Abstract
Five strains of anaerobic, Gram-negative bacilli isolated from the human oral cavity were subjected to a comprehensive range of phenotypic and genotypic tests and were found to comprise a homogeneous group. Phylogenetic analysis of full-length 16S rRNA gene sequences showed that these strains represented a novel group within the family Prevotellaceae, and the most closely related species was Prevotella tannerae. P. tannerae and the novel taxon are deeply branched from the genus Prevotella, with sequence identities to the type strain of the type species of Prevotella, Prevotella melaninogenica, of 82.2 and 85.6 %, respectively. The novel genus Alloprevotella gen. nov. is proposed to accommodate the novel species Alloprevotella rava gen. nov., sp. nov. and the previously named Prevotella tannerae Moore et al. 1994 as Alloprevotella tannerae gen. nov., comb. nov. The type species is Alloprevotella tannerae. The type strain of Alloprevotella rava is 81/4-12T ( = DSM 22548T = CCUG 58091T) and the type strain of Alloprevotella tannerae is ATCC 51259T = CCUG 34292T = CIP 104476T = NCTC 13073T. Alloprevotella rava is weakly to moderately saccharolytic and produces moderate amounts of acetic acid and major amounts of succinic acid as end products of fermentation. Strains are sensitive to 20 % bile and hydrolyse gelatin. The principal cellular long-chain fatty acids are anteiso-C15 : 0, iso-C15 : 0, C16 : 0, iso-C17 : 0 and iso-C17 : 0 3-OH. The G+C content of the DNA of the type strain is 47 mol%.
The genus Prevotella includes a number of species commonly found in the human mouth, some of which are associated with oral infections (Kononen et al., 2010). Despite the numerical importance of the genus in the human mouth, many ‘species’ remain undescribed; release 10 of the Human Oral Microbiome Database (HOMD) (Chen et al., 2010; Dewhirst et al., 2010) lists 28 unnamed taxa at species level. In this study, a group of five strains related to Prevotella tannerae were subjected to a range of phenotypic and genetic tests.
Strains 81/4-12T and P4P-62 and P. tannerae 104/4-8 were isolated from subgingival plaque in subjects with periodontitis, and strain W6968 was isolated from supragingival plaque from a periodontally healthy subject. Strains F0020 and F0323 were subcultured from strains D079N-07 and D082T-14, which were isolated from subgingival plaque in subjects with periodontitis and were among the collection of W. E. C. Moore and L. V. Holdeman Moore, formerly of the Virginia Polytechnic Institute. P. tannerae strains D083Z-15, D063A-11, D079E-11A and D146L-01 also came from the Moores’ collection, while the type strain, ATCC 51259T, was obtained from the ATCC.
Strains were grown at 37 °C on fastidious anaerobe agar (FAA; LabM) supplemented with 5 % horse blood under anaerobic conditions (80 % N2, 10 % H2, 10 % CO2) in an anaerobic workstation (Don Whitley Scientific). Colonial morphologies were determined using a dissecting microscope after 3 days of incubation. Pigmentation was examined on FAA supplemented with 5 % rabbit blood following up to 21 days of incubation. Cellular morphology was recorded after Gram-staining of smears prepared from 2-day FAA plate cultures. Hanging-drop preparations of 18-h cultures of peptone/yeast extract/glucose (PYG) broth were examined by phase-contrast microscopy for cellular motility. The range and optimum temperature for growth were determined after 5 days of incubation in pre-reduced PYG broth (Holdeman et al., 1977). The range and optimum pH for growth were determined in pre-reduced peptone/yeast extract (PY) broth incubated at 35 °C for 5 days with the initial pH obtained by adding HCl (0.2 M) or Na2CO3 (10 % w/v) to the PY broth.
Biochemical and physiological tests were performed using standard methods (Holdeman et al., 1977; Jousimies-Somer et al., 2002). Fermentation tests were performed using pre-reduced, anaerobically sterilized (PRAS) sugars prepared in-house in an anaerobic workstation (Holdeman et al., 1977). Susceptibility to special-potency antibiotic discs containing vancomycin (5 µg), kanamycin (1 mg) and colistin (10 µg) was determined on FAA (Jousimies-Somer et al., 2002). Bacterial strains were grown in PY and PYG, and short-chain volatile and non-volatile fatty acids produced as metabolic end products were extracted by standard methods and analysed by gas chromatography (Holdeman et al., 1977). Enzyme profiles were generated with the Rapid ID 32A anaerobe identification kit (bioMérieux), according to the manufacturer’s instructions. The G+C content of the DNA was determined by an HPLC method as described previously (Wade et al., 1999).
