Abstract
Sensory coding strategies within vertebrates involve the expression of a limited number of receptor types per sensory cell. In mice, each vomeronasal sensory neuron transcribes monoallelically a single V1R pheromone receptor gene, chosen from a large V1R repertoire. The nature of the signals leading to this strict receptor expression is unknown, but is apparently based on a negative feedback mechanism initiated by the transcription of the first randomly chosen functional V1R gene. We show, in vivo, that the genetic replacement of the V1rb2 pheromone receptor coding sequence by an unrelated one from the odorant receptor gene M71, maintains gene exclusion. The expression of this exogenous odorant receptor in vomeronasal neurons does not trigger the transcription of odorant receptor-associated signaling molecules. These results strongly suggests that despite the very divergent odorant and vomeronasal receptor localization, function and transduction cascades, a common mechanism is used by these chemoreceptors to regulate their expression.
Keywords: pheromone receptor, olfaction, gene regulation, mouse, monogenic expression
Introduction
In order to survive and reproduce, mammals use tools to extract information from the outside world. Evolution provided vertebrates with a remarkably wide variety of chemoreceptors, mostly seven transmembrane proteins. These are expressed by sensory neurons located in specific organs or tissues, mainly in the nasal cavity. The two major mammalian olfactory chemical sensors, the main olfactory and vomeronasal epithelia, are physically, molecularly and functionally separated. Sensory neurons of the main olfactory system are located in the dorsal recess of the nasal cavity and are responsible for odor detection. Vomeronasal sensory neurons lie in a blunt ended elongated bony capsule, the vomeronasal organ, located at the base of the nasal septum (Halpern & Martinez-Marcos, 2003); these neurons are apparently specialized in pheromone detection. A clean functional dichotomy is however not entirely true, since some pheromones can be perceived by the main olfactory system, and molecules with no known pheromonal effects can activate vomeronasal sensory neurons.
Based on molecular markers which likely reflect functional specializations, the vomeronasal neuroepithelium can be divided into two main populations. At the base of the vomeronasal pseudostratified sensory epithelium are the soma of neurons expressing members of th V2R receptor family (Herrada & Dulac, 1997; Matsunami & Buck, 1997; Ryba & Tirindelli, 1997), while cells transcribing chemoreceptors of the V1R family are located more apically (Herrada & Dulac, 1997). V1R receptors are part of a large superfamily, which in mice contains over 150 different members (Dulac & Axel, 1995; Rodriguez et al., 2002; Zhang et al., 2004).
Each V1R-expressing vomeronasal sensory neuron transcribes a single V1R gene (Dulac & Axel, 1995; Roppolo et al., 2007) from a single allele (Rodriguez et al., 1999). V1R genes are very divergent from each other and are highly polymorphic across alleles in outbred and wild type mice. A monogenic and monoallelic expression strategy thus ensures narrow functional properties to each vomeronasal neuron. Axonal projections of neurons expressing the same V1R receptor coalesce into the brain in the accessory olfactory bulb, and form numerous spherical structures called glomeruli, where they synapse with second order neurons (Belluscio et al., 1999; Rodriguez et al., 1999; Wagner et al., 2006).
V1R receptors are thought to play multiple roles in vomeronasal neurons. Without surprise, they are involved in chemodetection (Boschat et al., 2002; Del Punta et al., 2002). Less expectedly, they are also critical in axon guidance processes (Belluscio et al., 1999; Rodriguez et al., 1999) and in the regulation of their own expression (Roppolo et al., 2007).
Our current understanding of V1R expression involves a negative feedback mechanism driven by (or at least dependent on) the transcription of a functional V1R gene (Roppolo et al., 2007). Thus, as soon as a V1R receptor is expressed in a given vomeronasal neuron, an unknown signal apparently prevents the potential transcription of other V1R receptor genes. This mechanism is reminiscent of the one, also unexplained, regulating odorant receptor expression in the main olfactory system. How related (if at all) these two mechanisms are is unknown. The identification of a common receptor-mediated signaling pathway is not obvious since ligand-dependent odorant and vomeronasal receptor transduction cascades are different: the first one involves cAMP and the cyclic nucleotide-gated channel CNGA2 (Ronnett & Moon, 2002) while the second is based on the lipid messenger diacylglycerol and the transient receptor potential channel TRPC2 (Stowers et al., 2002; Lucas et al., 2003).
