Abstract
A polyphasic analysis was undertaken of seven independent isolates of Gram-negative cocci collected from pathological clinical samples from New York, Louisiana, Florida and Illinois and healthy subgingival plaque from a patient in Virginia, USA. The 16S rRNA gene sequence similarity among these isolates was 99.7–100 %, and the closest species with a validly published name was Neisseria lactamica (96.9 % similarity to the type strain). DNA–DNA hybridization confirmed that these isolates are of the same species and are distinct from their nearest phylogenetic neighbour, N. lactamica. Phylogenetic analysis of 16S and 23S rRNA gene sequences indicated that the novel species belongs in the genus Neisseria. The predominant cellular fatty acids were C16 : 0, summed feature 3 (C16 : 1ω7c and/or iso-C15 : 0 2-OH) and C18 : 1ω7c. The cellular fatty acid profile, together with other phenotypic characters, further supports the inclusion of the novel species in the genus Neisseria. The name Neisseria oralis sp. nov. (type strain 6332T = DSM 25276T = LMG 26725T) is proposed.
In 2005, the Wadsworth Center Bacteriology Laboratory received two independent isolates of bacteria from clinical specimens as part of their reference testing for New York State laboratories (designated 6332T and 8261) that had identical nearly full-length 16S rRNA gene sequences. The most closely related species, based on sequence similarity, was Neisseria lactamica (96.9 % similarity to the type strain returned by EzTaxon; Chun et al., 2007), suggesting that the isolates may represent a novel species (Tindall et al., 2010). A blast search of GenBank revealed an additional isolate with 100 % sequence similarity, Neisseria sp. oral taxon 014 strain F0314 (GenBank accession no. GQ131417), which we acquired (Dewhirst et al., 2010). Four additional isolates were acquired from Associated Regional and University Pathologists (ARUP) Laboratories and found to have 99.7–99.9 % 16S rRNA gene sequence similarity to the three other isolates. Suggesting a widespread prevalence for this organism, the GenBank blast search also identified other studies in which the same 16S rRNA gene sequence was detected (sequence similarity >99.6 %) by culture-independent methodologies. The sources of these clones were subgingival plaque (healthy and diseased), healthy skin and respiratory samples from cystic fibrosis patients, (Dewhirst et al., 2010; Grice et al., 2009; Paster et al., 2001; Rylev et al., 2011; Staudinger et al., 2011; van der Gast et al., 2011). In the polyphasic study described here, the seven acquired isolates were found to belong to a single novel species, for which we propose the name Neisseria oralis sp. nov.
The seven isolates were maintained at 37 °C in a 5 % CO2 atmosphere on trypticase soy agar (TSA) plates (Becton Dickinson) supplemented with 5 % sheep blood. Four of the isolates were from blood cultures; one was from urine, one was from paracentesis fluid and one was from subgingival oral biofilm at a healthy site (Table 1). A representative strain, 6332T, was chosen as the type strain.
Table 1. Strains used in this study.
| Strain | Isolation date | Source | GenBank accession no. | |
| 16S rRNA gene | 23S rRNA gene | |||
| 6332T ( = LMG 26725T = DSM 25276T) | July 2005 | Blood, 55-year-old male; Dutchess County, NY, USA | JN104029 | JN104031 |
| 8261 ( = LMG 26726 = DSM 25277) | October 2005 | Blood, 56-year-old female; Saratoga County, NY, USA | JN104030 | JN104032 |
| F0314 ( = 11-24509) | June 1982 | Subgingival oral biofilm, 23-year-old female; Blacksburg, VA, USA | GQ131417 | JN986585 |
| 11-26358 | nk | Urine, 56-year-old; FL, USA | JN986581 | JN986586 |
| 11-26359 | nk | Blood, >89-year-old; OH, USA | JN986582 | JN986587 |
| 11-26360 | nk | Blood, 19-month-old; LA, USA | JN986583 | JN986588 |
| 11-26361 | nk | Paracentesis fluid, 80-year-old; IL, USA | JN986584 | JN986589 |
nk, Not known.
16S rRNA gene sequences (1463–1515 nt) were determined for the isolates as described previously (Dewhirst et al., 2010; Wolfgang et al., 2011); see Table 1 for GenBank accession numbers. Additionally, for strain F0314, the whole-genome shotgun sequence (accession no. ADEA01000039) had been deposited previously at GenBank as part of the Human Microbiome Project (Dewhirst et al., 2010; Nelson et al., 2010).
