Abstract
A novel anaerobic, mesophilic, amino-acid-fermenting bacterium, designated strain CL-84T, was isolated from the swine intestinal tract on mucin-based media. Cells were curved rods (0.8–1.2×3.5–5.0 µm), stained Gram-negative and were non-motile with no evidence of spores. Strain CL-84T produced acetate, propionate, formate and butyrate as the end products of metabolism when grown on serine. Optimum growth occurred at 39 °C and pH 6.5. The major cellular fatty acids were iso-C15 : 0, iso-C15 : 0 3-OH, iso-C17 : 0 and C16 : 0, distinguishing strain CL-84T from closely related species. The DNA G+C content of strain CL-84T was 55.1 mol%. 16S rRNA gene sequence analysis showed that strain CL-84T shared 90–95 % similarity with characterized genera within the phylum Synergistetes, family Synergistaceae. Phylogenetic analysis showed that strain CL-84T was related to, but distinct from, Cloacibacillus evryensis. Based on these findings, we propose that strain CL-84T represents a novel species of the genus Cloacibacillus. We further propose the name Cloacibacillus porcorum sp. nov. be designated for this species. The type strain is CL-84T ( = DSM 25858T = CCUG 62631T). An emended description of the genus Cloacibacillus is provided.
Members of the recently described phylum Synergistetes have been identified in a diverse range of anaerobic environments, including anaerobic digesters (Ganesan et al., 2008) and some infections in the human body (e.g. peritoneal fluid, soft tissues, blood and periodontal pockets) (Vartoukian et al., 2007). Despite their culture-independent identification in a wide range of environments, there are few cultured representatives of this phylum. Synergistes jonesii, the first characterized Synergistetes species, was isolated from a goat rumen. S. jonesii degrades toxic pyridinediols in the animals’ diet, and in turn the animal’s gut provides required nutrients (Allison et al., 1992). In this paper we describe the isolation and characterization of a mucin-degrading bacterium, strain CL-84T, from the swine intestine. We suggest that strain CL-84T represents a novel species of the genus Cloacibacillus, for which the name Cloacibacillus porcorum sp. nov. is proposed. How many taxa may share this trait is unclear, but to our knowledge, this is the first description of a member of the phylum Synergistetes that can use mucin as a sole source of carbon and energy.
Strain CL-84T was one of eight Synergistetes strains isolated during the characterization of mucosa-associated and mucin-degrading micro-organisms from the swine intestinal tract. The gently rinsed mucosal surface of a pig caecum was scraped with a sterile microscope slide and inoculated into minimal medium containing mucin. A series of three enrichments (10 days each) in broth, containing a basal medium, described below, and 1 % (w/v) hog gastric mucin (HGM) (Sigma-Aldrich), were used to enhance the growth of mucolytic bacteria before inoculation on solid media. Mucin-degrading bacteria were isolated on solid basal medium supplemented with 1 % (w/v) HGM after incubation at 39 °C for 5 days. Pure cultures were obtained after isolates were streaked for isolation three times. All cultures were inoculated and incubated (39 °C) in a Coy anaerobic chamber inflated with an atmosphere of N2 (85 %), CO2 (5 %) and H2 (10 %). The basal medium contained (per litre) 0.45 g CaCl2, 0.45 g MgSO4, 2.25 g KH2PO4, 2.25 g K2HPO4, 4.5 g NaCl, 4.5 g (NH4)2SO4, 0.05 % cysteine, 0.05 g haemin, 0.0001 % resazurin and 1.6 % Noble agar.