Using gene-targeted mice, we show here that the exogenous transcription of an odorant receptor coding sequence from a V1R promoter in vomeronasal sensory neurons (coding for M71 instead of the V1R receptor V1RB2) is able to prevent the transcription of all endogenous V1R genes, and therefore to substitute for V1RB2. The ability of an odorant receptor to provide a negative feedback signal in neurons lacking all elements known to mediate the odorant-induced cascade points not only to a strategy but also to a mechanism shared by two different systems.
Materials and Methods
Animals
Animals were housed and handled in accordance with the guidelines and regulations of the institution and of the state of Geneva. When not indicated otherwise, animals were 4-5 week-old male or female mice. All mouse lines were on a mixed 129xC57BL/6 background (they were initially generated using E14 ES cells and backcrossed at least 8 times with C57BL/6).
Sequence alignments
All potentially functional V1R amino acid sequences of mouse, rat and dog genomes were extracted from the Ensembl database and were aligned at the protein level from the first to the last transmembrane segment using ClustalX (Thompson et al., 1994). The alignment was then manually refined using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and was used to generate a sequence logo via the Jan Gorodkin’s web script (Gorodkin et al., 1997). A consensus V1R sequence was derived from the sequence logo and was used as a template in prediction programs for 7TM domains, including TMPred, HMMTOP (Tusnady & Simon, 2001) and DAS (Cserzo et al., 1997).
Immunohistochemistry
Tissues were fixed for 3 hours in 4% paraformaldehyde, incubated overnight in 25% sucrose in 1x PBS, and embedded in OCT. 10-14 micrometers cryostat slices were mounted on Superfrost plus slides (Menzel-Glaser). Slides were preincubated in 0.5% Triton, 10% FCS, 1x PBS for 30′ at room temperature. Rabbit anti-beta-galactosidase antibodies were used (1:400, MP Biomedicals or Cappel). Guinea pig anti-M71 antibody (Lomvardas et al., 2006) was used at a 1:5000-1:10000 dilution. This antibody does recognize specifically M71 and M72-expressing neurons in the main olfactory epithelium (tested on M71 and M72 knock-in lines, data not shown), and does not stain wild type vomeronasal sensory neurons (not shown). After overnight incubation at 4°C, slides were washed 3 times with 0.5% Triton, 1x PBS for 10′. Primary antibodies were revealed with Cy3-conjugated donkey anti-rabbit (1:800) or Cy5-conjugated anti-guinea pig (1:200) secondary antibodies (Jackson Laboratory). Slides were mounted with antifade reagent (DABCO, Sigma).
Whole-mount analyses
Animals were dissected, the heads fixed by immersion in 4% paraformaldehye at 4°C for 5′ and stained with X-Gal as previously described (Rodriguez et al., 1999). Whole-mount images were taken on a Leica MZFLIII or a Zeiss SteREO Lumar.V12 fluorescence-equipped binoculars.
In situ hybridizations
14 micrometers cryostat slices of adult mouse vomeronasal organs were arranged on Superfrost plus slides, dried for 40′, fixed for 20′ at 4°C with 4% paraformaldehyde and hybridized overnight at 65°C in the following buffer: 1x salt buffer (0.2 M NaCl, 10 mM Tris, 5 mM NaH2PO4·H2O, 5 mM Na2HPO4·2H2O, 5 mM EDTA, pH 7.5), 50% formamide, 10% dextran sulphate, 1 microgram/microliter tRNA, 1x Denhardt’s, with 20 nanograms/microliter cRNA probe(s). Fluorescein and digoxigenin-labeled RNA probes were prepared using the DIG RNA labeling kit (Roche) following the manufacturer instructions. Probes for V1ra3, V1ra9, V1rb2, V1rb9, V1rc32, V1rd16, V1re4, V1rf4, V1rg1, V1rh5, V1ri1, V1ri8, V1rj3, V1rk1, V1rl1, Gαi2, LacZ and eGFP were previously described (Roppolo et al., 2007). Probes from different V1R families did not crosshybridize and sense probes did not show labeling (not shown). Probes for M71/M72, CNGA2, TRPC2, and Gαolf spanned nucleotides nt 68-920, nt 261-571, nt 3302-3527 and nt 464-993 respectively. Following hybridizations, slides were washed twice at 65°C and once at RT for 30′ with 1x SSC, 50% formamide and 0.1% Tween20. Slides were preincubated in 1x MABT, 2% blocking reagent (Roche) for 30′ followed by an hour incubation with alkaline phosphatase anti-digoxigenin antibody (1:1000, Roche) or/and peroxidase anti-fluorescein antibody (1:100-500, Roche). Peroxidase activity was revealed by washing slides 3 times with TNT (Tris 100 mM, NaCl 150 mM, Tween20 0.05%, pH 7.5), incubation for 30′ with a biotinyl-tyramide solution (PerkinElmer), 3x washes with TNT and incubation for 30′ with streptavidin-Alexa488 (Molecular Probes). Alkaline phosphatase activity was detected by incubating slides with Fast Red substrate (DAKO) for 30′. Sections were mounted in DABCO mounting medium.