Analysis of the cellular fatty acids was carried out by the Identification Service of the DSMZ (Braunschweig, Germany) on strain 81/4-12T and P. tannerae ATCC 51259T. Fatty acid methyl esters were obtained from 50 mg (dry weight) cells by saponification, methylation and extraction using minor modifications of previously described methods (Kuykendall et al., 1988; Miller, 1982). Fatty acid methyl ester mixtures were separated using the Sherlock Microbial Identification System (MIS) (MIDI, Microbial ID), which consisted of an Agilent model 6890N gas chromatograph fitted with a 5 % phenyl-methyl silicone capillary column (0.2 mm×25 m), a flame-ionization detector, an Agilent model 7683A automatic sampler and an HP computer with MIDI database (Hewlett Packard). Peaks were automatically integrated and fatty acid names and percentages were calculated by the MIS standard software (Sherlock version 6.1; Microbial ID). The gas chromatographic parameters were as follows: carrier gas, ultrahigh-purity hydrogen; column head pressure, 60 kPa; injection volume, 2 µl; column split ratio, 100 : 1; septum purge, 5 ml min−1; column temperature, 170–270 °C at 5 °C min−1; injection port temperature, 240 °C; and detector temperature, 300 °C.
16S rRNA genes were sequenced as described previously (Downes et al., 2005). Sequences were connected using the BioEdit program (Hall, 2004) and their closest relatives were identified by blast interrogation of the GenBank database (Altschul et al., 1990). Sequences were aligned by clustal w within BioEdit. Phylogenetic trees were constructed using mega version 5, by the neighbour-joining method from distance matrices prepared using the Jukes–Cantor correction.
The results of the phenotypic tests for the five novel strains are summarized in the species description. The five strains were obligately anaerobic, non-motile, Gram-negative bacilli, 0.5–0.6 µm wide by 1.0–5.0 µm long, that were arranged singly or in short chains, with occasional cells up to 10 µm long. After 3 days of incubation on FAA plates, colonies were 0.8–1.4 mm in diameter, circular, entire, convex, grey and opaque with an internal appearance of concentric rings of opacity and translucency when viewed under a plate microscope. No pigmentation was observed on FAA plates supplemented with 5 % rabbit blood after 21 days of incubation. Strains were resistant to special-potency discs containing colistin, kanamycin and vancomycin. The optimum temperature for growth was 35 °C, with moderate growth at 30 °C, marginal growth at 42 °C and no growth at 25 or 45 °C. The optimum pH for growth was pH 7, with marginal growth at pH 6 and no growth at pH 5 or 8. Growth in broth media was variable, and generally produced a light to moderate turbidity (2 to 3+ on a scale of 0 to 4+) and occasionally a moderate to heavy growth (3 to 4+), with slight stimulation of growth by the addition of fermentable carbohydrates. Growth in PY broth with 1 % lactose, even with the addition of 10 % sterile serum, was particularly variable, and it was often difficult to obtain sufficient growth to enable the determination of lactose fermentation. However, when growth of 3 to 4+ turbidity was achieved, the strains proved to be positive for lactose fermentation. Strains were weakly to moderately saccharolytic (see species description for sugar reactions), and moderate amounts of acetic acid and major amounts of succinic acid were produced as end products of metabolism when cells were grown in PYG broth. When cells were grown in PY broth, without glucose, trace to minor amounts of isovaleric acid were also produced in addition to acetic and succinic acids. Gelatin hydrolysis was positive but other biochemical tests were negative (see species description). The G+C content of the DNA of both strains 81/4-12T and W6968 was 47 mol%.
All five strains gave positive reactions in the Rapid ID 32A panel for α-galactosidase, β-galactosidase, 6-phospho-β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-fucosidase, mannose fermentation, raffinose fermentation, alkaline phosphatase, leucyl glycine arylamidase and alanine arylamidase. Reactions for glutamyl glutamic acid arylamidase were weak and variable. Negative reactions were obtained for the remaining 17 enzymes, resulting in a profile of 4707 4402 22/0. The six strains of P. tannerae gave similar enzymic reactions, except that 6-phospho-β-galactosidase was negative and N-acetyl-β-glucosaminidase and mannose fermentation were variable, resulting in a profile of 4507/4 4402 22.
The principal cellular long-chain fatty acids identified in strain 81/4-12T were anteiso-C15 : 0, iso-C15 : 0, C16 : 0, iso-C17 : 0 and iso-C17 : 0 3-OH. P. tannerae ATCC 51259T showed a similar profile apart from iso-C17 : 0, which was not a major component in this strain (Table 1).
