Abstract
An increasingly important role for the ErbB3 receptor in the genesis and progression of breast cancer is emerging. ErbB3 is frequently overexpressed in breast cancer and coexpression of ErbB2/3 is a poor prognostic indicator. ErbB3 has also been implicated in the development of resistance to antiestrogens such as tamoxifen and ErbB tyrosine kinase inhibitors such as gefitinib. Persistent activation of the AKT pathway has been postulated to contribute to ErbB3 –mediated resistance to these therapies. This activation may be due in part to the inappropriate production of the ErbB3 ligand heregulin. ErbB3 binding proteins, which negatively regulate ErbB3 protein levels and the ability of ErbB3 to transmit proliferative signals, also contribute to breast cancer progression and treatment resistance. These proteins include the intracellular RING finger E3 ubiquitin ligase Nrdp1 and the leucine-rich protein LRIG-1 that mediate receptor degradation. Ebp1, another ErbB3 binding protein, suppresses HRG driven breast cancer cell growth and contributes to tamoxifen sensitivity. These studies point to the importance of the evaluation of protein levels and functional activity of ErbB3 and its binding proteins in breast cancer prognosis and prediction of clinical response to treatment.
Keywords: ErbB3, Nrdp1, Ebp1, tamoxifen, gefitinib
The importance of the ErbB3 Receptor in breast cancer
The ErbB family of tyrosine kinase receptors [Human EGF Receptor 1 (HER1, ErbB1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4)] regulates the growth, differentiation and survival of human breast epithelial cells (1). Ligand binding to ErbB receptors results in the formation of homo or heterodimers leading to activation of receptor tyrosine kinases(2). ErbB3 is the only member of the ErbB receptor family that lacks tyrosine kinase activity due to amino acid substitutions in the conserved kinase domain (3). Thus the interaction of ErbB3 with its binding partners is essential for its biological activity. Tyrosine phosphorylation of the ErbB3 receptor carboxy terminal domain provides docking sites for cytoplasmic effectors of intracellular signaling (2). Of note, tyrosine phosphorylated ErbB3 has six consensus sites for the p85 adapter subunit of PI-3 kinase (4;5). Both SH2 groups of each of the three PI -3 kinase adapter isoforms bind with moderate or high affinity at multiple sites in ErbB3(6). Only ErbB3, of all ErbB family members, has three pairs of tyrosine residues separated by a single glutamic acid residue (YEY motif). The p85 subunit of PI 3-kinase p85 binds to the first tyrosine residues of each of these three sites (7), providing an explanation for the strong binding of p85 to ErbB3. After ligand binding, ErbB receptors activate MAPK and AKT signaling pathways. AKT stimulation of mTOR activity also results in activation of the p70S6 kinase and the 4E-binding protein 1, which in turn regulate transition through the G1-S phase of the cell cycle (8). Thus, evasion of apoptosis and cell growth inhibition via the AKT pathway play a major role in the ability of ErbB3 to stimulate cell growth and overcome clinical treatment.
A wealth of clinical data demonstrate the aberrant expression of ErbB2 and EGFR in breast cancer (1) (9). Although ErbB3 overexpression has been noted in breast cancer since the 1990s (10) (11-13), the role ErbB3 in breast cancer progression has only more recently been appreciated. Overexpression of ErbB3 is found in the absence of gene amplification or mutation (10). The prevalence of ErbB3 expression as judged by analysis of mRNA(14;15) or protein levels by immunohistochemistry (11;16;17) has been reported to be between 17-52%. High expression of ErbB3 was positively associated with metastasis (10), histological grade (11), and tumor size and recurrence (12). ErbB3 overexpression has been found to be an independent predictor of survival in some (18) (14) but not all studies (12). However, at present, a simple determination of ErbB3 mRNA or protein levels in breast cancer cannot be used by itself for biological or clinical predictions due in part to many factors that will influence ErbB3 expression, localization or activity.
There is also a growing awareness of the importance of the ErbB2/3 heterodimer in breast cancer progression. The majority of ErbB2 positive tumors are strongly positive for ErbB3 (19). Co expression of ErbB2 and ErbB3 is significantly associated with decreased patient survival (19). The role of the ErbB2/3 heterodimer as a growth stimulatory signal transduction unit was first demonstrated in vitro by Koland’s group that found that HRG stimulation caused ErbB2/3 heterodimerization (20). Since then, many studies have demonstrated that the ErbB2/ErbB3 receptor pair forms the most potent mitogenic and transforming receptor complex in vitro (21). The ErbB2/3 heterodimer has been proposed to be an “oncogenic unit” and key to the proliferation of human breast cancer cells (22). Holbro et al (22) demonstrated that ErbB2 required Erbb3 to drive breast tumor proliferation. Loss of functional ErbB3 by a designer transcription factor (E3) or loss of ErbB2 by an intracellular trapping antibody in breast cancer cell lines that overexpress ErbB2 led to the same degree of inhibition of cell proliferation. In addition, activity of downsteam signaling pathways such as MAPK or AKT was decreased when either functional ErbB3 or ErbB2 was ablated. A constitutively activated AKT could rescue the proliferative block induced by loss of either ErbB2 or ErbB3. The results demonstrated that ErbB2 overexpression and activity alone were insufficient to promote breast cancer cell division and point out the importance of ErbB3 in the design of ErbB directed therapies.
Further support for the role of ErbB3 as a target in breast cancer therapies comes from studies using ErbB3 directed antibodies that determined whether ErbB3 had a physiologically relevant ErbB2-independent role in transmitting proliferative and migratory signals in cell lines that have low levels of ErbB2. Such cell lines and patients that have low levels of ErbB2 expression are insensitive to the clinically approved ErbB2 directed antibody Herceptin (trastuzumab). Treatment of an MCF-7 adriamcyin resistant cell line and MDA-MB-468 cells with an antibody to the extracellular domain of ErbB3 decreased cellular migration and proliferation. The inhibition of cell growth was accompanied by a substantial decrease in HRG-induced ErbB2 tyrosine phosphorylation and ErbB2/3 heterodimerization. Activation of the PI 3-kinase and JNK pathways was also decreased. Finally, treatment with the ErbB3 targeted antibody resulted in a decrease in ErbB3 recycling to the cell surface after HRG stimulation. These studies suggested the utility of anti-ErbB3 antibodies in breast cancer therapy especially for patients whose tumors do not overexpress ErbB2 (23).
