Abstract
Cholera, caused by Vibrio cholerae, results in significant morbidity and mortality worldwide, including Thailand. Representative V. cholerae strains associated with endemic cholera (n = 32), including strains (n = 3) from surface water sources, in Khon Kaen, Thailand (2003–2011), were subjected to microbiological, molecular and phylogenetic analyses. According to phenotypic and related genetic data, all tested V. cholerae strains belonged to serogroup O1, biotype El Tor (ET), Inaba (IN) or Ogawa (OG). All of the strains were sensitive to gentamicin and ciprofloxacin, while multidrug-resistant (MDR) strains showing resistance to erythromycin, tetracycline, trimethoprim/sulfamethoxazole and ampicillin were predominant in 2007. V. cholerae strains isolated before and after 2007 were non-MDR. All except six diarrhoeal strains possessed ctxA and ctxB genes and were toxigenic altered ET, confirmed by MAMA-PCR and DNA sequencing. Year-wise data revealed that V. cholerae INET strains isolated between 2003 and 2004, plus one strain isolated in 2007, lacked the RS1 sequence (rstC) and toxin-linked cryptic plasmid (TLC)-specific genetic marker, but possessed CTXCL prophage genes ctxBCL and rstRCL. A sharp genetic transition was noted, namely the majority of V. cholerae strains in 2007 and all in 2010 and 2011 were not repressor genotype rstRCL but instead were rstRET, and all ctx+ strains possessed RS1 and TLC-specific genetic markers. DNA sequencing data revealed that strains isolated since 2007 had a mutation in the tcpA gene at amino acid position 64 (N→S). Four clonal types, mostly of environmental origin, including subtypes, reflected genetic diversity, while distinct signatures were observed for clonally related, altered ET from Thailand, Vietnam and Bangladesh, confirmed by distinct subclustering patterns observed in the PFGE (NotI)-based dendrogram, suggesting that endemic cholera is caused by V. cholerae indigenous to Khon Kaen.
Introduction
Cholera is a severe form of acute diarrhoea, caused by the gamma-proteobacterium Vibrio cholerae. V. cholerae comprises more than 200 ‘O’ serogroups even though the disease historically has been considered to be caused only by V. cholerae serogroups O1 and O139 (Alam et al., 2007). Serogroup O1 has two biotypes, Classical (CL) and El Tor (ET), of which the former is associated with the first six pandemics starting in 1817 before the latter was isolated at the beginning of the current ongoing seventh cholera pandemic in the 1960s (Kaper et al., 1995). The CL biotype differs from the ET biotype in some phenotypic traits, primarily haemolysis of sheep erythrocytes, agglutination of chicken erythrocytes, Voges–Proskauer reaction, sensitivity to polymyxin B and sensitivity to specific phages (Kaper et al., 1995). Additionally, genotypic tests have been used to determine biotypes of V. cholerae, such as presence of tcpA (encoding toxin co-regulated pilin A), ctxB (cholera toxin B), rstR (repeat sequence transcriptional regulator) and presence or absence of rtxC (repeat in the toxin gene), and epitope analysis of one of the two subunits of cholera toxin (CTB epityping) has been performed (Safa et al., 2010). The VSP-I and -II gene clusters are unique to ET strains of the seventh pandemic, in that they are absent from both the pre-seventh pandemic ET strains and CL biotype strains (Dziejman et al., 2002). The seventh and current cholera pandemic is ascribed to the ET biotype, while the fifth and sixth pandemics are associated with the CL biotype (Alam et al., 2010).
A significant and relatively recent development is the emergence of altered ET V. cholerae strains harbouring cholera toxin (CTX) and certain related traits of the CL biotype, which was first isolated in Asia in 2001, displacing the prototype ET in frequency of isolation from its Asian habitats (Nair et al., 2006). The ET biotype associated with cholera in Africa in the 1970s was similarly replaced by an altered ET (Morita et al., 2010). Results of genetic analysis revealed that these different ET strains vary in type of CTX prophage and flanking RS1 elements in the genome of the toxigenic strains. V. cholerae altered ET strains in Africa were shown to be different from those in Asia since they harboured the entire CL CTX prophage in an ET biotype background (Ansaruzzaman et al., 2004). According to a recent report, altered ET strains were predominant among CL and ET biotype progenitors associated with endemic cholera in Mexico between 1991 and 1997 (Alam et al., 2010). Such altered ET strains were found to cause a more severe disease in Asia (Siddique et al., 2010), and are being reported globally (Chin et al., 2011; Na-Ubol et al., 2011; Okada et al., 2010; Goel et al., 2011).