Microscopy
Images were acquired with confocal microscopy (Zeiss LSM 510 or Leica SP2) or with a standard Zeiss Axioplan 2. An HCX PL APO CS 40.0×1.25 OIL UV objective was used for the Leica SP2. Image were 512×512 pixels, on a thickness of 15 μm or 50 μm for vomeronasal epithelium and accessory bulb respectively, with a stepsize of 0.16 μm.
Results
Exogenous expression of an odorant receptor in vomeronasal neurons
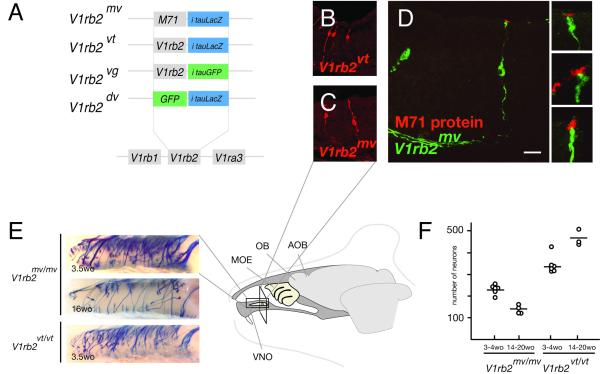
Mice in which the V1rb2 coding sequence was replaced with the class II M71 odorant receptor coding sequence (MOR171-2; Olfr151) along with IRES-tauLacZ (the V1rb2mv line, M71->V1rb2-IRES-tauLacZ, Figure 2A) were previously described (Rodriguez et al., 1999).
Figure 2.
Forced expression of the odorant receptor M71 in V1rb2mv-expressing vomeronasal neurons. (A) Schematic illustrating the mouse knock-in lines used in this study. (B, C) Coronal section of vomeronasal organs from the V1rb2vt and V1rb2mv lines. LacZ expression, which indirectly reflects the expression of V1RB2 or M71, is in red. (D) Coronal section of a vomeronasal organ from the V1rb2mv line. Exogenous expression of the odorant receptor M71 in vomeronasal neurons is shown in red, and is restricted to the dendritic endings. (E) Whole-mount X-Gal staining of vomeronasal organs showing punctate transcription of the V1rb2mv and V1rb2vt alleles in vomeronasal sensory neurons. Note the decreased number of labeled cells in 4 month-old V1rb2mv/mv mice (middle panel) relative to 3.5 week-old animals (upper panel). (F) Graph showing the decrease of labeled cells observed between 3.5 and 14-20 weeks in V1rb2mv mice. Each circle corresponds to a vomeronasal organ and horizontal lines indicate means (n=19). VNO, vomeronasal organ, MOE, main olfactory epithelium, OB, olfactory bulb, AOB, accessory olfactory bulb. Scale bar: 30 micrometers.
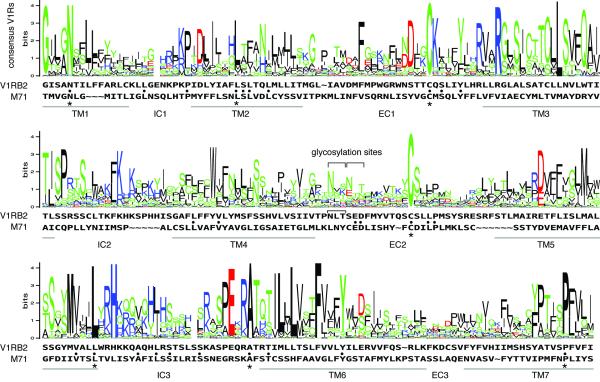
An amino acid alignment between a sequence logo representing all mouse, rat and dog V1Rs, and the V1RB2 and M71 sequences (see Materials and Methods) shows that most of the conserved residues in V1Rs are absent from the M71 sequence, including a highly conserved putative glycosylation site in extracellular loop 2 (Figure 1). M71 does however contain a glycosylation site conserved by members of the odorant receptor repertoire and localized at its N-terminus. Based on our alignment (Figure 1), in which the homology positions of the M71 sequence with V1Rs corresponding to positions of conserved residues in all mouse odorant receptors (Zhang & Firestein, 2002) were favored, M71 shares 10% identity with the V1RB2 protein. This level of homology corresponds to the identity observed between two random proteins. Among the identical residues found between M71 and V1RB2, only 7 are conserved in the V1R superfamily (Figure 1). Two of these residues are extracellular cysteines which form a bridge supposed to be necessary for proper protein folding and one is a proline required for receptor shuttling to the cell surface (Hwa et al., 2001).