The foregoing studies indicate a complex role for ErbB3 in the progression and development of breast cancer. They suggest that ErbB3 may be an attractive therapeutic target in its own right and an essential part of multi modality therapies focused on ErbB pathways. Unfortunately, ErbB3 is not an easily “druggable” target due to its lack of tyrosine kinase activity(3). Several therapeutic approaches are being developed such as the use of RNA aptamers to the extracellular domain of ErbB3(24), synthetic designer zinc transcription factors that inhibit ErbB3 gene expression (25), and micro RNAs directed towards ErbB3 (26).
The Role of ErbB3 in Treatment Resistance
As outlined above, the role of ErbB3 in the genesis and progression of breast cancer has been studied for many years. However, recently a role has emerged for ErbB3 in the development of resistance to newer forms of cancer therapies. I would like to discuss the role of ErbB3 in development of hormone resistance and of resistance to tyrosine kinase inhibitors (TKIs).
Resistance to antiestrogens
De novo and acquired resistance to antiestrogens such as tamoxifen is a significant problem in the treatment of breast cancer patients (27). Mechanisms postulated to contribute to the acquisition of the resistant phenotype include loss or mutation of Estrogen Receptor (ER), alterations in the intracellular pharmacology of breast tumor cells, ligand independent ER mediated transcription and perturbation of the interaction of ER with transcriptional corepressors (28). More recently, activation of ErbB family members has been linked to resistance to antiestrogens such as tamoxifen as the enhanced expression of members of the ErbB receptor family leads to the ability of cells to bypass normal endocrine responsiveness (29) (30). A large literature exists that demonstrate cross talk between the ER and ErbB2 and EGFR signaling pathways (31) (32) (33). The importance of ErbB3 in development of the hormone resistant phenotype is emerging. For example, clinical studies examining a large retrospective group of tamoxifen treated, ER-positive breast cancer patients have demonstrated that ErbB2/3 positive patients were significantly more likely to relapse on tamoxifen (34). In addition, it has been known for many years that tamoxifen sensitive MCF-7 cells transfected with a HRGßB-2 cDNA, become estrogen independent and resistant to antiestrogens both in vitro and in vivo (32).
Direct demonstration of the involvement of ErbB3 in tamoxifen resistance comes from studies of Liu et al who showed that downregulation of ErbB3 by siRNA abrogates ErbB2 mediated tamoxifen resistance in breast cancer cells (35). MCF-7 cells which express low levels of ErbB2 and MCF-7 cells transfected with ErbB2 (MCF-7/erbB2) were transfected with siRNA directed against ErbB3. Elimination of ErbB3 expression had no effect on expression or phosphorylation status of ERα. Downregulation of ErbB3 significantly enhanced tamoxifen associated growth inhibition not only in parental MCF-7 cells, but also in MCF-7/erbB2 cells. The increased growth inhibition was due to enhanced apoptosis. These data indicated that ErbB2 associated tamoxifen resistance can be abrogated by downregulation of ErbB3. The molecular mechanism responsible for the increased sensitivity to tamoxifen after abrogation of ErbB3 expression is not known. However, significantly decreased levels of phosphorylated AKT were observed when ErbB3 expression was abrogated. Tamoxifen resistance is associated with activation of PI-3 kinase/AKT. For example, MCF-7 cells expressing a constitutively active AKT proliferate under reduced estrogen conditions and are resistant to the growth inhibitory effects of tamoxifen (36). Overexpression of AKT pathways is associated with tamoxifen resistance in a clinical setting (36-38). Therefore, Liu et al (35) suggested that ErbB3 activation of AKT may contribute to tamoxifen resistance. In summary, this study suggests that ErbB3 and its cognate ligands may have a profound influence on tamoxifen resistance. The ErbB3 receptor may serve as a useful molecular target for modulating tamoxifen resistance in ER positive, ErbB2 overexpressing breast cancers.
Resistance to Tyrosine Kinase Inhibitors
Studies of in vitro and preclinical models that suggested the importance of ErbB family members in breast cancer progression led to the development of drugs targeted to ErbB receptors. Several small molecular weight tyrosine kinase inhibitors (TKIs) have undergone clinical trials in breast cancer. Unfortunately, initial phase II studies did not demonstrate high efficacy of the EGFR inhibitor gefitinib in heavily pretreated patients with metastatic breast cancer (39). Although TKIs against the ErbB family inhibit ErbB2 and EGFR activity both in vitro and in animal models, they show only limited clinical activity against ErbB2 driven breast cancers. One possible explanation for this lack of effect may be the inability of TKIs to inhibit ErbB3 activation of downstream signaling pathways. The importance of ErbB3 expression in resistance to TKIs in breast cancer has been recently highlighted by the demonstration that continued oncogenic signaling through ErbB3 in human breast cancer cell lines results in the failure of the EGFR/ErbB2 kinase inhibitor gefitinib to completely inhibit the kinase activity of ErbB2. While EGFR and ErbB2 phosphorylation and activation of downstream pathways such as AKT were durably inhibited by TKIs, dephosphorylation of ErbB3 was transient, leading to reactivation of AKT pathways (40). As mentioned previously, ErbB3 has six binding sites for PI-3 Kinase and is principally responsible for AKT activation after treatment of cells with EGFR or HRG like ligands (5). The resulting constitutive phosphorylation of AKT in the face of gefitinib treatment allowed cells to evade apoptosis. Persistent ErbB3 activation in the face of gefitinib treatment was due to a forward shift in the equilibrium of the ErbB3 phosphorylation-dephosphorylation reaction, establishing a new steady state ErbB3 phosphorylation level despite significant inhibition of ErbB2 kinase. The TKI induced forward shift in ErbB3 steady state phosphorylation was driven by increased ErbB3 substrate concentration in the forward reaction and decreased phosphatase activity impeding the reverse reaction. The increase in ErbB3 substrate at the plasma membrane was due to ErbB3 relocalization to the plasma membrane and was suppressed by inhibitors of vesicular trafficking. Thus, the failure to durably suppress ErbB3 activity significantly attenuates the anti-tumor effect of TKIs and is consistent with the limited clinical activities of these agents in ErbB2 amplified breast cancers. It should be noted that mono-specific EGFR reversible TKIs (gefitinib and erlotinib) and pan-ErbB irreversible inhibitors (CI-1033,canertinib) were used in these studies. These TKIs may be suboptimal for management of ErbB2 overexpressing breast cancer. Whether the therapeutic effects of ErbB1/2 reversible TKIs such as lapatinib can be overcome via ErbB3 transphosphorylation remains to be elucidated (41). Sergina et al (40) suggested that TKI therapy will remain ineffective until drugs that completely inactive ErbB kinase function are available. In addition, measures of ErbB2 and EGFR phosphorylation may overstate the efficacy of TKIs and are poor in vivo biological markers. ErbB3 transphosphorylation would serve as a better marker of TKI efficacy to guide therapeutic decisions (42).