Cholera is endemic and a major public health concern in Thailand (Bureau of Epidemiology, 2010), especially for lower socio-economic groups in the north-east region (Tangkanakul & Hanpanjakit, 2007). In Thailand, endemic cholera causes significant morbidity and mortality each year. For example, 986 cases of cholera were reported by the Department of Disease Control, Ministry of Public Health of Thailand in 2007, of which seven cases were fatal (Bureau of Epidemiology, 2010). At the beginning of 2010, outbreaks of cholera were reported in 15 provinces, including Khon Kaen, the largest provincial city in north-east Thailand. Although treatment for cholera includes a 3 day course of effective antibiotics and rehydration therapy, a progressive increase in drug resistance makes cholera treatment very difficult, not only in Thailand but worldwide (Ang et al., 2010; Jain et al., 2011; Kumar et al., 2010a; Quilici et al., 2010). A recent study reported limited phenotypic, genotypic and virulence characteristics of V. cholerae O1 strains associated with endemic cholera in Thailand (Na-Ubol et al., 2011; Okada et al., 2012). However, the study did not address the antibiotic response of V. cholerae or the source of cholera. To understand drug response and molecular and phylogenetic trends of V. cholerae associated with endemic cholera in north-eastern Thailand, representative V. cholerae strains (n = 35) were isolated between 2003 and 2011 from diarrhoea patients (n = 32) and environmental (n = 3) sources in Khon Kaen and all isolates were subjected to microbiological, molecular and phylogenetic analyses.
Methods
Bacterial strains.
A total of 35 V. cholerae O1 strains, 32 of which were associated with cholera and 3 from natural surface water sources in Khon Kaen, Thailand, between 2003 and 2011, were examined for phenotypic characteristics, namely antimicrobial response and virulence, and for molecular traits, including phylogenetic characteristics. V. cholerae O1 strains N16961 (ET biotype) and O395 (CL biotype) served as controls. All were cultured overnight on selective bacteriological media, including taurocholate tellurite gelatin (TTGA) and thiosulfate citrate bile salts sucrose agar. V. cholerae colonies were confirmed using a combination of biochemical, serological and molecular methods, as described previously (Alam et al., 2007).
Antibiotic susceptibility.
Susceptibility to antibiotics was determined by disc diffusion, as described by Bauer et al. (1966) and the Clinical and Laboratory Standards Institute (CLSI, 2010), using commercial antibiotic discs. Six antibiotics (Oxoid) were employed: erythromycin (15 µg), gentamicin (10 µg), trimethoprim/sulfamethoxazole (30 µg), tetracycline (30 µg), ampicillin (30 µg) and ciprofloxacin (5 µg). The resistance or susceptibility profiles of the isolates were determined by measuring the inhibitory zone and comparing it with an interpretative chart to determine sensitivity to the antibiotics.
Serogroup and biotype.
Serogroups of the V. cholerae isolates identified using biochemical and molecular methods were confirmed by slide agglutination using specific polyvalent antisera for V. cholerae O1 and O139, followed by screening with a monoclonal antibody specific for each serogroup (Alam et al., 2007). Biotyping primarily involved selective phenotypic tests, including chicken erythrocyte agglutination, and sensitivity to polymyxin B, Mukherjee CL phage IV and Mukherjee ET phage V (Kaper et al., 1995). Serogrouping and biotyping were further confirmed by PCR as described below.
Genomic DNA preparation.
Genomic DNA extraction was done by described methods (Nusrin et al., 2009).
PCR assays for serogroup and biotype determination.
Subtypes of all strains were reconfirmed using V. cholerae species-specific ompW PCR (Nandi et al., 2000). Serogroups were reconfirmed using multiplex PCR targeted at O1- (wbeO1) and O139- (wbfO139) specific O biosynthetic genes and the CTX gene (ctxA) (Hoshino et al., 1998). Biotype-specific characteristics were determined using PCR assays targeted to tcpA (CL and ET) (Rivera et al., 2001), rstR, encoding phage transcriptional regulator (Mwansa et al., 2007), presence of the repeat in toxin (rtxC) (Chow et al., 2001), rstC, encoding an anti-repressor protein, and tlc, encoding the toxin-linked cryptic plasmid (O’Shea et al., 2004).
Primers used in this study are shown in Table 1.
Table 1. PCR primers used in this study.