Strain CL-84T grew optimally on brain heart infusion broth with 0.05 % cysteine and 0.0001 % resazurin, and supplemented with 20 mM arginine and histidine (BHIAH). This medium was used to maintain cultures. After 3 days growth at 39 °C on BHIAH medium, strain CL-84T reached a terminal OD620 of 1.2, representing 1.5×109 c.f.u. ml−1. The calculated doubling time was 8 h. Cells of strain CL-84T cultured in BHIAH broth had a curved-rod shape, were non-motile (determined with motility test medium; Greene et al., 1951) and spores were not seen (Fig. 1a). On BHIAH plates after 5 days growth, strain CL-84T produced small semi-translucent brown colonies that were 1 mm in diameter. Gram strain was negative. Ultrathin sections were prepared for transmission electron microscopy (TEM) from 4-day-old cultures, stained with uranyl acetate and lead citrate, and examined with a Tecnai 12 G2 Biotwin microscope (FEI). Micrographs revealed a cell outer envelope structure consistent with Gram-negative cells with a thin peptidoglycan layer surrounded by an outer membrane (Fig. 1b). No spores or inclusion bodies were seen. Cells were variable in size (0.8–1.2 µm × 3.5–5.0 µm). The TEM appearance and Gram staining characteristics were consistent with the Gram-negative envelope characteristics of other Synergistetes spp.
Fig. 1.
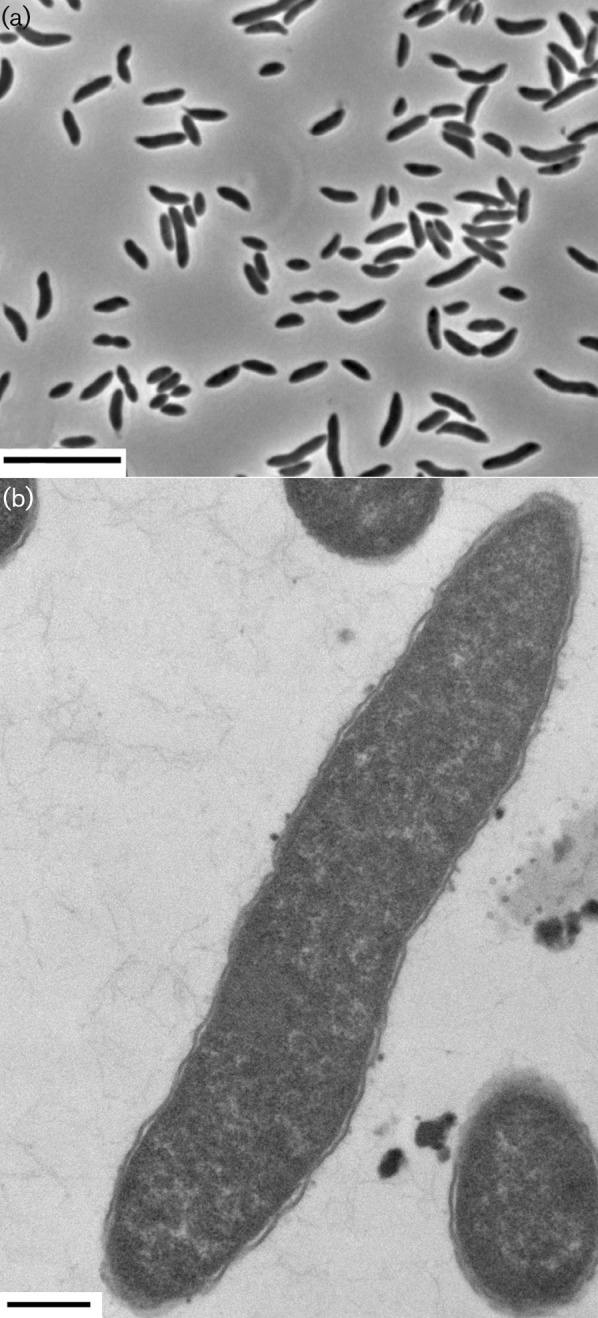
(a) Phase-contrast micrograph of cells of strain CL-84T grown in BHIAH medium. Bar, 10 µm. (b) Transmission electron micrograph of ultrathin sections of cells of strain CL-84T, showing a loose outer cell membrane consistent with a Gram-negative outer cell envelope and a negative Gram-staining reaction. Bar, 500 nm.