Further, the efficacy of therapies directed against a single ErbB receptor may be blunted by ErbB related ligands that interact with other members of the ErbB family. Paracine, autocrine or juxtacrine produced HRG can overcome the growth inhibitory effects of EGFR/ErbB2 TKIs. Motoyama et al (43) were the first to demonstrate that the growth inhibitory effects of EGFR directed TKIs were attenuated in the presence of HRG. The degree of inhibition of the TKIs’ therapeutic effect was dependent on the level of ErbB3 expression. The influence of HRG on the efficacy of anti-EGFR therapies in tamoxifen resistant MCF-7 cells, whose growth is driven by autocrine release of the EGFR ligand amphiregulin, was also recently demonstrated (44). HRGß1 was able to overcome the inhibitory effects of gefitinib on growth and invasion of these cells. HRGß1 promoted ErbB2/3 heterodimizeration and activation of PI-3-kinase/AKT signaling pathways in the face of gefitinib treatment. However, a combination of gefitinib and the PI-3 Kinase inhibitor LY294002 effectively blocked HRG mediated intracellular signaling, growth and invasiveness. Similarly, targeting ErbB2 with trastuzumab in combination with gefitinib reduced HRG induced ErbB2 kinase activity and MAPK activation. However HRGß1- driven AKT activity and cell growth were not inhibited and surprisingly, cellular invasion was enhanced. All clinical breast cancer samples examined demonstrated HRG ß1 in the cytoplasm with expression correlating with activation of AKT and MAPK. Thus, HRGß1 could overcome the inhibitory effects of gefitinib on cell growth and invasion in tamoxifen resistant cells through promotion of ErbB2/3 heterodimerization and activation of AKT. The fact that all samples tested expressed HRG in the cytoplasm suggests that the effectiveness of anti-EGFR therapies in breast cancer may be limited. In keeping with the importance of expression of HRG-like ligands in determining gefitinib sensitivity in breast cancer cells, another study demonstrated that gefitinib induced changes in endogenous levels of EGF like ligands such as HRG correlated with the cellular sensitivity to this drug. Cells resistant to gefitinib upregulated HRG upon gefitinib treatment accounting for the increased resistance. HRG accumulated in the nucleus in gefitnib resistant cells. Thus, an intracrine ligand dependent feedback mechanism involving EGF -like ligands may play a major role in the development of resistance to gefitinib in breast cancer (45)
Another possible mechanism of HRG’s ability to overcome resistance to TKIs may be related to its ability to increase ErbB3 plasma membrane substrate concentration. HRG induced nucleolar and nuclear export of ErbB3 , leading to a slow enrichment of ErbB3 in the cytoplasm/membrane compartment (46). Thus, HRG’s enhancement of ErbB3 plasma membrane concentrations may result in persistent phosphorylation of ErbB3 and sustained activation of AKT.
These studies demonstrate that ErbB3 may impede both hormone and TKI directed therapies in breast cancer. ErbB3 can contribute to tamoxifen resistance by unknown mechanisms involving AKT. In addition, persistent activation of ErbB3 results in an escape from growth inhibition induced by several TKIs. Therefore, increased interest in ErbB3 as a therapeutic target as discussed previously is warranted. In addition, incorporation of phosphorylated ErbB3, or the ErbB2/3 dimer as a marker to allow identification of patients who may respond to ErbB directed therapies as has previously been proposed for prediction of sensitivity to ErbB2 directed therapies (47) should be supported. Finally, increased attention to ErbB3 ligands such as HRG that are involved in the efficacy of ErbB directed TKIs are important for the design of future clinical trials.
Contributions of ErbB3 Binding Proteins to breast cancer progression and therapeutic resistance
Although many proteins can associate with ErbB3, I will focus here on the relatively limited group of binding proteins that have been demonstrated to down regulate ErbB3 activity and its response to HRG. Increasing data suggest that these proteins, which may be endogenous negative regulators of HRG signaling, play a role in breast cancer progression and treatment resistance.