| Primer | Sequence (5′–3′) | Targeted gene | Annealing temp. (°C) | Amplicon size (bp) | Reference |
| ompW F | CAC CAA GAA GGT GAC TTT ATT GTG | ompW | 64 | 304 | Nandi et al. (2000) |
| ompW R | GGT TTG TCG AAT TAG CTT CAC C | ||||
| rfbO1 F | GTT TCA CTG AAC AGA TGG G | rfbO1 | 55 | 192 | Hoshino et al. (1998) |
| rfbO1 R | GGT CAT CTG TAA GTA CAA C | ||||
| rfbO139 F | AGC CTC TTT ATT ACG GGT GG | rfbO139 | 55 | 449 | Hoshino et al. (1998) |
| rfbO139 R | GTC AAA CCC GAT CGT AAA GG | ||||
| ctxA F | ACA GAG TGA GTA CTT TGA CC | ctxA | 55 | 308 | Hoshino et al. (1998) |
| ctxA R | ATA CCA TCC ATA TAT TTG GGA G | ||||
| tcpA ET R | CGA AAG CAC CTT CTT TCA CAC GTT G | tcpA ET | 60 | 453 | Rivera et al. (2001) |
| tcpA F | CAC GAT AAG AAA ACC GGT CAA GAG | ||||
| tcpA class R | TTA CCA AAT GCA ACG CCG AAT G | tcpA CL | 60 | 620 | Rivera et al. (2001) |
| tcpA F | CAC GAT AAG AAA ACC GGT CAA GAG | ||||
| MAMA ElTor F | ACT ATC TTC AGC ATA TGC ACA TGG | ctxB ET | 55 | 186 | Morita et al. (2008) |
| MAMA ElTor R | CCT GGT ACT TCT ACT TGA AAC A | ||||
| MAMA class F | ACT ATC TTC AGC ATA TGC ACA TGG | ctxB CL | 55 | 186 | Morita et al. (2008) |
| MAMA class R | CCT GGT ACT TCT ACT TGA AAC G | ||||
| rstR1 F | CTT CTC ATC AGC AAA GCC TCC ATC | rstR CL | 50 | 500 | Mwansa et al. (2007) |
| rstR3A R | TCG AGT TGT AAT TCA TCA AGA GTG | ||||
| rstR2 F | GCA CCA TGA TTT AAG ATG CTC | rstR ET | 50 | 500 | Mwansa et al. (2007) |
| rstR3A R | TCG AGT TGT AAT TCA TCA AGA GTG | ||||
| rtxC F | CGA CGA AGA TCA TTG ACG AC | rtxC | 55/56 | 265 | Chow et al. (2001) |
| rtxC R | CAT CGT CGT TAT GTG GTT GC | ||||
| rstC1 | AAC AGC TAC GGG CTT ATT C | rstC | 52.4 | 238 | O’Shea et al. (2004) |
| rstC2 | TGA GTT GCG GAT TTA GGC | ||||
| tlc3 | GGG AAT GTT GAG TTC TCA GTG | tlc | 55.5 | 1548 | O’Shea et al. (2004) |
| tlc4 | GTT GCG AAG TGG ATT TTG TG | ||||
| tcpA-F | ATG CAA TTA TTA AAA CAG CTT TTT AAG | tcpA | 59 | 675 | Kumar et al. (2010b) |
| tcpA-R | TTA GCT GTT ACC AAA TGC AAC AG | ||||
| CTX7 | GGT TGC TTC TCA TCA TCG AAC CAC | ctxB | 55 | 460 | Olsvik et al. (1993) |
| CTX9B | GAT ACA CAT AAT AGA ATT AAG GAT |
Determination of ctxB genotype by MAMA-PCR.
The mismatch amplification mutation assay (MAMA) was recently developed to detect sequence polymorphism between the CL and ET ctxB genes (ctxBCL and ctxBET, respectively) by focusing on nucleotide position 203 of the ctxB gene (Morita et al., 2008). MAMA-PCR was used to test for presence of ctxB specific for CL and ET biotypes. A conserved forward primer (Fw-con, 5′-ACTATCTTCAGCATATGCACATGG-3′) and two allele-specific polymorphism detection primers, Rv-cl (5′-CCTGGTACTTCTACTTGAAACG-3′) and Rv-et (5′-CCTGGTACTTCTACTTGAAACA-3′), were used. PCR conditions were as follows: initial denaturation at 96 °C for 2 min, 25 cycles of denaturation at 96 °C for 10 s, annealing at 50 °C for 10 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 2 min. V. cholerae O1 CL O395 and ET N16961 served as reference strains.
Nucleotide sequencing of ctxB and tcpA.
The ctxB and tcpA genes of representative V. cholerae O1 strains from each year (2003–2011) were sequenced following conditions described elsewhere (Olsvik et al., 1993). PCR amplification of ctxB and tcpA was performed in a 25 µl reaction mixture in an automated Peltier thermal cycler (PTC-200, M. J. Research). PCR products were purified with a Microcon centrifugal filter device (Millipore) and sequenced using an ABI PRISM Big Dye Terminator Cycle Sequencing Reaction kit (Applied Biosystems) on an ABI PRISM 310 automated sequencer (Applied Biosystems). The sequences of the respective genes for other V. cholerae O1 ET and CL strains listed in Fig. 1 were retrieved from GenBank (accession numbers NC_002505, U25679, EU496278). The deduced amino acid sequences of the respective genes from all strains were aligned using clustal w.
Fig. 1.
clustal w alignment of amino acids encoded by the tcpA gene of the test V. cholerae O1 strains with the sequence from the ET reference strain N16961. Dots indicate identical amino acids. The amino acid sequences of TcpA of V. cholerae O1 strains isolated in Thailand between 2003 and 2004 showed 100 % identity with those of TcpA of reference strain N16961. Strains isolated in 2007 and thereafter showed a mutation at amino acid position 64 (N→S) of the tcpA gene.
PFGE.
Whole agarose-embedded genomic DNA from the V. cholerae isolates was prepared. PFGE was carried out using a contour-clamped homogeneous electrical field (CHEF-DRII) apparatus (Bio-Rad), according to procedures described elsewhere (Cameron et al., 1994). Conditions for separation were as follows: 2 to 10 s for 13 h, followed by 20 to 25 s for 6 h. An electrical field of 6 V cm−1 was applied at an included field angle of 120°. Genomic DNA of the test strains was digested using NotI (Gibco-BRL), and Salmonella enterica serovar Braenderup was digested using XbaI, with fragments employed as molecular size markers. Restriction fragments were separated in 1 % pulsed-field-certified agarose in 0.5× TBE (Tris/borate/EDTA) buffer. Post-electrophoresis gel treatment included gel staining and de-staining. The DNA was visualized using a UV transilluminator, and images were digitized via a one-dimensional gel documentation system (Bio-Rad).