To identify the type strains most closely related to 6332T for inclusion in our polyphasic analysis, the blast (Altschul et al., 1997) and megablast (Zhang et al., 2000) search programs were used with the 16S rRNA gene sequence against the database of type strains of prokaryotic species with validly published names (Chun et al., 2007). The 50 sequences with the highest scores were then selected for the calculation of pairwise sequence similarity using the global alignment algorithm implemented at the EzTaxon server (http://www.eztaxon.org/; Chun et al., 2007). Five of the most similar type strains by sequence similarity (96.1–96.9 %) were acquired for this study, Neisseria lactamica LMG 26610T, N. animalis LMG 26609T, N. macacae LMG 26611T, N. elongata subsp. elongata LMG 5124T and N. sicca LMG 5290T.
Previous studies indicate that, for organisms with more than 97 % 16S rRNA gene sequence similarity, alternative methods must be employed, such as DNA–DNA hybridization, to determine inclusion with or exclusion from closely related species (Tindall et al., 2010). Although none of the most closely related organisms crossed this threshold, N. lactamica LMG 26610T, with a similarity of 96.9 %, was the closest, and so was selected for DNA–DNA hybridization analysis. DNA was purified as described by Logan et al. (2000) and hybridization was performed using the microplate method described by Ezaki et al. (1989), with modifications described by Willems et al. (2001). Means of two reciprocal hybridization values were determined for all pairwise combinations of strains 6332T, 8261 and 11-26358 and N. lactamica LMG 26610T (Table S1, available in IJSEM Online). Based on a 70 % cut-off value for defining species (Wayne et al., 1987), strains 6332T, 8261 and 11-26358 belong to the same species, and that species is distinct from N. lactamica.
For cellular fatty acid (CFA) analysis, the seven strains of N. oralis sp. nov. and the five most closely related type strains were cultured aerobically for 48 h on trypticase soy broth agar (TSBA) at 37 °C and harvested in late-exponential growth phase. Fatty acid methyl esters were prepared according to the manufacturer’s instructions (Sherlock Microbial Identification Systems; MIDI, Inc.) and analysed on an Agilent Technologies 6890N gas chromatograph. For the type strain 6332T, the predominant CFAs were C16 : 0, summed feature 3 (C16 : 1ω7c and/or iso-C15 : 0 2-OH) and C18 : 1ω7c. These were the same as the predominant CFAs of the most closely related type strains (Table 2). The variation seen among these strains in Table 2 was similar to that seen among the seven strains of N. oralis (Table S2). Hence, the CFA composition is concordant with assigning the novel species to the genus Neisseria, but is not useful in distinguishing among the species within the genus.
Table 2. CFA compositions of Neisseria type strains.
Strains: 1, 6332T; 2, N. elongata subsp. elongata LMG 5124T; 3, N. sicca LMG 5290T; 4, N. animalis LMG 26609T; 5, N. lactamica LMG 26610T; 6, N. macacae LMG 26611T. Values are percentages of total fatty acids, and were determined in this study. Fatty acid methyl esters for which no values were greater than 1 % were omitted. The three most prevalent fatty acids for each strain are in bold. nd, Not detected.
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 |
| Saturated | ||||||
| C12 : 0 | 7.8 | 5.8 | 8.9 | 6.9 | 9.6 | 5.2 |
| C14 : 0 | 3.5 | 3.6 | 2.1 | 3.7 | 5.7 | 3.4 |
| C16 : 0 | 33.7 | 33.7 | 19.4 | 34.7 | 34.3 | 31.4 |
| C17 : 0 | nd | 1.1 | nd | nd | nd | nd |
| C18 : 0 | 0.4 | 0.7 | 0.3 | 1.0 | 0.4 | 0.3 |
| Unsaturated | ||||||
| C16 : 1ω5c | 0.2 | nd | 1.2 | 0.5 | 0.7 | 1.1 |
| C17 : 1ω6c | nd | 0.3 | 0.2 | 1.3 | nd | nd |
| C18 : 1ω7c | 17.4 | 21.4 | 20.8 | 19.0 | 11.2 | 14.6 |
| Hydroxy | ||||||
| C12 : 0 3-OH | 5.3 | 4.2 | 6.1 | 0.3 | 6.1 | 3.3 |
| C16 : 0 2-OH | nd | nd | 1.1 | nd | nd | 1.9 |
| Summed features* | ||||||
| Summed feature 2 | 3.3 | 2.9 | 4.0 | 7.9 | 3.6 | 2.6 |
| Summed feature 3 | 27.5 | 25.4 | 33.8 | 24.2 | 27.6 | 35.3 |
Summed features are groups of two or three fatty acids that cannot be separated by GLC with the MIDI System. Summed feature 2 contained one or more of C12 : 0 ALDE?, C14 : 0 3-OH and iso-C16 : 1 I. Summed feature 3 contained C16 : 1ω7c and/or iso-C15 : 0 2-OH.