NRDP-1
Nrdp1 (neuregulin receptor degradation protein-1) was originally identified as a RING finger domain containing protein that interacts with ErbB3 in a ligand and phosphotyrosine independent manner. Nrdp1 binds both ErbB3 and ErbB4 receptors, but not ErbB2 and EGFR (48). Ectopic expression of Ndrp1 results in decreased levels of ErbB3 protein. Nrdp1 stimulates ErbB3 ubiquitination and its rapid degradation by proteasomses. The carboxy- terminal domain of Nrdp1 associates with the cytoplasmic tail of ErbB3 and the amino- terminal RING finger domain promotes ErbB3 degradation and ubiquitination (49). Recently, the 1.95 angstrom crystal structure of the C terminal domain of Nrdp1 has been solved. Pull-down binding assays indicated that the C terminal domain was sufficient to mediate ErbB3 binding. The Nrdp1 cytoplasmic domain crystal structure was used to guide selection of a cluster of residues that map to discrete regions. Residues in each group were changed to alanine to evaluate their importance for ErbB3 binding and ErbB3 degradation. The ErbB3 binding sites localized to a region of Nrdp1 that is conserved from invertebrates to vertebrates (50). Binding studies indicated that the cytoplasmic domain of Ndrp1 mediates ubiquitinylation of ErbB3 in vitro and induces relocalization of ErbB3 from the cell surface into intracelluar compartments, suggesting its role in facilitating ErbB3 degradation.
Nrdp1 itself is highly labile as it undergoes self ubiquitination and degradation through a proteosomal pathway (51). Of interest, it has recently been found that Nrdp1 levels are regulated by HRG suggesting a feedback mechanism for maintenance of ErbB3 levels (52). HRG, which activates ErbB3, also stimulates the stabilization of Nrdp1 which results in ligand stimulated down regulation of ErbB3. HRG1 stimulation of MCF-7 cells resulted in two-to five fold increases in Nrdp1 protein levels. At the same time, ErbB3 levels were dramatically suppressed. These observations suggested that Nrdp1 is a posttranscriptional target of HRG signaling. Thus, HRG stimulation of ErbB2/3 signaling augments Nrdp1 stability, attenuating ErbB3 downstream responses.
A functional role for Nrpd-1 in breast cancer has recently been demonstrated (53). Overexpression of Nrdp1 in three human breast cancer cell lines resulted in the suppression of ErbB3 protein levels and inhibition of HRG mediated cell growth. In addition, the upregulation of AKT and MAPK pathways by HRG was attenuated. Overexpression of Nrdp1 resulted in impairment of HRG- induced migration of MCF-7 cells. In contrast, either Nrdp1 knockdown or overexpression of a dominant negative form enhanced ErbB3 levels and HRG-induced cell proliferation. Nrdp1 expression levels inversely correlated with ErbB3 levels in primary human breast cancers as determined by Western blotting. These studies suggest the role of Nrdp1 in the maintenance of steady state levels of ErbB3 protein in human breast cancer cells. Thus, Nrdp1 is a potential physiological regulator of HRG-induced signaling in breast epithelium. A loss of Nrdp1 may contribute to breast tumor progression through upregulation of ErbB3 signaling. These authors suggested the development of therapeutic methods to introduce Nrdp1 into tumors of patients presenting with ErbB2/3 positive breast cancer.
LRIG-1
ErbB3 is also negatively regulated by the leucine-rich repeat protein LRIG1 which colocalizes with ErbB receptors and enhances their ubiquitination. LRIG1 is a 140kDa transmembrane protein containing 15 leucine rich repeat domains and 3 Ig domains. LRIG1 is located at chromosome 3p14.3. Loss of heterozygosity at the LRIG1 locus was found in 41% of breast cancers examined (54). Similarly, Sweeney et al observed that the majority of primary human breast tumors examined displayed decreased LRIG protein expression compared to matched normal tissue (55). In contrast, another study showed an increase in copy number at 3p14.3 in breast tumors and an increase in LRIG1 protein (56).
LRIG bound each of the ErbB receptors in a ligand independent manner. LRIG1 overexpression enhanced ligand stimulated receptor ubiquitination (57). Inducible expression of LRIG1 resulted in suppression of HRG stimulated growth of prostate cancer cells. These results suggested that LRIG1-mediated receptor ubiquitination and degradation may contribute to the suppression of ErbB receptor function.
A possible link between LRIG1 activity and gefitinib resistance is suggested by recent studies showing that lung cancer cells with EGFR mutations that are resistant to gefitinib have the MET receptor gene amplified. MET is a transmembrane tyrosine kinase that is activated by the hepatocyte growth factor. MET and ErbB3 physically associated and the MET/ErbB3 interaction resulted in PI 3-kinase and AKT activation leading to gefitinib resistance (58). Interactions of ErbB3 with the MET receptor have not yet been demonstrated in gefitinib resistant breast cancer cells. LRIG1 has been shown to also down regulate the MET receptor in human breast cancer cells (59). Thus, a loss of LRIG1 may contribute to increased levels of MET and ErbB3 protein leading to gefitinib resistance.
EBP1
A protein isolated in our laboratory by its binding to ErbB3, Ebp1 (60), is also a negative regulator of ErbB signal transduction. Ebp1, encoded by the PA2G4 gene, is the human homologue of the cell cycle regulated mouse protein p38-2G4 (61). Ebp1 is widely expressed (62) and highly conserved throughout evolution (63). The EBP1 (PA2G4) gene is located adjacent to ErbB3 in both the mouse (at chromosome 10 D3) and human (at chromosome 12q13.2) genomes. Ebp1 binds to the first fifteen amino acids of the juxtamembrane domain of unphosphorylated ErbB3 (60).
A functional story is emerging for Ebp1 in human breast cancer. Overexpression of Ebp1 in ErbB2/3 positive breast cancer cells inhibited cell growth, while promoting G2/M cell cycle arrest and cellular differentiation (64). Ectopic expression of Ebp1 in MCF-7 and AU565 breast cancer cell lines specifically inhibited HRG, but not EGF, induced proliferation. HRG-induced AKT activation was attenuated in EBP1 stable transfectants and transfection of a constitutively activated AKT partially restored the growth response to HRG (Zhang et al, Cancer Letters in press).