Genomic DNA was extracted from pelleted cells of strain CL-84T by following the protocol of Dale & Greenaway (1985). PCR amplification was carried out as described by Downes et al. (2005), with the conserved bacterial primers 8F (Wilmotte et al., 1993) and 1492R (Stackebrandt & Goodfellow, 1991). Purified PCR products were sequenced, yielding nearly full-length (1447 bp) sequences for the 16S rRNA gene of strain CL-84T. Taxonomic assessments of the 16S rRNA gene sequences were made using the Ribosomal Database project (RDP) web tools (http://rdp.cme.msu.edu/) (Cole et al., 2009), which placed strain CL-84T within the phylum Synergistetes. The closest matches were to Cloacibacillus evryensis 158T (95 % sequence similarity) and S. jonesii 78-1T (90 %). C. evryensis was isolated from a municipal anaerobic waste digester (Ganesan et al., 2008) and S. jonesii was isolated from a goat rumen (Allison et al., 1992). Support for other Synergistetes bacteria being associated with mucus and possibly utilizing mucin can be found from their identification from subgingival plaque samples, and optimized growth (in co-culture) by the addition of mucin to the media (Vartoukian et al., 2010).
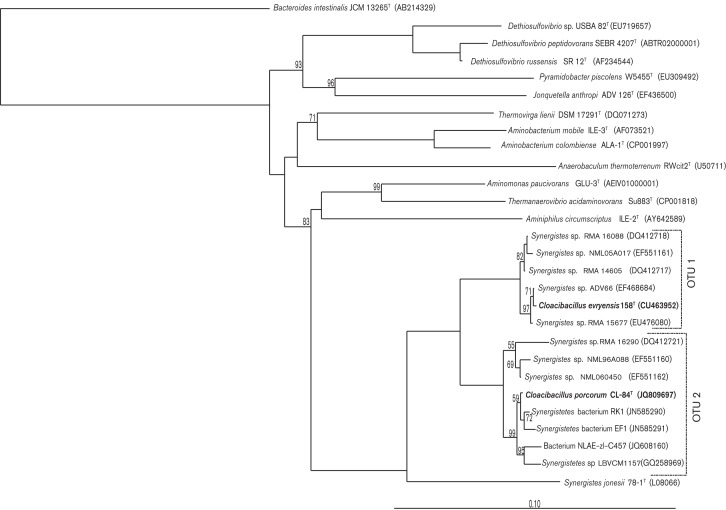
Neighbour-joining, maximum-parsimony and maximum-likelihood phylogenetic analyses were performed based on the alignment of the 16S rRNA gene sequence from strain CL-84T with Silva’s SINA web aligner (Pruesse et al., 2007) in the software package arb (Ludwig et al., 2004). The methods used to construct the tree were arb neighbour-joining (10 000 bootstrap replicates), maximum-parsimony with dnapars v3.6 (1000 bootstraps) and maximum-likelihood with RAxML (‘advanced bootstrap+refinement of BS tree’ algorithm, GTRGAMMA model, 1000 bootstraps). All three analyses produced trees with the same topology, and therefore only the neighbour-joining tree is presented (Fig. 2). Strain CL-84T grouped with uncharacterized isolates from infected human blood (GenBank accession numbers GQ258969, EF551162 and EF551160) (Marchandin et al., 2010) and peritoneal fluid samples (DQ412721) (Horz et al., 2006). These sequences were approximately 99 % similar to each other and to that of strain CL-84T, forming a cluster previously designated OTU cluster 2 by Ganesan et al. (2008). In contrast, the 16S rRNA gene sequences from C. evryensis 158T and S. jonesii 78-1T were only 95 and 90 % similar to the sequences in OTU cluster 2, respectively. Strain CL-84T and related sequences formed a branch distinct from C. evryensis (OTU cluster 1), within the genus Cloacibacillus.
Fig. 2.