Table 1. Fatty acid methyl ester (FAME) data for Alloprevotella rava sp. nov. 81/4-12T and Alloprevotella tannerae comb. nov. ATCC 51259T.
Values are percentages of total fatty acids.
| FAME | 81/4-12T | ATCC 51259T |
| iso-C13 : 0 | 3.4 | 2.3 |
| anteiso-C13 : 0 | 1.3 | 0.4 |
| iso-C14 : 0 | 3.1 | 2.4 |
| C14 : 0 | 1.8 | 3.9 |
| iso-C15 : 0 | 10.6 | 18.5 |
| anteiso-C15 : 0 | 30.4 | 25.9 |
| iso-C16 : 0 | 2.3 | 1.1 |
| C16 : 0 | 9.2 | 6.1 |
| iso-C17 : 0 | 11.8 | 1.2 |
| anteiso-C17 : 0 | 3.6 | 1.0 |
| C16 : 0 3-OH | 0.8 | 5.5 |
| C18 : 1ω9c | 0.6 | 3.0 |
| iso-C17 : 0 3-OH | 12.4 | 18.6 |
| C17 : 0 2-OH | 0.6 | 1.7 |
Phylogenetic analysis of full-length 16S rRNA gene sequences showed that the five strains represented a novel group within the family Prevotellaceae that is clearly distinct from any species with validly published names (Fig. 1). A fuller phylogenetic tree is available as Fig. S1 in IJSEM Online. Full-length sequences of the 16S rRNA gene for strains 81/4-12T, P4P-62, W6968, F0020 and F0323 showed 98.8 % sequence identity or more to each other over 1455 unambiguously aligned bases. The five strains were identified as Prevotella oral taxon 302 in the HOMD. The most closely related species with a validly published name was P. tannerae. P. tannerae ATCC 51259T and strain 81/4-12T exhibited 82.2 and 85.6 % sequence identity to the type strain of the type species of Prevotella, Prevotella melaninogenica. This degree of sequence divergence and the clustering observed in Fig. S1 suggest that P. tannerae and related taxa form a novel taxon at the genus level. We therefore propose the novel genus Alloprevotella gen. nov., to accommodate the group of novel strains studied here as the novel species Alloprevotella rava gen. nov., sp. nov., and the previously named Prevotella tannerae as Alloprevotella tannerae gen. nov., comb. nov., with Alloprevotella tannerae as the type species.
Fig. 1.
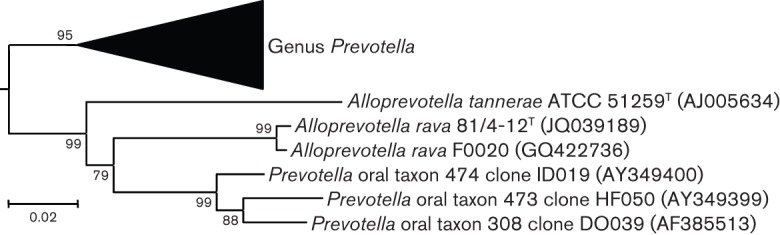
Phylogenetic tree based on 16S rRNA gene sequence comparisons over 1354 aligned bases showing the relationship between Alloprevotella rava sp. nov., Alloprevotella tannerae comb. nov. and the genus Prevotella. The tree was reconstructed using the neighbour-joining method from a distance matrix constructed from aligned sequences using the Jukes–Cantor correction. Numbers represent bootstrap percentages for each branch based on data for 500 trees. Bar, 0.02 nucleotide substitutions per site. A fuller version of this tree is available as Fig. S1.
High-coverage genomes of A. rava F0323 and A. tannerae ATCC 51259T are available as NCBI genome survey sequence accession IDs ACZK00000000 and ACIJ00000000, respectively. The genomes can also be viewed at the HOMD as A. rava oral taxon 302 (www.homd.org/taxon=302) and A. tannerae oral taxon 466 (www.homd.org/taxon=466). A partial genome sequence of about 361 550 bases is also available for A. rava D079N-07 at HOMD.
The two species could be differentiated by the production of H2S and pigmentation by A. tannerae but not by A. rava, and by the test for 6-phospho-β-galactosidase in the Rapid 32A kit, which was positive for A. rava but negative for A. tannerae. A. tannerae also formed smaller colonies than A. rava, and the fatty acid iso-C17 : 0 was a major component in A. rava but was insignificant in A. tannerae. The key phenotypic features of Alloprevotella and selected related genera are shown in Table 2.