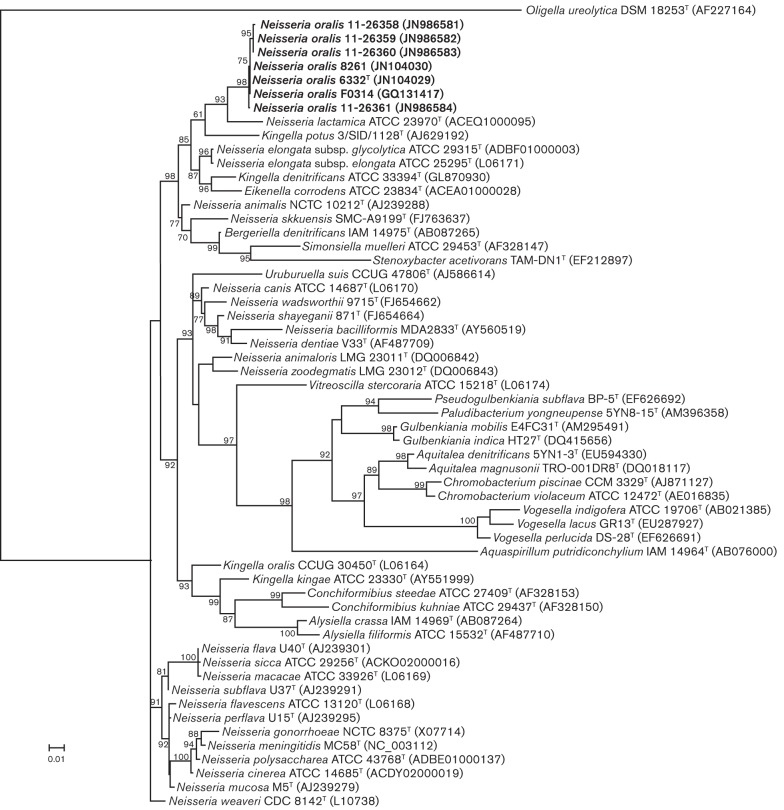
Phylogenetic analysis was performed using nearly full-length 16S rRNA gene sequences from the seven strains of N. oralis and the top 50 blast hits returned for type strain 6332T from the EzTaxon database (Chun et al., 2007). These sequences were aligned using the clustal w utility in mega 4.0.2 (Tamura et al., 2007) and exported to PhyML 3.0 (Guindon & Gascuel, 2003). A rooted maximum-likelihood tree was recreated using the general time reversible model, six substitution rate categories and five random starting trees (Guindon & Gascuel, 2003). Tree reliability was evaluated using the approximate likelihood ratio test (aLRT) (Anisimova & Gascuel, 2006) (Fig. 1). The seven novel strains reside in a well-supported clade that harbours N. lactamica as its closest relative as well as N. elongata, two species of Kingella and the one species of Eikenella. The other most closely related species (based on sequence distance) reside in other, distinct clades (Fig. 1).
Fig. 1.
Maximum-likelihood phylogenetic tree reconstructed from 16S rRNA gene sequences of strains of N. oralis sp. nov. and the 50 closest taxa returned from a blast search of the EzTaxon database. The tree was rooted with Oligella ureolytica DSM 18253T. Branch probability is indicated at nodes and was determined by the aLRT; probabilities below 50 % are not shown. Bar, 0.01 expected changes per site.
To seek further support for the novel species designation, 23S rRNA gene sequences were determined for the seven new isolates as well as the five most closely related type strains. Sequencing was performed as described previously (Wolfgang et al., 2011). Sequence similarity for the seven novel strains ranged from 99.4 to 100 %. A maximum-likelihood tree was reconstructed from sequences generated in this study as well as sequences from closely related species for which data were available at GenBank (Fig. S1) (note that the 23S rRNA gene sequence was not available for the type strain of some species; e.g. the sequence of strain MS11 was used for Neisseria gonorrhoeae). The seven isolates of N. oralis formed a clade that is distinct from all other Neisseria species, further supporting the novel species designation.