Neighbour-joining phylogentic tree of the proposed species Cloacibacillus porcorum sp. nov. (strain CL-84T) and selected reference type strains based on partial 16S rRNA gene sequences (all ≥1334 bp). The tree is rooted by Bacteroides intestinalis JCM 13265T. Numbers by the branches of the tree represent percentage bootstrap values of 10 000 resamplings, and are only noted if the percentage was greater than 50 %. OTU 1 and 2, previously described by Ganesan et al. (2008), are shown and represent a 16S sequence divergence of ~5 %. Bar, 10 nt substitutions per 100 nt.
As part of a larger survey of swine intestinal microbes, the genome of strain CL-84T was sequenced using a 454 titanium pyrosequencing platform (454 Life Sciences). The gyrase B gene sequence of strain CL-84T (GenBank accession no. JX443487) was obtained from the genome sequence and compared with that of C. evryensis, obtained from the finished genome sequence (IMG/GEBA database: 6858). The gyrase B nucleotide sequence from strain CL-84T showed only 90 % similarity to C. evryensis, which supports the conclusion that these are different species. Sequence analysis revealed that strain CL-84T has a DNA G+C content of 55.1 mol%.
Mucins are glycoproteins with carbohydrate side chains connected to a protein backbone by O-glycosidic links (Nataro, 2005) and are the major component of mucus. The carbohydrate side groups are made up of the sugars galactose, fucose, N-acetyl-d-galactosamine, N-acetyl-d-glucosamine, sialic acid and mannose (Allen et al., 1998). Mucolytic bacteria use proteases and glycosidases to degrade host mucin at polypeptide and glycosidic bonds, respectively (Bradshaw et al., 1994). Growth on different mucin components was examined by preparing basal medium containing 0.2 % (w/v) yeast extract with 0.5 % (w/v) of each of the following: chondroitin sulfate sodium salt, N-acetyl-d-glucosamine, N-acetyl-d-galactosamine, hyaluronan biotin sodium salt, mannose, N-acetylneuraminic acid, d-galactose or l-fucose. Strain CL-84T was further evaluated for its ability to degrade mucin O-linked glycans, purified from the HGM, as described by Martens et al. (2008). Growth on a variety of amino acids was examined by supplementing basal medium containing 0.2 % (w/v) yeast extract with 10 mM of each of the following: arginine, histidine, lysine, serine, tryptophan, alanine, glutamate, aspartate, proline, glycine, cysteine, phenylalanine, isoleucine, leucine, valine, threonine, methionine, glutamine, asparagine or tyrosine. Strain CL-84T grew on BHI and Casamino acids broth (both used for growth of C. evryensis), but grew faster on BHIAH. Growth was observed on a limited number of amino acids and whole mucin and mucin components. Growth on whole mucin and mucin O-linked glycans was also tested and observed with C. evryensis (OD620 of 0.8 and 0.06, respectively). Growth results for strain CL-84T are summarized in Table 1 as OD620 values.
Table 1. Fermentation products from substrates that support growth of strain CL-84T in basal medium.
No growth was observed on methionine, aspartate, valine, proline, isoleucine, lysine, alanine, phenylalanine, leucine, glucose, glutamine, glycine, asparagine, glutamate or tyrosine.
| Substrate | Formic acid* | Acetic acid* | Propionic acid* | Butyric acid* | OD620† |
| BHIAH | − | ++ | + | − | +++ |
| Histidine | + | + | + | − | ++ |
| Arginine | + | + | + | − | ++ |
| Tryptophan | − | + | − | − | + |
| Cysteine | + | ++ | + | − | ++ |
| Threonine | − | + | − | − | + |
| Serine | + | + | − | + | + |
| Serine, threonine, proline | + | ++ | + | + | ++ |
| (−)-N-Acetylneuraminic acid | + | − | + | − | + |
| Hyaluronan biotin sodium salt | + | + | + | − | ++ |
| Mannose | + | + | − | − | ++ |
| Chondroitin sulfate sodium salt | + | + | + | − | + |
| Fucose | − | + | + | − | + |
| d-Galactose | − | + | − | − | + |
| N-Acetyl-d-galactosamine | − | − | + | − | + |
| N-Acetyl-d-glucosamine | − | − | − | − | + |
| Mucin | + | + | + | − | + |
| Mucin O-linked glycans | + | ++ | + | − | + |
Measured by GC. All values were corrected for the small amount of short-chain fatty acids formed in the control tubes. Experiments were conducted in triplicate and mean values are given. −, <1.0 mM; +, 1.0–5.0 mM; ++, 5.1–10.0 mM.
