ) and NIH grant 1R01CA138583 (AWH). We thank Dr. M. Lin (University of Nebraska Medical Center) for permission to use the C81 cells and Dr. Yun Qiu (University of Maryland School of Medicine) for providing these cells.
References
- 1.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellinghoff IK, Vivanco I, Kwon A, Tran C, Wongvipat J, Sawyers CL. HER2/neu kinase-dependent modulation of androgen receptor function through effects on DNA binding and stability. Cancer Cell. 2004;6:517–527. doi: 10.1016/j.ccr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 4.Leung HY, Weston J, Gullick WJ, Williams G. A potential autocrine loop between heregulin-alpha and erbB-3 receptor in human prostatic adenocarcinoma. Br J Urol. 1997;79:212–216. doi: 10.1046/j.1464-410x.1997.30412.x. [DOI] [PubMed] [Google Scholar]
- 5.Koumakpayi IH, Diallo JS, Le PC, Lessard L, Gleave M, Begin LR, Mes-Masson AM, Saad F. Expression and nuclear localization of ErbB3 in prostate cancer. Clin Cancer Res. 2006;12:2730–2737. doi: 10.1158/1078-0432.CCR-05-2242. [DOI] [PubMed] [Google Scholar]
- 6.Cheng CJ, Ye XC, Vakar-Lopez F, Kim J, Tu SM, Chen DT, Navone NM, Yu-Lee LY, Lin SH, Hu MC. Bone microenvironment and androgen status modulate subcellular localization of ErbB3 in prostate cancer cells. Mol Cancer Res. 2007;5:675–684. doi: 10.1158/1541-7786.MCR-06-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Hamburger AW. Specificity and heregulin regulation of Ebp1 (ErbB3 binding protein 1)- mediated repression of androgen receptor signalling. Br J Cancer. 2005;92:140–146. doi: 10.1038/sj.bjc.6602257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, Goloubeva O, Ross DD, Brodie A, Hamburger AW. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc Natl Acad Sci USA. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YX, Fondell JD, Wang QB, Xia XM, Cheng AW, Lu ML, Hamburger AW. Repression of androgen receptor mediated transcription by the ErbB-3 binding protein, Ebp1. Oncogene. 2002;21:5609–5618. doi: 10.1038/sj.onc.1205638. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Linn D, Liu Z, Melamed J, Tavora F, Young CY, Burger AM, Hamburger AW. EBP1, an ErbB3-binding protein, is decreased in prostate cancer and implicated in hormone resistance. Mol Cancer Ther. 2008;7:3176–3186. doi: 10.1158/1535-7163.MCT-08-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Ali TZ, Zhou H, D'Souza DR, Lu Y, Jaffe J, Liu Z, Passaniti A, Hamburger AW. ErbB3 binding protein 1 represses metastasis-promoting gene anterior gradient protein 2 in prostate cancer. Cancer Res. 2010;70:240–248. doi: 10.1158/0008-5472.CAN-09-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Akinmade D, Hamburger AW. The ErbB3-binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 2005;33:6024–6033. doi: 10.1093/nar/gki903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Mazan-Mamczarz K, Martindale JL, Barker A, Liu Z, Gorospe M, Leedman PJ, Gartenhaus RB, Hamburger AW, Zhang Y. Post-transcriptional regulation of androgen receptor mRNA by an ErbB3- binding protein 1 in prostate cancer. Nucleic Acids Res. 2010;38:3619–3631. doi: 10.1093/nar/gkq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, Leedman PJ. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3'-untranslated region of the androgen receptor messenger RNA. J Biol Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 15.Yeap BB, Wilce JA, Leedman PJ. The androgen receptor mRNA. Bioessays. 2004;26:672–682. doi: 10.1002/bies.20051. [DOI] [PubMed] [Google Scholar]
- 16.Xia X, Lessor TJ, Zhang Y, Woodford N, Hamburger AW. Analysis of the expression pattern of Ebp1, an ErbB-3-binding protein. Biochem Biophys Res Commun. 2001;289:240–244. doi: 10.1006/bbrc.2001.5942. [DOI] [PubMed] [Google Scholar]
- 17.Squatrito M, Mancino M, Sala L, Draetta GF. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem. Biophys Res Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 18.Monick MM, Powers LS, Gross TJ, Flaherty DM, Barrett CW, Hunninghake GW. Active ERK contributes to protein translation by preventing JNK-dependent inhibition of protein phosphatase 1. J Immunol. 2006;177:1636–1645. doi: 10.4049/jimmunol.177.3.1636. [DOI] [PubMed] [Google Scholar]
- 19.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 20.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 22.Cai C, Portnoy DC, Wang H, Jiang X, Chen S, Balk SP. Androgen receptor expression in prostate cancer cells is suppressed by activation of epidermal growth factor receptor and ErbB2. Cancer Res. 2009;69:5202–5209. doi: 10.1158/0008-5472.CAN-09-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cinar B, De BA, Freeman MR. Post-transcriptional regulation of the androgen receptor by Mammalian target of rapamycin. Cancer Res. 2005;65:2547–2553. doi: 10.1158/0008-5472.CAN-04-3411. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay NK, Kim J, Cinar B, Ramachandran A, Hager MH, Di VD, Adam RM, Rubin MA, Raychaudhuri P, De BA, Freeman MR. Heterogeneous nuclear ribonucleoprotein K is a novel regulator of androgen receptor translation. Cancer Res. 2009;69:2210–2218. doi: 10.1158/0008-5472.CAN-08-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]