As in other studies, our phylogenetic analysis using the 16S rRNA gene sequence reveals that the genus Neisseria is polyphyletic (Vandamme et al., 2006; Wolfgang et al., 2011). We found that N. oralis resides in a well-supported clade that harbours two additional species of Neisseria, two species of Kingella and Eikenella corrodens (Fig. 1). This clade is distinct from the clade that harbours the type species of the genus, N. gonorrhoeae. As such, it is necessary to identify additional characters that support or reject the inclusion of the novel species in closely related genera. Characteristics that support inclusion of our isolates in the genus Neisseria are the formation of Gram-negative-staining cocci, the presence of catalase and oxidase activities, the absence of motility and the CFA profile (Tønjum, 2005; Weyant et al., 1984). Characteristics that support exclusion from closely related genera are the absence of catalase activity in the genera Bergeriella, Eikenella and Kingella, the formation of long filaments and gliding motility in the genera Alysiella, Simonsiella and Conchiformibium, which additionally reside in distinct clades (Tønjum, 2005), and the fact that the single species of the genus Uruburuella falls in a separate clade and has additional characters that distinguish it from our isolates (Vela et al., 2005).
Characteristics that aid in distinguishing all seven strains of N. oralis from the closely related type strains examined in this study are the ability to reduce nitrate, the absence of acid phosphatase activity and acid production from d-glucose, maltose and sucrose (Tables 3 and S3) [for methods, see Forbes et al. (1998) and Kohlerschmidt et al. (2009)]. Other species, not analysed in this study, that may also be excluded on the basis of the absence of nitrate reduction are N. gonorrhoeae, N. cinerea, N. dentiae, N. flavescens, N. meningitidis, N. subflava, N. weaveri and N. polysaccharea (Janda & Gaydos, 2007; Tønjum, 2005). For the remaining taxa that reduce nitrate, with the exception of Neisseria mucosa and N. elongata subsp. nitroreducens, the absence of acid production from glucose, maltose and/or sucrose is diagnostic. For N. mucosa and N. elongata subsp. nitroreducens, the presence of large mucoid colonies or rod-shaped cells, respectively, are diagnostic.
Table 3. Characteristics that distinguish N. oralis sp. nov. from closely related species.
Strains/species: 1, N. oralis sp. nov. 6332T; 2, N. elongata subsp. elongata LMG 5124T; 3, N. sicca LMG 5290T; 4, N. animalis LMG 26609T; 5, N. lactamica LMG 26610T; 6, N. macacae LMG 26611T; 7, N. gonorrhoeae [data from Janda & Gaydos (2007) and the bioMérieux package insert]. Data were obtained in this study unless indicated otherwise. nd, No data available. Characteristics listed for 6332T were the same for all seven strains of N. oralis sp. nov. (see Table S3).
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Acid production (peptone water) from: | |||||||
| d-Glucose | + | − | − | − | + | + | + |
| Maltose | + | + | + | − | + | + | − |
| Nitrate reduction | + | − | − | − | − | − | − |
| Catalase reaction | + | + | + | + | + | − | + |
| Acid production (API NH) from: | |||||||
| d-Glucose | + | − | + | − | + | − | + |
| Maltose | + | − | − | − | − | − | − |
| Sucrose | + | − | − | + | − | − | − |
| Enzyme activity (API NH) | |||||||
| Proline 4-methoxy-β-naphthylamide | + | + | + | − | + | + | + |
| γ-Glutamyl 4-methoxy-β-naphthylamide | + | − | − | − | − | + | − |
| Enzyme activity (API ZYM) | |||||||
| Esterase (C4) | − | + | − | + | + | + | nd |
| Acid phosphatase | − | + | + | + | + | + | nd |
Based on the 16S and 23S rRNA gene sequence comparisons, DNA–DNA hybridization, CFA composition and phenotypic and phylogenetic analyses, we propose that the seven novel strains (Table 1) represent a single novel species, Neisseria oralis sp. nov.
Description of Neisseria oralis sp. nov.
Neisseria oralis (o.ra′lis. N.L. fem. adj. oralis of the mouth, the source of the first isolate).