In AU565 breast cancer cells, binding of Ebp1 to ErbB3 is dependent on constitutive phosphorylation of PKC (65) and the required phosphorylation site has been mapped to Ser360 (66). HRG treatment induces dissociation of Ebp1 from ErbB3 and increases its nuclear localization (60). In the nucleus, Ebp1 interacts with proteins important in transcriptional repression such as Rb (67), the histone deacetylase HDAC2 (68), and the transcriptional repressor Sin3A (69). Ebp1 inhibits transcription of E2F1 regulated endogenous and exogenous proliferation-associated genes such as E2F1, Cyclin D1 and cyclin E (67) (68). The interactions of Ebp1 with histone deacetylase 2 (HDAC2) , Rb and Sin3A are necessary for its ability to repress transcription (68;70) (69) and depend on phosphorylation of Ser363 (71). Ebp1 binds E2F1 at E2F1 regulated promoters (70) in vivo and HRG increases binding of Ebp1 to the E2F1 promoter complex and enhances Ebp1- mediated repression of E2F1 regulated gene transcription (70). Thus Ebp1 is a potential critical effector of Erb3 signaling.
Ebp1 also has properties of an RNA binding protein. Squaritto et al. (72) demonstrated that a pool of Ebp1 localized to the nucleolus is a component of ribonucleoprotein complexes and associates with different rRNA species via ds RNA and sigma 70 like binding domains. Furthermore, in the cytoplasm Ebp1 associates with mature ribosomes and potentially influences protein translation via inhibition of phosphorylation of eukaryotic initiation factor 2α (73). The crystal structures of human (74) and murine (75) Ebp1 have recently been solved and demonstrate that the C terminal region of Ebp1 was needed for RNA binding, in keeping with functional studies indicating the importance of this domain for inhibition of both transcriptional repression and cell growth (67).
As Ebp1 phosphorylation, binding to the ErbB3 receptor and transcriptional activity are affected by HRG treatment, we inquired whether Ebp1 could be a substrate for PAK1, an HRG-regulated protein implicated in breast cancer progression (76) and tamoxifen resistance (77) (78). We examined the ability of PAK1 to phosphorylate and regulate the function of Ebp1. We found that PAK1 phosphorylated Ebp1 in vitro and mapped the phosphorylation site to Threonine 261. Both HRG treatment and expression of a constitutively activated PAK1 in MCF-7 breast cancer cells enhanced threonine phosphorylation of Ebp1. In MCF-7 cells, ectopically expressed Ebp1 bound endogenous PAK1 and this association was enhanced by treatment with HRG. Mutation of the PAK1 phosphorylation site to glutamic acid, mimicking a phosphorylated state, completely abrogated the ability of Ebp1 to repress transcription and inhibit growth of breast cancer cell lines. As PAK1 inhibits tamoxifen action in MCF-7 cells (77), we postulated that inactivation of Ebp1 by PAK1 may also contribute to tamoxifen resistance in hormone dependent cells. We first tested the contribution of Ebp1 to tamoxifen sensitivity by knockdown of endogenous Ebp1 expression via transduction of MCF-7 cells with an shRNA vector. Growth of the Ebp1 knock down cells was significantly increased, rather than decreased, by tamoxifen treatment. The inactive T261E Ebp1 mutant could function as a dominant negative to inhibit the ability of wild type Ebp1 to contribute to tamoxifen sensitivity (79). Paradoxically, in one study EBP1 gene expression was reported to be upregulated in breast cancer and the protein was slightly decreased by tamoxifen treatment (80). Our studies demonstrate that Ebp1 is a substrate of PAK1 and the importance of the PAK1 phosphorylation site for the functional activity of Ebp1 in breast cancer cells. We suggest that inactivation of Ebp1 by PAK1 can contribute to resistance to antiestrogens by causing sustained ErbB downstream signaling. Use of drugs that inhibit Pak1 (81) may restore Ebp1 function, leading to a therapy in ErbB2/3 overexpressing breast cancer.
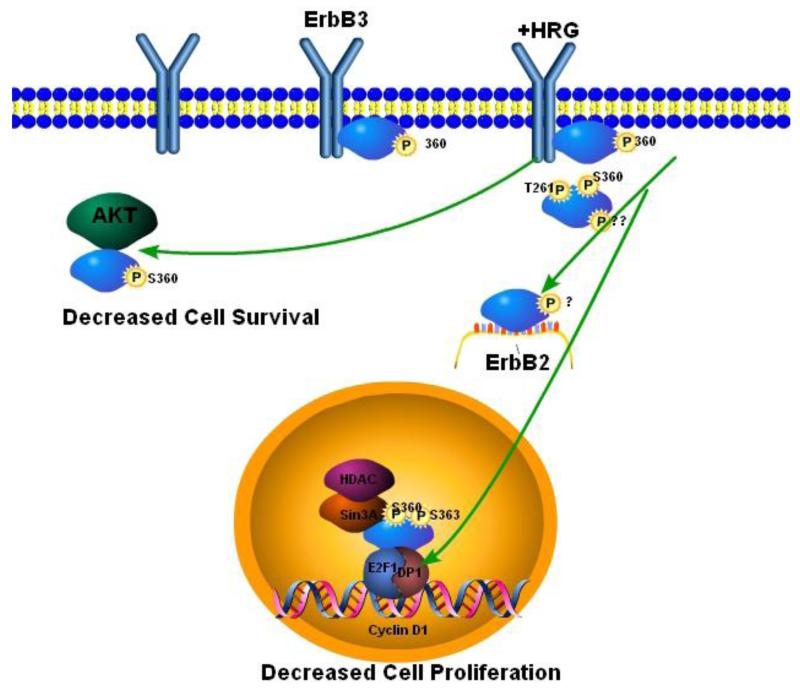
In summary, ErbB3 binding proteins may serve as endogenous suppressors of tumor cell growth and invasion by controlling the physiological levels of ErbB3 and ErbB3 activation (Fig.1). While the involvement of ErbB3 binding proteins in receptor degradation has been most intensively studied, several other mechanisms that modulate the magnitude or duration of receptor signaling may also be involved. Loss of these binding proteins could contribute to ErbB3 overexpression and constitutive activation of downstream effectors such as AKT. Restoration or augmentation of the function of these binding proteins could provide a novel means of suppressing ErbB3 activity in breast cancer cells. It will be important to determine whether loss of expression or functional activity of these ErbB3 binding proteins plays a role in breast cancer progression and therapy resistance.