Figure 1.
Sequence homology between M71 and V1R receptors. A sequence logo corresponding to all mouse, rat and dog V1R sequences is aligned with V1RB2 and M71. This type of analysis provides the consensus sequence and the frequency of a given amino acid at a given position. The size of the residues correlates with their conservation in rat, mouse and dog. Green, black, red and blue colors represent, respectively, uncharged polar (except for glycine and cysteine), nonpolar, acidic and basic residues. Black dots represent amino acid identities between M71 and V1RB2. Asterisks indicate residues shared between the conserved amino acids in the V1R sequence logo and M71.
As expected, we found the distribution in the vomeronasal organ of V1rb2mv allele-expressing neurons restricted to the apical part of the neuroepithelium, in a similar location as V1rb2vt-transcribing neurons (V1rb2-IRES-tauLacZ, Figure 2A, B and C).
We then evaluated the localization of the M71 receptor protein in vomeronasal sections of the V1rb2mv line, since our previous analysis relied solely on the expression of beta-galactosidase (the product of the second cistron of the allele). Intense staining of vomeronasal dendritic endings was found in sensory neurons expressing the V1rb2mv allele (Figure 2D), while only weak labeling of the corresponding somata was visible.
When observed by whole mount analyses the expression pattern in vomeronasal neurons of the V1rb2mv allele did not show obvious differences with the one characteristic of the endogenous V1rb2vt allele, i.e. punctate expression in the vomeronasal sensory epithelium (Figure 2E). We evaluated the number of sensory neurons expressing the V1rb2mv or V1rb2vt alleles in 3-4 week-old animals, and found about 1.5 times fewer neurons transcribing the V1rb2mv allele (228+/-21 (mean +/-SD), 6 mice analyzed, vs. 338+/-41 (mean +/-SD), 6 mice analyzed, Student’s t test, df=10, p=0.0008, Figure 2E and F), in accordance to our previous report (Rodriguez et al., 1999). We then analyzed older animals (3.5-5 month-old) and observed a sharp decrease in the number of V1rb2mv-expressing neurons relative to the numbers observed in younger mice, or to age-matched V1rb2vt mice (V1rb2mv: 149+/-21 (mean +/-SD), 4 mice analyzed, V1rb2vt: 470+/−30 (mean +/-SD), 3 mice analyzed, Student’s t test, df=5, p<0.0001, Figure 2F).
We previously showed that axons of vomeronasal neurons expressing the V1rb2 gene in the V1rb2vt and V1rb2vg mouse lines (V1rb2-IRES-tauGFP, Figure 2A), converge and form homogenous glomerular structures in the accessory olfactory bulb (Rodriguez et al., 1999). Axons from vomeronasal sensory neurons expressing a non-functional V1rb2 allele (V1rb2dv line, GFPΔV1rb2-IRES-tauLacZ, Figure 2A) fail to form homogenous glomeruli and innervate multiple glomeruli (Rodriguez et al., 1999). By contrast, expression in vomeronasal neurons of the odorant receptor coding sequence M71 (the V1rb2mv line) leads axons emanating from neurons expressing the exogenous receptor to surprisingly regain the ability to converge and to form multiple homogenous glomeruli in the accessory olfactory bulb (Rodriguez et al., 1999). These glomerular formation data could be explained by three potential mechanisms. First, neurons expressing the V1rb2mv allele may coexpress one or a few V1R genes; expression of a specific V1R receptor in a given vomeronasal neuron is thought to play a major role in the ability of its axon to find like-axons and form glomeruli in the accessory olfactory bulb. Second, expression of the odorant receptor M71 may transform a vomeronasal sensory neuron into an odorant-like sensory neuron, triggering the expression of odorant sensory neuron-specific transduction molecules. A last hypothesis would involve directly the M71 receptor, where it would be recognized by vomeronasal sensory neurons as if it were a V1R receptor, and direct axon guidance, but importantly also prevent the transcription of V1R receptor genes.