Image analysis.
The fingerprint pattern in the gel was analysed using the Bionumeric software (Applied Maths). After background subtraction and gel normalization, the fingerprint patterns were subjected to typing on the basis of banding similarity and dissimilarity, using the Dice similarity coefficient and unweighted-pair group method employing average linkage (UPGMA) clustering, as recommended by the manufacturer. The results were graphically represented as dendrograms.
Results
Microbiology and serology
The V. cholerae strains included in the present study are shown in Table 2, with source and year of isolation. All V. cholerae O1 strains [clinical patient (n = 32) and environmental (n = 3) isolates] included in this study produced colonies typical of V. cholerae on TTGA and gave biochemical reactions characteristic for V. cholerae. All reacted positively to V. cholerae O1 monoclonal antibody, but not to O139. They also were positive for either Inaba or Ogawa serotype-specific monovalent antisera, confirming that all belonged to serogroup O1.
Table 2. Phenotypic, genotypic and drug resistance properties of V. cholerae O1 isolated in Thailand (n = 35) between 2003 and 2011.
| Strains | Year of isolation | No. of isolates | Source* | Serotype | rfbO1 | Phenotypic properties† | Genetic screening by PCR | Resistance pattern§ | |||||||||
| CCA | Sensitivity | ctxA | tcpA type | ctxB type‡ | rstR type | rtxC | rstC | tlc | |||||||||
| PMB (50 U) | CL-specific phage IV | ET-specific phage V | |||||||||||||||
| Thai isolates | 2003 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E, AMP |
| 2003 | 1 | Env | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E, AMP | |
| 2003 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E | |
| 2003 | 2 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E | |
| 2004 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E, AMP | |
| 2004 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E | |
| 2004 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | CL | + | − | − | E | |
| 2007 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | CL | + | − | − | E, TE, SXT, AMP | |
| 2007 | 2 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | E, TE, SXT, AMP | |
| 2007 | 2 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | E, TE, SXT | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | TE, SXT | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | E | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | TE, SXT | |
| 2010 | 3 | Clin | Inaba | + | + | R | R | S | + | ET | CL | ET | + | + | + | E, SXT | |
| 2010 | 3 | Clin | Inaba | + | + | R | R | S | + | ET | CL | ET | + | + | + | SXT | |
| 2010 | 1 | Clin | Inaba | + | + | R | R | S | − | ET | − | − | + | − | + | E | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | E, TE, SXT | |
| 2010 | 2 | Clin | Ogawa | + | + | R | R | S | − | ET | − | − | + | − | + | E, AMP | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | − | ET | − | − | + | − | + | Sensitive to all | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | SXT | |
| 2010 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | ET | + | + | + | SXT | |
| 2010 | 1 | Clin | Ogawa | + | + | R | R | S | − | ET | − | − | + | − | + | Sensitive to all | |
| 2010 | 2 | Env | Inaba | + | + | R | R | S | + | ET | CL | ET | + | + | + | E | |
| 2010 | 1 | Clin | Inaba | + | + | R | R | S | + | ET | CL | ET | + | + | + | E, AMP, SXT | |
| 2011 | 1 | Clin | Ogawa | + | + | R | R | S | + | ET | CL | ET | + | + | + | AMP, SXT | |
| 2011 | 1 | Clin | Ogawa | + | + | R | R | S | − | − | − | − | + | − | − | E, AMP, SXT | |
| Reference strains | |||||||||||||||||
| O395 | 1965 | – | Clin | Ogawa | + | − | S | S | R | + | CL | CL | CL | − | − | + | – |
| N16961 | 1971 | – | Clin | Inaba | + | + | R | R | S | + | ET | ET | ET | + | + | + | – |
Clin, Clinical; Env, environmental.
CCA, Chicken cell agglutination; PMB, polymyxin B; R, resistant; S, sensitive.
Determined by MAMA-PCR.
AMP, ampicillin; E, erythromycin; SXT, trimethoprim/sulfamethoxazole; TE, tetracycline.
Serotyping results showed that both environmental and clinical strains of V. cholerae isolated between 2003 and 2004 were Inaba. All V. cholerae O1 clinical strains isolated in 2007 and 2011 and a majority of clinical strains isolated in 2010 were Ogawa. Two strains isolated from surface water sources in 2010 were Inaba (Table 2).
Amplification of primers specific for V. cholerae serogroup O1 and ctxA by PCR
All strains amplified primers for V. cholerae species-specific ompW and all amplified primers specific for the O biosynthetic gene wbe of V. cholerae O1, but not wbf, which is specific for serogroup O139. In addition, all except five clinical strains associated with cholera during 2010, plus one strain isolated in 2011, amplified primers for the CTX gene ctxA, confirming that the strains were toxigenic V. cholerae O1. Six V. cholerae O1 strains that did not amplify primers for ctxA were concluded to be ctx negative (Table 2).