Fig. 1. Negative regulation of ErbB3 by the ErbB3 interacting proteins Ebp1 and Nrdp1.
Ebp1 phosphorylated at Ser 360 binds ErbB3 in the absence of HRG. After HRG stimulation, Ebp1 translocates to the nucleus where it becomes phosphorylated at Ser 363 and can then bind Sin3A and HDAC2 leading to repression of Cyclin D1 transcription and decreased cell proliferation. Inactivation of Ebp1 by phosphorylation at Thr 261 by Pak1 inhibits its ability to repress cyclin D1 transcription and leads to tamoxifen resistance. Ebp1 may also associate with AKT in the cytoplasm, inhibiting its activation leading to decreased cell survival. Nrdp1 can bind unphosphorylated or phosphorylated ErbB3 at the plasma membrane. Both phosphorylated and unphosphorylated receptors can be internalized. Ubiquitylation of ErbB3 in the endosome after Nrdp1 binding marks the receptors for degradation. Decreased levels of ErbB3 result in decreases in cell proliferation, AKT activation and cellular invasion. ErbB3-induced sustained activation of AKT can lead to resistance to TKIs and hormonal therapies.
Concluding Remarks
The purpose of this review was to emphasize the role of ErbB3 and its binding proteins in both the progression of breast cancer and the development of resistance to currently available therapies. Although EGFR and ErbB2 have received enormous attention as targets for therapeutic intervention, both functional and immunohistochemical data over the past 15 years have indicated the importance of ErbB3 in breast cancer progression. Therefore, increased attention to both the level and phsophorylation status of ErbB3 will be critical in devising better prognostic models. Further work will also be needed to evaluate ErbB3 as a marker for breast cancer progression both in the context of other ErbB receptors and the expression of its ligands. In addition, the ability of ErbB3 through sustained activation of AKT to circumvent both hormonal and TKI based therapies presents an important clinical problem. Recent data indicating that ErbB3 binding proteins may play a role in both the expression and functional activity of ErbB3 suggest that these proteins may be novel targets for therapy. Understanding the complex interactions between the ErbB3, its ligands, and its binding proteins will be needed to devise more effective therapies for breast cancer.
Acknowledgments
This work was supported by NIH grants R01 CA76047 and R21 088882-01 and a grant from the Department of Pathology, University of Maryland School of Medicine (to AWH).
Grant Information NIH grants R01 CA76047 and R21 088882-01 and a grant from the Department of Pathology, University of Maryland School of Medicine (to AWH).
Abbreviations
- HRG1
heregulin-1
- Nrdp1
neuregulin receptor degradation protein-1
- Ebp1
ErbB3 receptor binding protein 1
- ER
Estrogen Receptor
References
- 1.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284(1):99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 3.Kraus MH, Issing W, Miki T, Popescu NC, Aaronson SA. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci U S A. 1989;86(23):9193–7. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedi P, Pierce JH, di Fiore PP, Kraus MH. Efficient coupling with phosphatidylinositol 3-kinase, but not phospholipase C gamma or GTPase-activating protein, distinguishes ErbB-3 signaling from that of other ErbB/EGFR family members. Mol Cell Biol. 1994;14(1):492–500. doi: 10.1128/mcb.14.1.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellyer NJ, Cheng K, Koland JG. ErbB3 (HER3 interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J. 1998;333(Pt 3):757–63. doi: 10.1042/bj3330757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soltoff SP, Carraway KL, III, Prigent SA, Gullick WG, Cantley LC. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol. 1994;14(6):3550–8. doi: 10.1128/mcb.14.6.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4(4) doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 9.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 10.Lemoine NR, Barnes DM, Hollywood DP, Hughes CM, Smith P, Dublin E, et al. Expression of the ERBB3 gene product in breast cancer. Br J Cancer. 1992;66(6):1116–21. doi: 10.1038/bjc.1992.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naidu R, Yadav M, Nair S, Kutty MK. Expression of c-erbB3 protein in primary breast carcinomas. Br J Cancer. 1998;78(10):1385–90. doi: 10.1038/bjc.1998.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travis A, Pinder SE, Robertson JF, Bell JA, Wencyk P, Gullick WJ, et al. C-erbB-3 in human breast carcinoma: expression and relation to prognosis and established prognostic indicators. Br J Cancer. 1996;74(2):229–33. doi: 10.1038/bjc.1996.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasparini G, Gullick WJ, Maluta S, Dalla PP, Caffo O, Leonardi E, et al. c-erbB-3 and c-erbB-2 protein expression in node-negative breast carcinoma--an immunocytochemical study. Eur J Cancer. 1994;30A(1):16–22. doi: 10.1016/s0959-8049(05)80010-3. [DOI] [PubMed] [Google Scholar]
- 14.Bieche I, Onody P, Tozlu S, Driouch K, Vidaud M, Lidereau R. Prognostic value of ERBB family mRNA expression in breast carcinomas. Int J Cancer. 2003;106(5):758–65. doi: 10.1002/ijc.11273. [DOI] [PubMed] [Google Scholar]
- 15.Pawlowski V, Revillion F, Hebbar M, Hornez L, Peyrat JP. Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res. 2000;6(11):4217–25. [PubMed] [Google Scholar]
- 16.Barnes NL, Khavari S, Boland GP, Cramer A, Knox WF, Bundred NJ. Absence of HER4 expression predicts recurrence of ductal carcinoma in situ of the breast. Clin Cancer Res. 2005;11(6):2163–8. doi: 10.1158/1078-0432.CCR-04-1633. [DOI] [PubMed] [Google Scholar]
- 17.Quinn CM, Ostrowski JL, Lane SA, Loney DP, Teasdale J, Benson FA. c-erbB-3 protein expression in human breast cancer: comparison with other tumour variables and survival. Histopathology. 1994;25(3):247–52. doi: 10.1111/j.1365-2559.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 18.Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200(3):290–7. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 19.Wiseman SM, Makretsov N, Nielsen TO, Gilks B, Yorida E, Cheang M, et al. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer. 2005;103(9):1770–7. doi: 10.1002/cncr.20970. [DOI] [PubMed] [Google Scholar]