Allelic exclusion between V1rb2mv and V1rb2vg-expressing vomeronasal sensory neurons
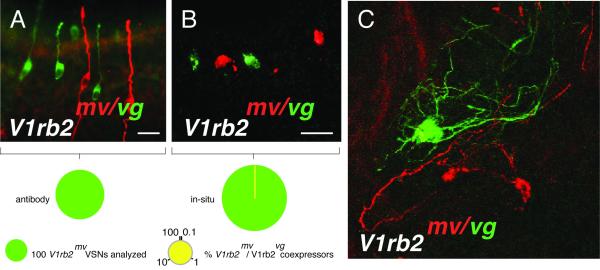
We investigated the possibility that the expression of the V1rb2mv allele was concomitant with the transcription of the other parental V1rb2 allele. We took three parallel approaches. First, we analyzed by immunohistochemistry the potential colocalization of the beta-galactosidase and GFP markers associated with the expression of the V1rb2mv and V1rb2vg alleles respectively. No coexpression of the two alleles was observed after scoring 684 and 1593 neurons expressing the V1rb2mv and V1rb2vg alleles respectively (n=10 mice analyzed, Figure 3A). Next, we hybridized vomeronasal coronal sections from V1rb2mv/vg compound heterozygous mice with probe pairs specific for the two alleles. A single neuron transcribing both alleles was observed among 1064 V1rb2mv and 2445 V1rb2vg-expressing neurons (n=10 mice analyzed, Figure 3B). Finally, we analyzed the axonal projections of neurons expressing the V1rb2mv and V1rb2vg alleles in V1rb2mv/vg compound heterozygous mice. We counted 149 beta-galactosidase and 141 GFP positive glomeruli (6 mice analyzed). None of the 290 analyzed glomeruli received mixed inputs, i.e. fibers pertaining to neurons expressing the V1rb2mv and V1rb2vg alleles (Figure 3C).
Figure 3.
Allelic exclusion between V1rb2mv and V1rb2vg-expressing vomeronasal sensory neurons. (A, B) Coronal sections of V1rb2mv/vg vomeronasal organs. Expression of the V1rb2vg and V1rb2mv alleles corresponds to the green and red colors respectively. (A) Coexpression of the V1rb2vg and V1rb2mv alleles evaluated by immunohistochemistry. (B) Coexpression of the V1rb2vg and V1rb2mv alleles evaluated by in situ hybridizations. The area of the green pies correspond to the number of analyzed neurons expressing the V1rb2mv allele. The yellow color represents the percentage of neurons coexpressing the V1rbvg and V1rb2mv alleles. (C) Axonal projections of vomeronasal neurons expressing the V1rb2vg and V1rb2mv alleles in the accessory olfactory bulb form independent glomeruli. Glomeruli corresponding to the V1rb2vg and V1rb2mv alleles are in green and red respectively. Scale bar: 30 micrometers.
Taken as a whole, and despite a single observed event of biallelic transcription, our data strongly support a monoallelic expression of the V1rb2mv and V1rb2vg alleles.
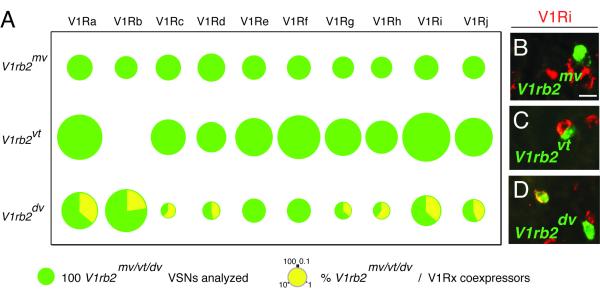
One could however interpret this result differently: monoallelic expression of V1Rs may simply reflect monogenic V1R expression, where all V1R genes, allelic or not, are considered as different entities. With this in mind, the observation of rare coexpression events between the V1rb2mv and V1rb2vg alleles could represent a second V1R choice event taking place after the choice of the V1rb2mv allele, which would be expected in vomeronasal neurons expressing a non-functional V1rb2 gene (the V1rb2dv allele, Figure 2A, Figure 4A) (Roppolo et al., 2007). To test this hypothesis, we decided to evaluate the degree of monogenic expression of the V1rb2mv allele by in situ mRNA hybridizations using probes covering the V1R repertoire.
Figure 4.
V1rb2mv mediated negative feedback. (A) Pies indicating the percentage of vomeronasal neurons coexpressing a V1rb2mv, V1rb2vt orV1rb2dv allele with a member of the V1Ra-j families (data not shown for V1Rk and V1Rl families). (B-D) In situ hybridizations of vomeronasal coronal sections with probes corresponding to V1Ri genes (red) and V1rb2mv (green), V1rb2vt (green) and V1rb2dv (green), alleles. A sensory neuron coexpressing the V1rb2dv allele and a member of the V1Ri family is visible in (D) (yellow). Numbers from control V1rb2vt and V1rb2dv alleles correspond to an addition of novel and previous data (Roppolo et al., 2007). Scale bar: 20 micrometers.