Phenotypic and genetic characteristics
All V. cholerae strains showed biotype ET-specific phenotypic traits, such as chicken cell agglutination, sensitivity to ET-specific phage V, and resistance to both polymyxin B and CL-specific phage IV, and were recognized to be biotype ET (Table 2). All, except six ctx− strains, were phenotypic variants of ET with major ET traits, but failed to show all properties typical of V. cholerae ET reference strain N16961.
Phenotypically confirmed ET biotype strains amplified primers for the ET-specific marker gene rtxC, confirming their identification as V. cholerae ET. tcpA, the major virulence-associated gene of the VPI-I gene cluster, was present in all strains except the ctxA− strain isolated in 2011, and all amplified primers for tcpAET, but not tcpACL. The V. cholerae strains were analysed by MAMA-PCR using primers specific for CL or ET biotype, offering a precise and accurate method for determining the type of CTX. All V. cholerae O1 ctxA+ strains, including the V. cholerae O395 CL reference, amplified primers specific for ctxB1CL, an allele identified in CL biotype strains worldwide, including altered ET biotype strains from the US Gulf Coast. Among the toxigenic altered ET strains confirmed in this study, V. cholerae isolated from clinical sources and natural surface water between 2003 and 2004, plus a clinical strain isolated in 2007, carried the CL-biotype-specific repressor gene rstRCL, but not the ET-biotype-specific repressor gene rstRET, confirming that they carried CL-biotype CTX prophage. However, all of the atypical ET strains isolated in 2003–2004, plus one strain isolated in 2007, were devoid of rstC and tlc, suggesting a unique genetic characteristic of V. cholerae associated with endemic cholera in Thailand. These V. cholerae showed a sharp genetic transition, as a majority of the 2007 strains, and all strains (ctx+) isolated in Khon Kaen thereafter, until 2011, possessed rstC and tlc and all carried the repressor rstRET, although the ctxB gene of these strains was CL biotype, as was found in altered ET of contemporary cholera (Table 2). The six ctx− V. cholerae strains confirmed in this study also lacked ctxAB, rstR and rstC, suggesting that they did not carry the CTX prophage.
Antibiotic susceptibility assay
Antibiotic susceptibility patterns showed that among the 35 V. cholerae O1 strains isolated between 2003 and 2011, 71 % (n = 25) were resistant to erythromycin, 54 % (n = 19) to trimethoprim/sulfamethxazole, 23 % (n = 8) to tetracycline, and 31 % (n = 11) to ampicillin, whereas all were uniformly sensitive to gentamicin and ciprofloxacin. Overall, 23 % (n = 8) of the strains were multidrug resistant (MDR). Year-wise data analysis revealed that, in Thailand, V. cholerae showing resistance to four drugs, erythromycin, tetracycline, trimethoprim/sulfamethxazole and ampicillin, occurred among strains isolated in 2007 (Table 2). The majority of V. cholerae isolated before or after 2007 were sensitive, although strains varied in their patterns of response to the different drugs tested. Interestingly, two isolates from 2010 were sensitive to all six antibiotics tested (Table 2). Of the eight resistant strains showing resistance to at least three antibiotics, five were isolated in 2007, two in 2010, and only one in 2011. Overall, 54 % of the V. cholerae isolated between 2003 and 2011 were resistant to two or more antibiotics.
Sequencing of ctxB and tcpA
PCR-amplified genes ctxB (460 bp) and tcpA (675 bp) of randomly selected V. cholerae O1 strains (n = 9) representing each year between 2003 and 2011 were sequenced and the amino acid sequences were determined by employing bioinformatics tools. The results showed that the deduced amino acid sequence of CTXB of all the tested V. cholerae O1 strains was identical to that of the CL biotype CT (ctxB genotype 1), with histidine and threonine at positions 39 and 68, respectively. However, the strains isolated since 2007 had a mutation in the tcpA gene that resulted an amino acid substitution at position 64 (N→S) in the mature peptide of TcpA (Fig. 1).
PFGE and cluster analysis
The NotI-digested genomic DNAs of V. cholerae O1 strains from Thailand subjected to PFGE to determine genetic relatedness and clonal origin yielded 20 to 23 fragments (Fig. 2) and their molecular size ranged from 20.5 to 350 kb. All showed ET biotype PFGE patterns, with divergence in the number and position of the DNA fragments. Four major PFGE types, A–D, designated clonal types, and subtypes were determined from the overall PFGE patterns of the 35 V. cholerae O1 environmental and clinical strains (Table 2). Clonal type A, with seven different PFGE patterns (A1–A7), was predominantly associated with cholera in Khon Kaen between 2003 and 2011, while clonal type B was represented by a single strain associated with cholera in 2011; clonal types C and D were environmental isolates from surface water samples collected in 2010.