- 20.Kim HH, Vijapurkar U, Hellyer NJ, Bravo D, Koland JG. Signal transduction by epidermal growth factor and heregulin via the kinase-deficient ErbB3 protein. Biochem J. 1998;334(Pt 1):189–95. doi: 10.1042/bj3340189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15(10):2452–67. [PMC free article] [PubMed] [Google Scholar]
- 22.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, III, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100(15):8933–8. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Horst EH, Murgia M, Treder M, Ullrich A. Anti-HER-3 MAbs inhibit HER-3-mediated signaling in breast cancer cell lines resistant to anti-HER-2 antibodies. Int J Cancer. 2005;115(4):519–27. doi: 10.1002/ijc.20867. [DOI] [PubMed] [Google Scholar]
- 24.Chen CH, Chernis GA, Hoang VQ, Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc Natl Acad Sci U S A. 2003;100(16):9226–31. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lund CV, Popkov M, Magnenat L, Barbas CF., III Zinc finger transcription factors designed for bispecific coregulation of ErbB2 and ErbB3 receptors: insights into ErbB receptor biology. Mol Cell Biol. 2005;25(20):9082–91. doi: 10.1128/MCB.25.20.9082-9091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282(2):1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 27.Clarke R, Leonessa F, Welch JN, Skaar TC. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev. 2001;53(1):25–71. [PubMed] [Google Scholar]
- 28.Mouridsen HT, Rose C, Brodie AH, Smith IE. Challenges in the endocrine management of breast cancer. Breast. 2003;12(Suppl 2):S2–19. doi: 10.1016/s0960-9776(03)80158-3. [DOI] [PubMed] [Google Scholar]
- 29.Newby JC, Johnston SR, Smith IE, Dowsett M. Expression of epidermal growth factor receptor and c-erbB2 during the development of tamoxifen resistance in human breast cancer. Clin Cancer Res. 1997;3(9):1643–51. [PubMed] [Google Scholar]
- 30.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 31.Pietras RJ, Arboleda J, Reese DM, Wongvipat N, Pegram MD, Ramos L, et al. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10(12):2435–46. [PubMed] [Google Scholar]
- 32.Tang CK, Perez C, Grunt T, Waibel C, Cho C, Lupu R. Involvement of heregulin-beta2 in the acquisition of the hormone-independent phenotype of breast cancer cells. Cancer Res. 1996;56(14):3350–8. [PubMed] [Google Scholar]
- 33.Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, et al. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24(2):85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 34.Tovey S, Dunne B, Witton CJ, Forsyth A, Cooke TG, Bartlett JM. Can molecular markers predict when to implement treatment with aromatase inhibitors in invasive breast cancer? Clin Cancer Res. 2005;11(13):4835–42. doi: 10.1158/1078-0432.CCR-05-0196. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Ordonez-Ercan D, Fan Z, Edgerton SM, Yang X, Thor AD. Downregulation of erbB3 abrogates erbB2-mediated tamoxifen resistance in breast cancer cells. Int J Cancer. 2007;120(9):1874–82. doi: 10.1002/ijc.22423. [DOI] [PubMed] [Google Scholar]
- 36.deGraffenried LA, Friedrichs WE, Russell DH, Donzis EJ, Middleton AK, Silva JM, et al. Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant Akt Activity. Clin Cancer Res. 2004;10(23):8059–67. doi: 10.1158/1078-0432.CCR-04-0035. [DOI] [PubMed] [Google Scholar]
- 37.Jordan NJ, Gee JM, Barrow D, Wakeling AE, Nicholson RI. Increased constitutive activity of PKB/Akt in tamoxifen resistant breast cancer MCF-7 cells. Breast Cancer Res Treat. 2004;87(2):167–80. doi: 10.1023/B:BREA.0000041623.21338.47. [DOI] [PubMed] [Google Scholar]
- 38.Clark AS, West K, Streicher S, Dennis PA. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol Cancer Ther. 2002;1(9):707–17. [PubMed] [Google Scholar]
- 39.Agrawal A, Gutteridge E, Gee JM, Nicholson RI, Robertson JF. Overview of tyrosine kinase inhibitors in clinical breast cancer. Endocr Relat Cancer. 2005;12(Suppl 1):S135–S144. doi: 10.1677/erc.1.01059. [DOI] [PubMed] [Google Scholar]
- 40.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–41. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menendez JA, Lupu R. Transphosphorylation of kinase-dead HER3 and breast cancer progression: a new standpoint or an old concept revisited? Breast Cancer Res. 2007;9(5):111. doi: 10.1186/bcr1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh AC, Moasser MM. Targeting HER proteins in cancer therapy and the role of the non-target HER3. Br J Cancer. 2007;97(4):453–7. doi: 10.1038/sj.bjc.6603910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motoyama AB, Hynes NE, Lane HA. The efficacy of ErbB receptor-targeted anticancer therapeutics is influenced by the availability of epidermal growth factor-related peptides. Cancer Res. 2002;62(11):3151–8. [PubMed] [Google Scholar]
- 44.Hutcheson IR, Knowlden JM, Hiscox SE, Barrow D, Gee JM, Robertson JF, et al. Heregulin beta1 drives gefitinib-resistant growth and invasion in tamoxifen-resistant MCF-7 breast cancer cells. Breast Cancer Res. 2007;9(4):R50. doi: 10.1186/bcr1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrer-Soler L, Vazquez-Martin A, Brunet J, Menendez JA, De LR, Colomer R. An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review) Int J Mol Med. 2007;20(1):3–10. [PubMed] [Google Scholar]
- 46.Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW. c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157(6):929–39. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arteaga CL. Can trastuzumab be effective against tumors with low HER2/Neu (ErbB2 receptors? J Clin Oncol. 2006;24(23):3722–5. doi: 10.1200/JCO.2006.06.5268. [DOI] [PubMed] [Google Scholar]