Intrafamily monogenic expression of the V1rb2mv allele
The mouse V1R repertoire is composed of 12 subfamilies (V1Ra-l) (Rodriguez et al., 2002; Zhang et al., 2004). We recently showed that the transcription of the V1rb2vt allele is incompatible with the cotranscription of members of the V1Ra, and V1Rc-l families (Roppolo et al., 2007). Potential coexpression with members of the V1Rb family could not be assessed because of the high V1Rb intrafamily sequence homology, which prevented the use of gene-specific probes. Taking advantage of the M71 coding sequence present in the V1rb2mv allele and its lack of homology to any V1R gene, we assayed for monogenic expression between the V1rb2mv allele and the entire V1Rb repertoire. Analysis of 199 neurons expressing the V1rb2mv allele from heterozygous mice indicated no cohybridization with V1Rb probes (Figure 4A). Thus, the expression of the V1rb2 locus is monogenic relative to members of its own family.
Expression of M71 prevents the coexpression of all V1R receptors
We evaluated the possibility that the transcription of the V1rb2mv allele was concomitant with the expression of V1R genes pertaining to the other 11 V1R families. We hybridized vomeronasal sections from V1rb2mv heterozygous mice with probes specific for the odorant receptor M71 coding sequence and each mouse V1R gene family. In total, 2344 vomeronasal neurons expressing the V1rb2mv allele were scored (80 mice analyzed, Figure 4A and B). No coexpression between the vomeronasally-transcribed M71 and any V1R gene was observed.
The strict monogenic expression of the V1rb2mv allele is apparently indistinguishable from that of the V1rb2vt allele (which we previously reported (Roppolo et al., 2007)). It also contrasts with the lack of negative feedback associated with the transcription of a non-functional V1rb2 allele (V1rb2dv) (Roppolo et al., 2007) (Figure 4A and D). Thus, our data strongly argue that the M71 receptor is able to function as a V1R receptor, i.e. to prevent the expression of the entire V1R gene repertoire in vomeronasal sensory neurons.
Vomeronasal sensory neurons expressing the V1rb2mv allele do not acquire odorant sensory neuron characteristics
The lack of V1R transcription observed in V1rb2mv-expressing vomeronasal sensory neurons could result from the potential acquisition of odorant sensory neuron-specific characteristics by these neurons. It is not known if the expression of a specific chemoreceptor would impart neurons with their odorant/vomeronasal transcriptional identity. Olfactory chemoreceptor transduction cascades are relatively well known. Odorant receptors couple to a Gαs family subunit, Gαolf, which activates adenylyl cyclase 3, whose products trigger the opening of the multimeric cyclic nucleotide-gated channel CNGA2/CNGB1b leading to the entry of Ca++ ions. In contrast, V1R receptor transduction likely involves a Gαi/o family subunit, Gαi2, the messenger diacylglycerol, and a transient receptor potential cation channel (TRPC2). Most of these transduction elements, including Gαolf, Gαi2, CNGA2 or TRPC2, are specifically and exclusively expressed in either main olfactory or vomeronasal sensory neurons.
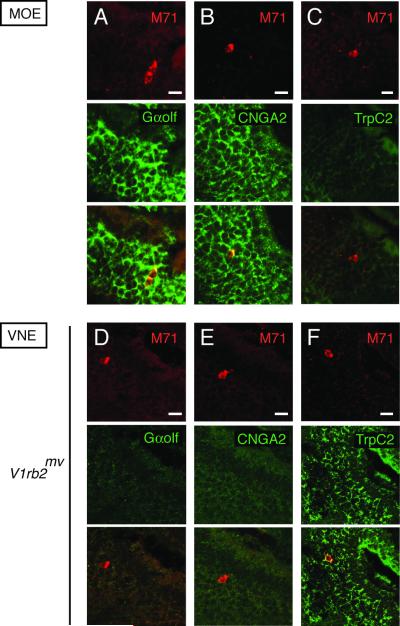
M71-expressing neurons in the main olfactory epithelium strongly coexpress Gαolf and CNGA2, whereas TRPC2 channel transcription is absent (Figure 5A, B and C). By contrast, expression of these genes is reversed in vomeronasal neurons. Similiarly, those transcribing the V1rb2mv allele do not express Gαolf or CNGA2, but strongly transcribe the TRPC2 gene (Figures 5D, E and F). Exogenous expression of an odorant receptor in vomeronasal neurons is thus not able to alter the transcription of critical signal transduction components.