OD620 values: +, 0.05–0.1; ++, 0.2–0.5; +++, 0.6–1.2.
The supernatants from cultures on each substrate were analysed for fermentation acids by GC of butyl esters (Salanitro & Muirhead, 1975; Stanton & Lebo, 1988). Major fermentation products were formate, acetate or propionate; however, growth on serine, both alone and with threonine and proline, also produced buyrate (Table 1). Strain CL-84T contained butyryl-CoA : acetate CoA-transferase (E.C. 2.8.3.8) activity [specific activity = 9.88±0.45 µmol min−1 (mg protein)−1] as determined using cells cultured on basal medium with serine. The assay for transferase activity was performed on French press extracts of strain CL-84T according to Buckel et al. (1981) except that it was evaluated at 412 nm, 39 °C, with 0.1 mM butyryl-CoA and 0.5 units (8.8 nkat) of citrate synthase. The most prevalent amino acids in mucin are serine and threonine (Schrager, 1970). The ability of strain CL-84T to produce butyrate when grown on serine may reflect its role in the gut, as butyrate has been shown to stimulate mucin synthesis in human colonic cell lines and animal mucosa (Finnie et al., 1995; Hatayama et al., 2007).
Additional biochemical characterizations were performed using Rapid ID 32 A and API ZYM test strips (bioMérieux). Strain CL-84T showed positive arginine dihydrolase and glutamic acid decarboxylase activities and weak alkaline phosphatase and proline arylamidase activities. Additionally, the API ZYM test showed that strain CL-84T had acid phosphatase, naphthol-AS-BI-phosphohydrolase, alkaline phosphatase, esterase (C4), esterase lipase (C8) and leucine arylamidase activities. Strain CL-84T tested negative for catalase.
Heat, pH and oxygen tolerance were tested in BHIAH medium. The pH range for growth of strain CL-84T was pH 4–8, with optimal growth (reaching an OD620 of >1.0) at pH 6–7. Growth between 20 and 45 °C was observed. No viable cells remained after heating cultures to 80 °C for 30 min. Growth of CL-84T was inhibited by NaCl concentrations above 1.4 % (w/v), with optimal growth at 0.6–0.8 % (w/v) NaCl. Strain CL-84T cells were aerotolerant and still viable after 24 h of exposure to oxygen. Growth of CL-84T was not observed on BHIAH plates, exposed to 1.0 % oxygen, after 1 week of incubation.
The closest related type strain to CL-84T, C. evryensis 158T, was obtained from the DSM culture collection (accession no.19522). Both strain CL-84T and C. evryensis 158T were grown in 10 ml BHIAH medium to an OD620 of 1.0, and cell pellets were used for cellular fatty acid analysis. The cellular fatty acid composition was determined by Microbial ID Inc. with the GC-based MIDI Sherlock Microbial Identification System. The major cellular fatty acids of strain CL-84T were iso-C15 : 0 (27.1 %), iso-C15 : 0 3-OH (15.0 %), iso-C17 : 0 (11.7 %) and C16 : 0 (9.7 %). The profile for strain CL-84T differed from that of C. evryensis 158T in both the fatty acid types and the proportions of each component. The top three fatty acids for strain CL-84T were iso-C15 : 0, iso-C15 : 0 3-OH and iso-C17 : 0 (27.1, 15 and 11.7 %, respectively), while the top three for C. evryensis 158T were iso-C15 : 0, iso-C15 : 0 3-OH and C17 : 1ω11c (14.2, 14 and 12.4 %, respectively) (Table 2).