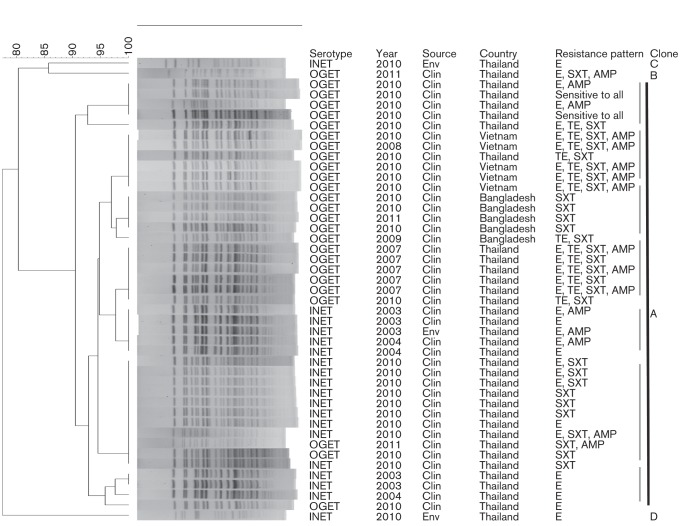
Fig. 2.
Genomic fingerprinting patterns of V. cholerae O1 strains isolated from diarrhoea and environmental sources in Khon Kaen, Thailand (2003–2011). The dendrogram was prepared by Dice similarity coefficient and UPGMA clustering using PFGE images of the NotI-digested genomic DNA. The scale-bar at the top left indicates similarity coefficient (%). The PFGE types (A–D) of the V. cholerae O1 strains are shown. The major cluster, A, comprised most of the Thai V. cholerae O1 strains (2003–2011) and all of the representative Bangladeshi (2009–2010) and Vietnamese (2008–2010) V. cholerae O1 strains showing clonal relatedness. The seven different PFGE pattern-based subclusters within cluster A are marked with grey bars. Distinct signatures for clonally related altered ET in Thailand, Vietnam, and Bangladesh, as confirmed by subclustering patterns in the dendrogram, suggest endemic cholera to be caused by V. cholerae population indigenous to Khon Kaen, Thailand. INET, Inaba El Tor; OGET, Ogawa El Tor; Clin, clinical; Env, environmental.
Only the environmental V. cholerae strain isolated in 2003 had a PFGE pattern common to clinical strains isolated in 2003 and 2004 (Fig. 2). Year-wise data revealed that all except three V. cholerae O1 strains isolated between 2003 and 2011 exhibited closely related PFGE patterns belonging to clonal type A (Fig. 2). The remaining three V. cholerae O1 strains, of which two had been isolated in 2010 from surface water and one from a clinical case in 2011, represented three additional clonal types (B–D) from Thailand.
In order to understand the clonal link between the V. cholerae O1 strains occurring in Khon Kaen, cluster analysis was performed using PFGE (NotI) images of genomic DNA of O1 strains isolated in Thailand (2003–2011), together with representative V. cholerae O1 isolates from Bangladesh (2009–2010) and Vietnam (2008–2010). Cluster analysis separated the distinct clonal types and the subtypes although the majority of Thai V. cholerae O1 strains constituted a major cluster (A) with all Bangladesh and Vietnam V. cholerae O1 isolates, suggesting clonality. None of the contemporary Bangladesh or Vietnam V. cholerae O1 strains belonged to clonal types B–D identified in Thailand, suggesting that they were distant clonally (Fig. 2). Although strains of cluster A were deemed ‘clonal’, based on similarity index, the minor, but consistent, divergence indicated seven subclusters, depending on serotypes (Inaba/Ogawa) and spatio-temporal origin. Country-specific subclustering of the recent Thai, Vietnam and Bangladesh V. cholerae O1 isolates reflected their different signatures.
Discussion
V. cholerae causes both epidemic and pandemic cholera and is a serious public health threat for low socio-economic groups in many countries, including Thailand. Research on V. cholerae at the molecular level has made enormous strides over the past 100 years (Chun et al., 2009; Kaper et al., 1995). A recent MLVA/PFGE study of V. cholerae O1 isolated in Thailand suggested that some clones with similar but distinctive genetic traits circulated during an outbreak (Okada et al., 2012). The present study provides data on antibiotic resistance patterns with evidence of genetic changes in V. cholerae O1 in endemic cholera, namely four major clones with distinct regional signatures in natural surface waters in Khon Kaen, Thailand.
Overall, the results of microbiological, biochemical and serological tests confirmed that 35 V. cholerae isolates from cholera outbreaks in north-eastern Thailand between 2003 and 2011 were serogroup O1. Year-wise phenotypic results revealed that V. cholerae O1 isolated between 2003 and 2011 showed temporal variation in serotype, Inaba or Ogawa, according to year of isolation. The microbiological results were complemented with simplex PCR assay for amplification of V. cholerae species-specific ompW (Nandi et al., 2000), together with multiplex PCR assay for ctxA (encoding subunit A of CTX) and wbe (encoding serogroup O1 antigen) (Hoshino et al., 1998). These tests confirmed that the majority of the V. cholerae isolates associated with cholera in north-eastern Thailand were toxigenic and belonged to serogroup O1. Six serologically confirmed O1 strains reacted to monovalent Inaba antiserum and amplified primers for ompW and wbe (Hoshino et al., 1998), but not ctxA, indicating that they were ctx−V. cholerae O1. The ctx−V. cholerae O1 occurs in the aquatic environment and has been shown to arise following loss of the CTX prophage (Alam et al., 2007; Alam et al., 2010).