Figure 5.
V1rb2mv-expressing vomeronasal sensory neurons do not acquire odorant sensory neuron characteristics. (A, B) Gαolf and CNGA2 expression (green) in M71/72-expressing neurons (red) in the main olfactory epithelium. (C) Absence of TRPC2 expression (green) in M71/M72-expressing neurons (red) in the main olfactory system. (D, E) Gαolf and CNGA2 are not expressed (green) in vomeronasal neurons expressing the V1rb2mv allele (red). (F) TRPC2 is expressed in vomeronasal neurons expressing the V1rb2mv allele (red). Scale bars: 20 micrometers.
Discussion
Despite the identification of odorant and vomeronasal receptors more than 10 years ago (Buck & Axel, 1991; Herrada & Dulac, 1997; Matsunami & Buck, 1997; Ryba & Tirindelli, 1997), the remarkable problem of their monoallelic or more precisely monogenic regulation remains unsolved.
We show here that two olfactory sensory systems share at least part of the molecular mechanisms which allow the transcription of a single olfactory receptor allele in each sensory neuron. This conclusion is based on the observation that the expression of an exogenous odorant receptor coding sequence in vomeronasal neurons prevents the coexpression of V1R genes. This substitution is functional in terms of negative feedback despite the lack of amino acid identity between the V1RB2 and M71 and the use of different transduction cascades by odorant and V1R receptors.
What signal is given by the transcription of the chemoreceptor to negatively affect the expression of other olfactory chemoreceptors? It likely involves the receptor protein and not the corresponding mRNA: the M4 odorant receptor coding sequence, when rendered untranslatable by the proximal insertion of an unrelated sequence and a mutated start ATG, looses its capacity to block the expression of other odorant receptors (Lewcock & Reed, 2004). This view is also supported by the inability of expressed odorant receptor pseudogenes to provide negative feedback (Serizawa et al., 2003).
Can an odorant receptor couple with the vomeronasal transduction cascade? Can it couple to promiscuous G proteins? Can it make use of a completely different mechanism, independent of G proteins? We report here an age-dependent sharp decrease in the number of vomeronasal neurons expressing the V1rb2mv allele. This decrease could reflect an age-dependent change in V1rb2mv allele choice frequency, or an extinction in the expression of the V1rb2mv allele. Alternatively, the relatively short half life of the neurons transcribing this allele could be consistent with the inability of vomeronasal neurons expressing the odorant receptor to adequately transduce chemosensory signals and/or maintain glomeruli, and as a consequence result in cell death. The existence of a negative feedback mechanism independent of the known odorant transduction cascade is in fact supported by two other observations. First, the exchange of an odorant receptor coding sequence by the one of a constitutively active Gα subunit does not induce negative feedback. Second, several mutations in multiple odorant receptors which impair their ability to couple with Gα subunits still allow for proper negative feedback (Imai et al., 2006; Chesler et al., 2007; Nguyen et al., 2007).
The existence of a still unidentified singnaling cascade responsible for odorant and V1R receptor-induced negative feedback does not explain how two non-homologous protein families could feed into a common cascade. V1Rs and odorant receptors indeed only share a few residues possibly linked to correct cellular targeting in addition to their belonging to the large superfamily of proteins containing seven putative transmembrane segments. Is it therefore a structural characteristic which odorant and V1R receptor proteins share? And are these characteristics limited to these two chemoreceptor superfamilies? This remains to be established.
Strict monogenic regulation is not encountered in the main olfactory system, where a small proportion of sensory neurons can coexpress multiple odorant receptor genes (Shykind et al., 2004; Tian & Ma, 2008). Data from both the the V1rb2mv or V1rb2vt alleles suggest that the very tight V1R transcriptional control observed in vomeronasal neurons is not dependent on the nature of the expressed chemoreceptor.
An absolute dichotomy between the types of receptors expressed by main olfactory versus vomeronasal neurons is in fact not entirely true. A few vomeronasal V1R receptor genes are expressed in the main olfactory system (Karunadasa et al., 2006; Wakabayashi et al., 2007); inversely, some odorant receptors are transcribed in vomeronasal sensory neurons (Levai et al., 2006). If these observations reflect aberrant transcriptional activity or not is unclear, but at least they point to potential common features shared by the odorant and V1R receptor gene transcriptional regulation mechanisms.