Colonies are small, circular, entire, raised, moist, yellow, weakly α-haemolytic (except 11-26358, which is β-haemolytic) and 1–1.5 mm in diameter after 48 h of growth at 37 °C in 5 % CO2. Facultative anaerobe. Growth is observed at 28 and 42 °C, with no growth at 10 °C. No growth is observed on MacConkey agar after 5 days. Cells are Gram-negative, 0.5 µm in diameter, may be present in chains and are non-motile. Produces catalase and cytochrome oxidase and reduces nitrate to nitrite. Negative for acid production using Hugh & Leifson O-F base with 1 % d-mannitol and d-xylose and displays interstrain variability (see Table S3 for details) for d-glucose (6/7 positive; type strain positive), maltose (6/7 positive; type strain positive), lactose (2/7 positive; type strain negative) and sucrose (5/7 positive; type strain positive). Positive for acid production in peptone water base with 1 % d-glucose and maltose and negative for acid production from adonitol, l-arabinose, dulcitol, inositol, d-mannitol, raffinose, d-rhamnose, d-salicin, d-sorbitol and d-xylose; displays interstrain variability for acid production from lactose (3/7 positive; type strain negative) and sucrose (6/7 positive; type strain positive). Negative for utilization of Simmons’ citrate, hydrolysis of aesculin, urea and gelatin, indole production, decarboxylation of arginine, lysine and ornithine using Moeller’s decarboxylase medium and production of H2S in triple-sugar iron agar [for methods, see Forbes et al. (1998) and Kohlerschmidt et al. (2009)]. Using the API NH system (bioMérieux), positive for acidification of d-glucose, maltose and sucrose and for the presence of proline arylamidase and γ-glutamyl transferase activities and negative for penicillinase, lipase, alkaline phosphatase and indole production; displays interstrain variability for acidification of d-fructose (6/7 positive; type strain negative), ornithine decarboxylase (1/7 positive; type strain negative), urease (2/7 positive; type strain negative) and β-galactosidase (2/7 positive; type strain negative). Using the API ZYM system (bioMérieux), displays interstrain variability for leucine arylamidase (3/7 positive; type strain positive) and β-galactosidase (2/7 positive; type strain negative) and is negative for all other activities tested in the kit. The DNA G+C content of the type strain is 52.6 mol%. The predominant CFAs are C16 : 0, summed feature 3 (C16 : 1ω7c and/or iso-C15 : 0 2-OH) and C18 : 1ω7c.
The type strain, 6332T ( = LMG 26725T = DSM 25276T), was isolated from whole blood of a 55-year-old male in New York, USA. Details of other strains are given in Table 1.
Acknowledgements
The authors wish to thank the Wadsworth Center Applied Genomic Technologies Core and the Broad Institute of MIT and Harvard for sequencing, the Wadsworth Center Orphan Bacterium Collection for curation of the novel strains described in this study, Margo Cnockaert for assistance with the DNA hybridization studies and Dr Andrea Habura for helpful discussions relating to the phylogenetic analysis of the novel species. A. C. received a grant from the research fund of University College Ghent. F. E. D. and J. I. were supported by NIDCR grant DE016937.
Abbreviations:
- CFA
cellular fatty acid
Footnotes
Three supplementary tables and a supplementary figure are available with the online version of this paper.
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55, 539–552 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Chun J., Lee J. H., Jung Y., Kim M., Kim S., Kim B. K., Lim Y. W. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57, 2259–2261 10.1099/ijs.0.64915-0 [DOI] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C., Yu W. H., Lakshmanan A., Wade W. G. (2010). The human oral microbiome. J Bacteriol 192, 5002–5017 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki T., Hashimoto Y., Yabuuchi E. (1989). Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39, 224–229 10.1099/00207713-39-3-224 [DOI] [Google Scholar]
- Forbes B. A., Sahm D. F., Weissfeld A. S. (1998). Overview of bacterial identification methods and strategies. In Bailey and Scott’s Diagnostic Microbiology, pp. 424–446 Edited by Roche J. St Louis, MO: Mosby [Google Scholar]
- Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C., Bouffard G. G., Blakesley R. W., Murray P. R., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Janda W. M., Gaydos C. A. (2007). Neisseria. In Manual of Clinical Microbiology, 9th edn, pp. 601–620 Edited by Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. Washington, DC: American Society for Microbiology [Google Scholar]