Table 2. Cellular fatty acid profiles (%) of strain CL-84T and C. evryensis 178T, grown on BHIAH medium.
Taxa: 1, CL-84T; 2, C. evryensis 178T.
| Fatty acid | Proportion of total fatty acids (%) | |
| 1 | 2 | |
| iso-C15 : 0 | 27.1 | 14.2 |
| iso-C15 : 0 3-OH | 15 | 14 |
| iso-C17 : 0 | 11.7 | 2.4 |
| C16 : 0 | 9.7 | 1.8 |
| iso-C13 : 0 | 7.6 | 5.8 |
| C17 : 0 | 4.2 | 12 |
| iso-C17 : 1 at 10 | 3.9 | 2.7 |
| C16 : 1ω7c | 3.7 | 1.6 |
| C18 : 0 | 2.8 | 0 |
| C18 : 1ω11c | 2.6 | 0 |
| C17 : 1ω11c | 2.3 | 12.4 |
| C15 : 0 | 1.9 | 4.8 |
| C16 : 1ω11c | 1.7 | 0 |
| iso-C17 : 1 at 9 | 1.6 | 0 |
| C13 : 0 | 0 | 3.4 |
| C15 : 0 3-OH | 0 | 7.7 |
| C15 : 1ω6c | 0 | 4.3 |
| C16 : 1ω5c | 0 | 1.8 |
| C17 : 1ω3c | 0 | 1.3 |
| C17 : 1ω6c | 0 | 5.3 |
| Summed features* | ||
| C14 : 0 3-OH/iso-C16 : 1 | 3.2 | 2.2 |
| C17 : 0 DMA/unknown at 17.49† | 0 | 1.8 |
Summed features represent two fatty acids that cannot be separated with the MIDI system.
Unknown fatty acids do not have a name listed in the MIDI system library; values are equivalent chain lengths.
Antibiotic minimum inhibitory concentration (MIC) assays were modified from Allen et al. (2009) by incubating anaerobically in BHIAH for 1 week. Strain CL-84T was susceptible to tylosin, lincomycin, chlortetracycline, penicillin, florphenicol, ceftiofur and carbadox (MIC <4 µg µl−1) and resistant to vancomycin and sulfathiazole (MIC 512 µg µl–1 for each). Other Synergistetes spp. have also been shown to be resistant to vancomycin (Allison et al., 1992; Downes et al., 2009; Ganesan et al., 2008).
Emended description of the genus Cloacibacillus Ganesan et al. 2008
The description of the genus Cloacibacillus follows that of Ganesan et al. (2008) with the following modifications and additions. The ability to grow on mucin and mucin components, as a sole carbon source, is a trait shared by members of this genus. Cells are glucose non-fermenters, but can grow on fucose. The predominant fatty acids include iso-C15 : 0 and iso-C15 : 0. Differential characteristics between strain CL-84T and C. evryensis 158T and S. jonesii 78-1T are given in Table 3.
Table 3. Differential characteristics between strain CL-84T and related species in the phylum Synergistetes.
Taxa: 1, CL-84T; 2, C. evryensis 158T; 3, S. jonesii 78-1T.
| Characteristic | 1 | 2* | 3† |
| Isolation source | Swine intestinal tract | Anaerobic digester | Goat rumen |
| Cell morphology | Slightly curved rods | Straight rods | Rods |
| Cell size (µm) | 0.8–1.2×3.5–5.0 | 0.8–1.0×2.0–3.0 | 0.6–0.8×1.2–1.8 |
| 16S rRNA similarity to CL-84T (%) | 100 | 95 | 90 |
| Optimum growth temperature (°C) | 39 | 35–40 | 39 |
| DNA G+C content (mol%) | 55.1 | 55.8 | 58 |
| Fermentation | Proteolytic, some carbohydrates | Proteolytic | Proteolytic |
| Short-chain fatty acids produced during fermentation | Acetate, propionate, formate; butyrate, only when serine is supplied | Acetate, propionate, butyrate, valerate | Acetate, propionate; formate only when histidine is supplied |
| Major cellular fatty acids | iso-C15 : 0, iso-C15 : 0 3-OH, iso-C17 : 0, C16 : 0 | iso-C15 : 0, iso-C15 : 0 3-OH, C17 : 1ω11c, C17 : 0 | C17 : 0, C20 cyclo, C17 : 1ω6c, C15 : 0 |
| Additional features | Degrades mucin | Degrades mucin | Degrades 3,4-dihydroxy pyridine |
Data from Ganesan et al. (2008).