Treatment of cholera patients with appropriate oral or intravenous rehydration, and a 1–3 day course of effective antibiotics (Saha et al., 2006) can reduce the severity of infection, hospitalization, and faecal–oral transmission of cholera bacteria. Tetracycline (TE) has long been the drug of choice for the treatment of cholera, except for young children and pregnant women (Greenough et al., 1964; Lindenbaum et al., 1967). Other effective drugs have included furazolidone, erythromycin (E), trimethoprim/sulfamethoxazole (SXT), and chloramphenicol (Greenough et al., 1964). However, antibiotic therapy has faced challenges related to the rapid emergence and spread of V. cholerae strains resistant to multiple antimicrobial agents, such as TE, ampicillin (AMP), kanamycin, streptomycin, SXT, nalidixic acid, E and most recently to ciprofloxacin and norfloxacin (Mhalu et al., 1979; Glass et al., 1980; Jain et al., 2011). In this study, V. cholerae O1 strains from north-eastern Thailand isolated between 2003 and 2010 showed temporal variation in their response to different antibiotics. Although all strains isolated in 2007, two strains isolated in 2010, and one strain isolated in 2011 were MDR, strains isolated before and after 2007 were sensitive to the antibiotics tested and their response to different antibiotics varied from being sensitive to all, resistant to one, either E or SXT, and a few to AMP. Supawat et al. (2009) showed that some V. cholerae O1 strains isolated in Thailand between 2000 and 2004 were resistant to TE and SXT. In this study, V. cholerae associated with cholera between 2003 and 2004 showed resistance only to E and a few were also resistant to AMP, but none to TE and SXT. However, V. cholerae O1 strains isolated during 2007 in north-east Thailand were all resistant to four antibiotics, E, TE, SXT and AMP, although the patterns changed in 2010–2011, as all except three strains were sensitive to TE, with the rest either sensitive to either all or resistant to one, two or three of the antibiotics.
MDR V. cholerae strains isolated from cholera patients in Vietnam between 2008 and 2010 were resistant to four antibiotics, namely E, AMP, TE and SXT (Tran et al., 2012), as occurred in Khon Kaen, Thailand, in 2007. Although Thailand shares its border with Vietnam and despite the fact that the resistant V. cholerae strains occurring in the two countries showed similar resistance patterns, it is highly unlikely that the resistant strains emerged in one of the two countries and were transmitted to the other, since resistant strains were present in Thailand in 2007 but not in 2010 when resistant strains were reported in Vietnam (Tran et al., 2012). Most of the V. cholerae O1 isolated in Thailand between 2003 and 2011 were resistant to SXT, a recommended first-line drug for children with acute diarrhoea. In Thailand, TE and norfloxacin were the most frequently used drugs for treatment of cholera (Supawat et al., 2009), but the data presented in this study on the emergence of MDR V. cholerae in Thailand, together with the increasing incidence of cholera caused by MDR V. cholerae in Africa, Asia, and South America (Glass et al., 1980; Goel et al. 2011; Ibarra & Alvarado, 2007; Mandomando et al., 2007; Tran et al., 2012) indicate that drugs should not be administered without having sensitivity patterns first determined, considering that drug response patterns change frequently and can vary spatio-temporally.
In recent years, V. cholerae O1 ET has shown a shift in ctxB from genotype 3 (found in ET strains of the seventh pandemic and the Latin American epidemic) to genotype 1 (found in strains of CL biotype worldwide and the US Gulf Coast ET strains). Atypical ET possessing CL biotype CTX was first reported in Asia at the start of this century (Nair et al., 2006; Raychoudhuri et al., 2008; Morita et al., 2010) and subsequently in Africa (Ansaruzzaman et al., 2007) and Central America (Alam et al., 2010), including Haiti (Chin et al., 2011; Hasan et al., 2012). Several genotypic variants of the Asian atypical (altered) ET were identified in Africa (Ansaruzzaman et al., 2007; Choi et al., 2010) and Central America (Alam et al., 2010). In this study, V. cholerae O1 strains associated with cholera in Khon Kaen, Thailand, were phenotypically and genetically confirmed to be ET, but possessing CTX of the CL biotype, as reported previously in Thailand (Okada et al., 2010; Na-Ubol et al. 2011). The data also show that, unlike the altered ET strains reported from Bangladesh (Nair et al., 2006) and India (Raychoudhuri et al., 2008), V. cholerae O1 ET strains isolated between 2003 and 2004, and one strain isolated in 2007 from a cholera outbreak in Khon Kaen, Thailand, did not carry rstC, a repressor gene sequence of the RS1 genetic element (Safa et al., 2010) or the TLC gene cluster, which encodes a filamentous phage replicase and is proposed to be a satellite phage. Besides, all of these strains contained the repressor rstR gene (allele) of CL biotype, and thus appeared indistinguishable from the Mozambique variant of ET carrying CL CTX prophage (Ansaruzzaman et al., 2004; Faruque et al., 2007). Interestingly, a sharp genetic transition of V. cholerae from the ET Mozambique variant to the Asian altered ET (possessing ctxB of the CL biotype) was observed in north-east Thailand in 2007, with all of the tested V. cholerae strains acquiring RS1 and TLC and switching the repressor rstR allele from rstRCL (CL) to rstRET (ET), as in the altered ET reported from Bangladesh (Nair et al., 2006) and India (Raychoudhuri et al., 2008). Our data on the temporal absence of virulence and related genetic elements and their acquisitions clearly indicate horizontal gene transfer, which has been proposed to provide V. cholerae strains with improved evolutionary fitness (Faruque et al., 2007).