As already mentioned, V1Rs play, in addition to their role in chemodetection and gene regulation, a function in axon guidance. Our data on monogenic expression of the V1rb2mv allele establishes that the odorant receptor M71 is apparently able to subtitute for V1RB2 in axon guidance and eventual glomerular formation within the accessory olfactory bulb. A similar common negative feedback system associated with a receptor’s ability to guide axons, may also explain a previous and inverse experiment in which the V1RB2 receptor was shown to partially substitute for the odorant M71 in the main olfactory bulb (Feinstein et al., 2004). It is naturally tempting to speculate that the mechanisms that allow for odorant/V1R monogenic expression and axon coalescence could share a single process, coupled to chemoreceptor expression.
Acknowledgements
We thank Gilad Barnea for providing anti-M71 antibodies, the NCCR “Frontiers in Genetics” bioimaging platform for technical help, and members of the laboratory for comments on the manuscript. This work was supported by grants from NIH (R03DC007340 and NCRRRCMI03037 to P.F.), the NCCR “Frontiers in Genetics” (L.C. and I.R.), and the Swiss National Science Foundation (I.R.).
References
- Belluscio L, Koentges G, Axel R, Dulac C. A map of pheromone receptor activation in the mammalian brain. Cell. 1999;97:209–220. doi: 10.1016/s0092-8674(00)80731-x. [DOI] [PubMed] [Google Scholar]
- Boschat C, Pelofi C, Randin O, Roppolo D, Luscher C, Broillet MC, Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Zou DJ, Le Pichon CE, Peterlin ZA, Matthews GA, Pei X, Miller MC, Firestein S. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proc Natl Acad Sci U S A. 2007;104:1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Del Punta K, Leinders-Zufall T, Rodriguez I, Jukam D, Wysocki CJ, Ogawa S, Zufall F, Mombaerts P. Deficient pheromone responses in mice lacking a cluster of vomeronasal receptor genes. Nature. 2002;419:70–74. doi: 10.1038/nature00955. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- Halpern M, Martinez-Marcos A. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 2003;70:245–318. doi: 10.1016/s0301-0082(03)00103-5. [DOI] [PubMed] [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Hwa J, Klein-Seetharaman J, Khorana HG. Structure and function in rhodopsin: Mass spectrometric identification of the abnormal intradiscal disulfide bond in misfolded retinitis pigmentosa mutants. Proc Natl Acad Sci U S A. 2001;98:4872–4876. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant Receptor-Derived cAMP Signals Direct Axonal Targeting. Science. 2006 doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Karunadasa DK, Chapman C, Bicknell RJ. Expression of pheromone receptor gene families during olfactory development in the mouse: expression of a V1 receptor in the main olfactory epithelium. Eur J Neurosci. 2006;23:2563–2572. doi: 10.1111/j.1460-9568.2006.04795.x. [DOI] [PubMed] [Google Scholar]
- Levai O, Feistel T, Breer H, Strotmann J. Cells in the vomeronasal organ express odorant receptors but project to the accessory olfactory bulb. J Comp Neurol. 2006;498:476–490. doi: 10.1002/cne.21067. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lucas P, Ukhanov K, Leinders-Zufall T, Zufall F. A diacylglycerol-gated cation channel in vomeronasal neuron dendrites is impaired in TRPC2 mutant mice: mechanism of pheromone transduction. Neuron. 2003;40:551–561. doi: 10.1016/s0896-6273(03)00675-5. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I, Del Punta K, Rothman A, Ishii T, Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat Neurosci. 2002;5:134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, Feinstein P, Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97:199–208. doi: 10.1016/s0092-8674(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Ronnett GV, Moon C. G proteins and olfactory signal transduction. Annu Rev Physiol. 2002;64:189–222. doi: 10.1146/annurev.physiol.64.082701.102219. [DOI] [PubMed] [Google Scholar]
- Roppolo D, Vollery S, Kan CD, Luscher C, Broillet MC, Rodriguez I. Gene cluster lock after pheromone receptor gene choice. Embo J. 2007;26:3423–3430. doi: 10.1038/sj.emboj.7601782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba NJ, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative Feedback Regulation Ensures the One Receptor-One Olfactory Neuron Rule in Mouse. Science. 2003 doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol Cell Neurosci. 2008 doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Wagner S, Gresser AL, Torello AT, Dulac C. A multireceptor genetic approach uncovers an ordered integration of VNO sensory inputs in the accessory olfactory bulb. Neuron. 2006;50:697–709. doi: 10.1016/j.neuron.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Ohkura S, Okamura H, Mori Y, Ichikawa M. Expression of a vomeronasal receptor gene (V1r) and G protein alpha subunits in goat, Capra hircus, olfactory receptor neurons. Journal of Comparative Neurology. 2007;503:371–380. doi: 10.1002/cne.21394. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]