- 48.Diamonti AJ, Guy PM, Ivanof C, Wong K, Sweeney C, Carraway KL., III An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc Natl Acad Sci U S A. 2002;99(5):2866–71. doi: 10.1073/pnas.052709799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A. 2002;99(23):14843–8. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouyain S, Leahy DJ. Structure-based mutagenesis of the substrate-recognition domain of Nrdp1/FLRF identifies the binding site for the receptor tyrosine kinase ErbB3. Protein Sci. 2007;16(4):654–61. doi: 10.1110/ps.062700307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., III Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24(17):7748–57. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Z, Wu X, Yen L, Sweeney C, Carraway KL., III Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol Cell Biol. 2007;27(6):2180–8. doi: 10.1128/MCB.01245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yen L, Cao Z, Wu X, Ingalla ER, Baron C, Young LJ, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 2006;66(23):11279–86. doi: 10.1158/0008-5472.CAN-06-2319. [DOI] [PubMed] [Google Scholar]
- 54.Maitra A, Wistuba, Washington C, Virmani AK, Ashfaq R, Milchgrub S, et al. High-resolution chromosome 3p allelotyping of breast carcinomas and precursor lesions demonstrates frequent loss of heterozygosity and a discontinuous pattern of allele loss. Am J Pathol. 2001;159(1):119–30. doi: 10.1016/S0002-9440(10)61679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sweeney C, Miller JK, Shattuck DL, Carraway KL., III ErbB receptor negative regulatory mechanisms: implications in cancer. J Mammary Gland Biol Neoplasia. 2006;11(1):89–99. doi: 10.1007/s10911-006-9015-3. [DOI] [PubMed] [Google Scholar]
- 56.Ljuslinder I, Malmer B, Golovleva I, Thomasson M, Grankvist K, Hockenstrom T, et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res. 2005;7(5):R719–R727. doi: 10.1186/bcr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, III, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279(45):47050–6. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 58.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 59.Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, III, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27(5):1934–46. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoo JY, Wang XW, Rishi AK, Lessor T, Xia XM, Gustafson TA, et al. Interaction of the PA2G4 (EBP1 protein with ErbB-3 and regulation of this binding by heregulin. Br J Cancer. 2000;82(3):683–90. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38-2G4, which varies with the cell cycle. Exp Cell Res. 1995;220(2):434–45. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 62.Xia X, Lessor TJ, Zhang Y, Woodford N, Hamburger AW. Analysis of the expression pattern of Ebp1, an ErbB-3-binding protein. Biochem Biophys Res Commun. 2001;289(1):240–4. doi: 10.1006/bbrc.2001.5942. [DOI] [PubMed] [Google Scholar]
- 63.Horvath BM, Magyar Z, Zhang Y, Hamburger AW, Bako L, Visser RG, et al. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006;25(20):4909–20. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lessor TJ, Yoo JY, Xia X, Woodford N, Hamburger AW. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J Cell Physiol. 2000;183(3):321–9. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 65.Lessor TJ, Hamburger AW. Regulation of the ErbB3 binding protein Ebp1 by protein kinase C. Mol Cell Endocrinol. 2001;175(1-2):185–91. doi: 10.1016/s0303-7207(01)00387-2. [DOI] [PubMed] [Google Scholar]
- 66.Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc Natl Acad Sci U S A. 2006;103(29):10917–22. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia X, Cheng A, Lessor T, Zhang Y, Hamburger AW. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J Cell Physiol. 2001;187(2):209–17. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- 68.Zhang YX, Woodford N, Xia XM, Hamburger AW. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Research. 2003;31(8):2168–77. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Akinmade D, Hamburger AW. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 2005;33(18):6024–33. doi: 10.1093/nar/gki903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Hamburger AW. Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription. J Biol Chem. 2004;279(25):26126–33. doi: 10.1074/jbc.M314305200. [DOI] [PubMed] [Google Scholar]
- 71.Akinmade D, Lee M, Zhang Y, Hamburger AW. Ebp1-mediated inhibition of cell growth requires serine 363 phosphorylation. Int J Oncol. 2007;31(4):851–8. [PubMed] [Google Scholar]
- 72.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23(25):4454–65. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 73.Squatrito M, Mancino M, Sala L, Draetta GF. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem Biophys Res Commun. 2006;344(3):859–68. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 74.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, et al. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007 doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kowalinski E, Bange G, Bradatsch B, Hurt E, Wild K, Sinning I. The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions. FEBS Lett. 2007;581(23):4450–4. doi: 10.1016/j.febslet.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 76.Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, et al. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275(46):36238–44. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 77.Rayala SK, Talukder AH, Balasenthil S, Tharakan R, Barnes CJ, Wang RA, et al. P21-activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 2006;66(3):1694–701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- 78.Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98(10):671–80. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 79.Akinmade D, Talukder AH, Zhang Y, Luo WM, Kumar R, Hamburger AW. Phosphorylation of the ErbB3 binding protein Ebp1 by p21-activated kinase 1 in breast cancer cells. Br J Cancer. 2008 doi: 10.1038/sj.bjc.6604261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ou K, Kesuma D, Ganesan K, Yu K, Soon SY, Lee SY, et al. Quantitative profiling of drug-associated proteomic alterations by combined 2-nitrobenzenesulfenyl chloride (NBS isotope labeling and 2DE/MS identification. J Proteome Res. 2006;5(9):2194–206. doi: 10.1021/pr060115n. [DOI] [PubMed] [Google Scholar]
- 81.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6(6):459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]