Data from Allison et al. (1992).
Description of Cloacibacillus porcorum sp. nov.
Cloacibacillus porcorum (por.co′rum. L. n. porcus swine, pig; L. masc. pl. n. porcorum of/from pigs).
Cells are obligately anaerobic, non-motile and curved-rod shaped. Cells ferment amino acids, and can use mucin and mucin components as a sole carbon source. Fermentation products are acetate, propionate and formate but only butyrate is produced from serine. This bacterium is an intestinal commensal of swine. Cells have a Gram-negative cell-wall structure and range in size from 0.8 to 1.2 µm wide and 3.5 to 5.0 µm long. Strong growth is obtained in BHI medium, and growth is enhanced by the addition of histidine and arginine, but not glucose. After 7 days incubation, colonies are 1 mm in diameter, circular, shiny, brown and semi-translucent. The main cellular fatty acids are iso-C15 : 0, iso-C15 : 0 3-OH, iso-C17 : 0 and C16 : 0. Cells are resistant to vancomycin and sulfathiazole but are susceptible to tylosin, lincomycin, chlortetracycline, penicillin, florphenicol, ceftiofur and carbadox. Optimal growth occurs at 39 °C and pH 6.5.
The type strain, CL-84T ( = DSM 25858T = CCUG 62631T), was isolated from the mucosal lining of a pig caecum in Ames, Iowa, USA. The DNA G+C content of the type strain is 55.1 mol%.
Acknowledgements
The authors thank Sam Humphrey, Deb Lebo, Darrell Bayles and David Alt for scientific advice and technical support. We thank Judi Stasko for the electron microscopy work, Floyd Dewhirst for taxonomic advice, Eric Martens for advice with our glycan purifications, Jonathan Eisen for access to the C. evryensis genome, and Heather Allen, Milt Allison and Nancy Cornick for helpful discussions and comments on the manuscript. Animals were raised in accordance with National Animal Disease Center Animal Care and Use Committee guidelines. This work was supported by the Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Abbreviations:
- TEM
transmission electron microscopy
References
- Allen A., Hutton D. A., Pearson J. P. (1998). The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol 30, 797–801 10.1016/S1357-2725(98)00028-4 [DOI] [PubMed] [Google Scholar]
- Allen H. K., Cloud-Hansen K. A., Wolinski J. M., Guan C., Greene S., Lu S., Boeyink M., Broderick N. A., Raffa K. F., Handelsman J. (2009). Resident microbiota of the gypsy moth midgut harbors antibiotic resistance determinants. DNA Cell Biol 28, 109–117 10.1089/dna.2008.0812 [DOI] [PubMed] [Google Scholar]
- Allison M. J., Mayberry W. R., McSweeney C. S., Stahl D. A. (1992). Synergistes jonesii, gen. nov., sp. nov.: a rumen bacterium that degrades toxic pyridinediols. Syst Appl Microbiol 15, 522–529 10.1016/S0723-2020(11)80111-6 [DOI] [Google Scholar]
- Bradshaw D. J., Homer K. A., Marsh P. D., Beighton D. (1994). Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 140, 3407–3412 10.1099/13500872-140-12-3407 [DOI] [PubMed] [Google Scholar]
- Buckel W., Dorn U., Semmler R. (1981). Glutaconate CoA-transferase from Acidaminococcus fermentans. Eur J Biochem 118, 315–321 10.1111/j.1432-1033.1981.tb06404.x [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., Kulam-Syed-Mohideen A. S., McGarrell D. M., Marsh T., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37 (Database issue), D141–D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Greenaway P. J. (1985). Preparation of chromosomal DNA from E. coli. Methods Mol Biol 2, 197–200 [DOI] [PubMed] [Google Scholar]
- Downes J., Sutcliffe I., Tanner A. C., Wade W. G. (2005). Prevotella marshii sp. nov. and Prevotella baroniae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 55, 1551–1555 10.1099/ijs.0.63634-0 [DOI] [PubMed] [Google Scholar]