Table 2. Differential characteristics of Alloprevotella gen. nov. and selected related genera of the order Bacteroidales.
Genera: 1, Alloprevotella gen. nov.; 2, Prevotella (data from Shah et al., 2010); 3, Paraprevotella (Morotomi et al., 2009); 4, Bacteroides (Sakamoto et al., 2010); 5, Porphyromonas (Sakamoto et al., 2010); 6, Tannerella (Sakamoto et al., 2010).
| Character | 1 | 2 | 3 | 4 | 5 | 6 |
| Habitat | Human oral cavity | GI tract, rumen and vagina of mammals; environment | GI tract of mammals | GI tract of mammals | GI tract, rumen and vagina of mammals | Oral cavity of mammals |
| Metabolic end products from glucose* | A, S | A, S | A, S | A, S | A, B, IV, P, PA, S | A, B, IV, P, PA |
| DNA G+C content (mol%) | 45–47 | 40–52 | 48–49 | 40–49 | 43–46 | 44–48 |
| Principal cell-wall FAME(s)† | ai-C15 : 0 | ai-C15 : 0 | i-C15 : 0, ai-C15 : 0, C18 : 1ω9c | ai-C15 : 0 | i-C15 : 0 | ai-C15 : 0 |
A, Acetate; B, butyrate; IV, isovalerate; P, propionate; PA, phenylacetate; S, succinate.
ai, anteiso-branched; i, iso-branched.
Description of Alloprevotella gen. nov.
Alloprevotella (Al.lo.pre.vo.tel′la. Gr. adj. allos different; N.L. fem. n. Prevotella a bacterial generic name; N.L. fem. n. Alloprevotella organism different from, but related to, the genus Prevotella).
Cells are obligately anaerobic, non-motile, Gram-negative bacilli. Strains are weakly to moderately saccharolytic and produce acetic and succinic acids as end products of fermentation. Gelatin is hydrolysed but not arginine or urea. Nitrate is not reduced and there is no growth in 20 % bile. The principal cellular long-chain fatty acids are anteiso-C15 : 0, iso-C15 : 0, C16 : 0 and iso-C17 : 0 3-OH. The G+C content of the DNA is 45–47 mol%. The type species is Alloprevotella tannerae (Moore et al. 1994).
Description of Alloprevotella tannerae (Moore et al. 1994) comb. nov.
Alloprevotella tannerae (tan′ne.rae. N.L. gen. fem. n. tannerae of Tanner, in honour of Anne C. R. Tanner, a US microbiologist).
Basonym: Prevotella tannerae Moore et al. 1994.
Characteristics are as described previously for P. tannerae by Moore et al. (1994), with the addition of positive reactions in the Rapid ID 32A panel for α-galactosidase, β-galactosidase, α-glucosidase, α-fucosidase, raffinose fermentation, alkaline phosphatase, leucyl glycine arylamidase, alanine arylamidase and glutamyl glutamic acid arylamidase and variable reactions for N-acetyl-β-glucosaminidase and mannose fermentation, resulting in a profile of 4507/4 4402 22. Hydrolysis of arginine and urea is negative.
The type strain is strain ATCC 51259T = CCUG 34292T = CIP 104476T = NCTC 13073T.
Description of Alloprevotella rava sp. nov.
Alloprevotella rava (ra′va. L. fem. adj. rava grey, referring to the colour of colonies of the organism growing on blood-containing agar media).
The description is based on five strains isolated from dental plaque. Cells are obligately anaerobic, non-motile, Gram-negative bacilli, 0.5–0.6 µm wide by 1.0–5.0 µm long, that are arranged singly or in short chains, with occasional cells up to 10 µm long After 3 days of incubation on FAA plates, colonies are 0.8–1.4 mm in diameter, circular, entire, convex, grey and opaque with an internal appearance of concentric rings of opacity and translucency. The optimum temperature for growth is 35 °C, with a range of 30–42 °C. The optimum pH for growth is pH 7, with marginal growth at pH 6 and no growth at pH 5 or 8. Growth in broth media is variable and produces a light or moderate turbidity. Cells are weakly to moderately saccharolytic and ferment fructose, glucose, lactose, maltose and mannose; arabinose, mannitol, melezitose, melibiose, raffinose, rhamnose, salicin, sorbitol, sucrose, trehalose and xylose are not fermented. Moderate amounts of acetic acid and major amounts of succinic acid are produced as end products of metabolism in PYG broth. Trace to minor amounts of isovaleric acid are also produced as a metabolic by-product in PY broth. Gelatin is hydrolysed; arginine, aesculin and urea are not hydrolysed. Indole, catalase and H2S are not produced and nitrate is not reduced. There is no growth in 20 % bile. Strains have positive enzymic reactions in the Rapid ID 32A panel for α-galactosidase, β-galactosidase, 6-phospho-β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-fucosidase, mannose fermentation, raffinose fermentation, alkaline phosphatase, leucyl glycine arylamidase and alanine arylamidase, with weak and variable reactions for glutamyl glutamic acid arylamidase, resulting in a profile of 4707 4402 20/2. The species is oral taxon 302 in the Human Oral Microbiome Database. The DNA G+C content of the type strain is 47 mol%.