- Kohlerschmidt D. J., Musser K. A., Dumas N. B. (2009). Identification of aerobic Gram-negative bacteria. In Practical Handbook of Microbiology, 2nd edn, pp. 67–80 Edited by Goldman E., Green L. H. Boca Raton, FL: CRC Press [Google Scholar]
- Logan N. A., Lebbe L., Hoste B., Goris J., Forsyth G., Heyndrickx M., Murray B. L., Syme N., Wynn-Williams D. D., De Vos P. (2000). Aerobic endospore-forming bacteria from geothermal environments in northern Victoria Land, Antarctica, and Candlemas Island, South Sandwich archipelago, with the proposal of Bacillus fumarioli sp. nov. Int J Syst Evol Microbiol 50, 1741–1753 [DOI] [PubMed] [Google Scholar]
- Nelson K. E., Weinstock G. M., Highlander S. K., Worley K. C., Creasy H. H., Wortman J. R., Rusch D. B., Mitreva M., Sodergren E., et al. (2010). A catalog of reference genomes from the human microbiome. Science 328, 994–999 10.1126/science.1183605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., Boches S. K., Galvin J. L., Ericson R. E., Lau C. N., Levanos V. A., Sahasrabudhe A., Dewhirst F. E. (2001). Bacterial diversity in human subgingival plaque. J Bacteriol 183, 3770–3783 10.1128/JB.183.12.3770-3783.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylev M., Bek-Thomsen M., Reinholdt J., Ennibi O. K., Kilian M. (2011). Microbiological and immunological characteristics of young Moroccan patients with aggressive periodontitis with and without detectable Aggregatibacter actinomycetemcomitans JP2 infection. Mol Oral Microbiol 26, 35–51 10.1111/j.2041-1014.2010.00593.x [DOI] [PubMed] [Google Scholar]
- Staudinger T., Pipal A., Redl B. (2011). Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up. J Appl Microbiol 110, 1381–1389 10.1111/j.1365-2672.2011.04991.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). mega4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tindall B. J., Rosselló-Móra R., Busse H. J., Ludwig W., Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60, 249–266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- Tønjum T. (2005). Family I. Neisseriaceae Prevot 1933, 119AL emend. Dewhirst, Paster and Bright 1989, 265. In Bergey’s Manual of Systematic Bacteriology, 2nd edn, vol. 2C, pp. 775–776 Edited by Garrity G. M., Brenner D. J., Krieg N. R., Staley J. T. New York: Springer [Google Scholar]
- van der Gast C. J., Walker A. W., Stressmann F. A., Rogers G. B., Scott P., Daniels T. W., Carroll M. P., Parkhill J., Bruce K. D. (2011). Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5, 780–791 10.1038/ismej.2010.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Holmes B., Bercovier H., Coenye T. (2006). Classification of Centers for Disease Control Group Eugonic Fermenter (EF)-4a and EF-4b as Neisseria animaloris sp. nov. and Neisseria zoodegmatis sp. nov., respectively. Int J Syst Evol Microbiol 56, 1801–1805 10.1099/ijs.0.64142-0 [DOI] [PubMed] [Google Scholar]
- Vela A. I., Collins M. D., Lawson P. A., García N., Domínguez L., Fernández-Garayzábal J. F. (2005). Uruburuella suis gen. nov., sp. nov., isolated from clinical specimens of pigs. Int J Syst Evol Microbiol 55, 643–647 10.1099/ijs.0.63346-0 [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Brenner D. J., Colwell R. R., Grimont P. A. D., Kandler O., Krichevsky M. I., Moore L. H., Moore W. E. C., Murray R. G. E., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37, 463–464 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- Weyant R. S., Moss C. W., Weaver R. E., Hollis D. G., Jordan J. G., Cook E. C., Daneshvar M. I. (1984). Identification of Unusual Pathogenic Gram-Negative Aerobic and Facultatively Anaerobic Bacteria, 2nd edn Baltimore: Williams & Wilkins [Google Scholar]
- Willems A., Doignon-Bourcier F., Goris J., Coopman R., de Lajudie P., De Vos P., Gillis M. (2001). DNA–DNA hybridization study of Bradyrhizobium strains. Int J Syst Evol Microbiol 51, 1315–1322 [DOI] [PubMed] [Google Scholar]
- Wolfgang W. J., Carpenter A. N., Cole J. A., Gronow S., Habura A., Jose S., Nazarian E. J., Kohlerschmidt D. J., Limberger R., et al. (2011). Neisseria wadsworthii sp. nov. and Neisseria shayeganii sp. nov., isolated from clinical specimens. Int J Syst Evol Microbiol 61, 91–98 10.1099/ijs.0.022426-0 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W. (2000). A greedy algorithm for aligning DNA sequences. J Comput Biol 7, 203–214 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]