- Downes J., Vartoukian S. R., Dewhirst F. E., Izard J., Chen T., Yu W. H., Sutcliffe I. C., Wade W. G. (2009). Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol 59, 972–980 10.1099/ijs.0.000364-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie I. A., Dwarakanath A. D., Taylor B. A., Rhodes J. M. (1995). Colonic mucin synthesis is increased by sodium butyrate. Gut 36, 93–99 10.1136/gut.36.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A., Chaussonnerie S., Tarrade A., Dauga C., Bouchez T., Pelletier E., Le Paslier D., Sghir A. (2008). Cloacibacillus evryensis gen. nov., sp. nov., a novel asaccharolytic, mesophilic, amino-acid-degrading bacterium within the phylum ‘Synergistetes’, isolated from an anaerobic sludge digester. Int J Syst Evol Microbiol 58, 2003–2012 10.1099/ijs.0.65645-0 [DOI] [PubMed] [Google Scholar]
- Greene R. A., Blum E. F., DeCoro C. T., Fairchild R. B., Kaplan M. T., Landau J. L., Sharp T. R. (1951). Rapid methods for the detection of motility. J Bacteriol 62, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatayama H., Iwashita J., Kuwajima A., Abe T. (2007). The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun 356, 599–603 10.1016/j.bbrc.2007.03.025 [DOI] [PubMed] [Google Scholar]
- Horz H. P., Citron D. M., Warren Y. A., Goldstein E. J., Conrads G. (2006). Synergistes group organisms of human origin. J Clin Microbiol 44, 2914–2920 10.1128/JCM.00568-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., et al. (2004). arb: a software environment for sequence data. Nucleic Acids Res 32, 1363–1371 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchandin H., Damay A., Roudière L., Teyssier C., Zorgniotti I., Dechaud H., Jean-Pierre H., Jumas-Bilak E. (2010). Phylogeny, diversity and host specialization in the phylum Synergistetes with emphasis on strains and clones of human origin. Res Microbiol 161, 91–100 10.1016/j.resmic.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Martens E. C., Chiang H. C., Gordon J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P. (2005). Colonization of Mucosal Surfaces. Washington, DC: American Society for Microbiology [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., Glöckner F. O. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanitro J. P., Muirhead P. A. (1975). Quantitative method for the gas chromatographic analysis of short-chain monocarboxylic and dicarboxylic acids in fermentation media. Appl Microbiol 29, 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager J. (1970). The chemical composition and function of gastrointestinal mucus. Gut 11, 450–456 10.1136/gut.11.5.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goodfellow M. (1991). Nucleic Acid Techniques in Bacterial Systematics. New York: Wiley [Google Scholar]
- Stanton T. B., Lebo D. F. (1988). Treponema hyodysenteriae growth under various culture conditions. Vet Microbiol 18, 177–190 10.1016/0378-1135(88)90063-6 [DOI] [PubMed] [Google Scholar]
- Vartoukian S. R., Palmer R. M., Wade W. G. (2007). The division “Synergistes”. Anaerobe 13, 99–106 10.1016/j.anaerobe.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Vartoukian S. R., Palmer R. M., Wade W. G. (2010). Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ Microbiol 12, 916–928 10.1111/j.1462-2920.2009.02135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmotte A., Van der Auwera G., De Wachter R. (1993). Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF‘) strain PCC7518, and phylogenetic analysis. FEBS Lett 317, 96–100 10.1016/0014-5793(93)81499-P [DOI] [PubMed] [Google Scholar]