The type strain is 81/4-12T ( = DSM 22548T = CCUG 58091T), isolated from the human oral cavity.
Acknowledgements
This research was supported by a grant from the Guy’s and St Thomas’ Charity (ref. R050724). The authors acknowledge financial support from the UK Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London. Investigators F. E. D., A. C. R. T. and W. G. W. were supported in part by National Institute for Dental Research grant DE016937.
Footnotes
A supplementary figure is available with the online version of this paper.
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Chen T., Yu W. H., Izard J., Baranova O. V., Lakshmanan A., Dewhirst F. E. (2010). The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010, baq013 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C., Yu W. H., Lakshmanan A., Wade W. G. (2010). The human oral microbiome. J Bacteriol 192, 5002–5017 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes J., Sutcliffe I., Tanner A. C., Wade W. G. (2005). Prevotella marshii sp. nov. and Prevotella baroniae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 55, 1551–1555 10.1099/ijs.0.63634-0 [DOI] [PubMed] [Google Scholar]
- Hall T. (2004). BioEdit: Biological Sequence Alignment Editor for Win95/98/NT/2K/XP. Carlsbad, CA: Ibis Biosciences [Google Scholar]
- Holdeman L. V., Cato E. P., Moore W. E. C. (1977). Anaerobe Laboratory Manual, 4th edn Blacksburg, VA: Virginia Polytechnic Institute and State University [Google Scholar]
- Jousimies-Somer H. R., Summanen P., Citron D. M., Baron E.-J., Wexler H. M., Finegold S. M. (2002). Wadsworth-KTL Anaerobic Bacteriology Manual. Belmont, CA: Star Publishing Company [Google Scholar]
- Kononen E., Wade W. G., Citron D. M. (2010). Bacteroides, Porphyromonas, Prevotella, Fusobacterium, and other anaerobic Gram-negative rods. In Manual of Clinical Microbiology, 10th edn, pp. 858–880 Edited by Versalovic J., Carroll K. C., Funke G., Jorgensen J. H., Landry M. L., Warnock D. W. Washington, DC: American Society for Microbiology [Google Scholar]
- Kuykendall L. D., Roy M. A., O’Neill J. J., Devine T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38, 358–361 10.1099/00207713-38-4-358 [DOI] [Google Scholar]
- Miller L. T. (1982). Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16, 584–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. V., Johnson J. L., Moore W. E. (1994). Descriptions of Prevotella tannerae sp. nov. and Prevotella enoeca sp. nov. from the human gingival crevice and emendation of the description of Prevotella zoogleoformans. Int J Syst Bacteriol 44, 599–602 10.1099/00207713-44-4-599 [DOI] [PubMed] [Google Scholar]
- Morotomi M., Nagai F., Sakon H., Tanaka R. (2009). Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’ isolated from human faeces. Int J Syst Evol Microbiol 59, 1895–1900 10.1099/ijs.0.008169-0 [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Tanner A. C. R., Benno Y. (2010). Genus VII. Tannerella. In Bergey’s Manual of Systematic Bacteriology, 2nd edn, vol. 4, pp. 78–82 Edited by Krieg N. R., Ludwig W., Whitman W. B., Hedlund B. P., Paster B. J., Staley J. T., Ward N., Brown D., Parte A. New York: Springer [Google Scholar]
- Shah H. N., Chattaway M. A., Rajakurana L., Gharbia S. E. (2010). Genus I. Prevotella. In Bergey’s Manual of Systematic Bacteriology, 2nd edn, vol. 4, pp. 86–102 Edited by Krieg N. R., Ludwig W., Whitman W. B., Hedlund B. P., Paster B. J., Staley J. T., Ward N., Brown D., Parte A. New York: Springer [Google Scholar]
- Wade W. G., Downes J., Dymock D., Hiom S. J., Weightman A. J., Dewhirst F. E., Paster B. J., Tzellas N., Coleman B. (1999). The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49, 595–600 10.1099/00207713-49-2-595 [DOI] [PubMed] [Google Scholar]