A few V. cholerae O1 ET carrying a tandem repeat of the CL CTX prophage on the small chromosome, indistinguishable from the Mozambique ET variant, were isolated in Vietnam between 1995 and 2004 (Nguyen et al., 2009). In a subsequent study, Tran et al. (2012) showed that V. cholerae O1 ET involved in recurrent cholera in Vietnam (2007–2010) was an ET variant carrying ctxBCL and rstRET genes, as observed for ET strains involved in recurrent cholera in Khon Kaen, Thailand. Although the source of V. cholerae O1 ET analogous to the Mozambique variant of ET causing outbreaks in Thailand between 2003 and 2007 is not known, the Vietnamese ET strains carrying a tandem repeat of the CL CTX prophage and not involved in any major cholera outbreak were proposed to have been recently introduced into Vietnam (Nguyen et al., 2009). Our observations of temporal fluctuation in resistance to antimicrobial agents and a genetic transition from Mozambique variant type ET to Asian altered ET suggest that V. cholerae has a natural reservoir (niche), which has allowed the bacterium to evolve locally in Khon Kaen, although the appearance of the V. cholerae O1 altered ET carrying ctxBCL and rstRET alleles has been hypothesized to be a recent event in Thailand (Okada et al., 2012).
Toxin co-regulated pilus (TCP), a type IV bundle-forming pilus of V. cholerae, is an essential colonization factor (Taylor et al., 1987) that also serves as receptor for CTX-Φ (Waldor & Mekalanos, 1996). The genes encoding the biosynthesis of TCP pilus were shown to be encoded on a Vibrio pathogenicity island (VPI), a novel filamentous bacteriophage (Karaolis et al., 1999). The TCP is a homopolymer of a 20.5 kDa major pilus protein, TcpA pilin (Taylor et al., 1987) encoded by tcpA. The DNA sequence of tcpA differs slightly in the C-terminal domain for CL and ET biotype strains (Safa et al., 2010). In the present study, the amino acid sequences of TcpA of V. cholerae O1 ET strains isolated in Thailand between 2003 and 2004 showed 100 % homology with the amino acid sequence of TcpA of ET reference strain N16961. However, the tcpA gene of V. cholerae O1 ET strains isolated in 2007 and thereafter had a mutation at amino acid position 64 (N→S). Although the change is subtle and it is not clear whether such genetic switching of the tcpA gene can provide V. cholerae O1 strains with increased environmental and/or epidemiological fitness (Faruque et al., 2007), a similar change of amino acid at position 64 of TcpA was found in ctx− V. cholerae O1 strains ZJ65 (2006) isolated in China (GenBank accession no. EU622532) and V. cholerae strain 2010EL1786 (GenBank accession no. CP003069) causing fatal epidemics in Haiti (Reimer et al., 2011).
Historically, cholera has been endemic for centuries in the Ganges delta of the Bay of Bengal. Epidemic cholera in Africa and Latin America is suggested to have been introduced from cholera endemic countries of Asia (Blake, 1994). V. cholerae O1 ET populations involved in the 1991 cholera epidemic in Latin America were homogeneous initially (Blake, 1994), although divergent strains were detected later (Dalsgaard et al., 1997; Beltrán et al., 1999; Popovic et al., 1993; Nusrin et al., 2009). In Thailand, PFGE of NotI-digested genomic DNA revealed the majority of the V. cholerae O1 altered ET strains involved in recurrent cholera to be of a single clonal type, A, between 2003 and 2011, although more clonal types (B–D) mainly of surface water origin, were isolated in 2010 and thereafter, reflecting genetic diversity in the V. cholerae population. Evidence of an aquatic reservoir for V. cholerae is very extensive (Alam et al., 2007), but the source and transmission of altered ET in Asia (Nair et al., 2006), Africa (Ansaruzzaman et al., 2004) and the Americas (Alam et al., 2010; Chin et al., 2011) remain enigmatic. Distinct signatures of clonally related altered ET strains in Thailand, Vietnam and Bangladesh, as revealed by PFGE (NotI)-based dendrograms, are presumed to be attributable to independent evolution of V. cholerae strains in different ecosystems. In any case, the findings of this study strongly suggest that cholera in north-east Thailand is endemic, caused by V. cholerae thriving locally and independent of cholera outbreaks in Vietnam and Bangladesh.
Acknowledgements
This study was partly supported by research grants from the Faculty of Medicine, Khon Kaen University and Department of Disease Control, Ministry of Public Health, Thailand National Institutes of Health Grant No. RO1AI039129, under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health, the University of Maryland, College Park, and the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B). ICDDR,B acknowledges its major donors: Australian Agency for International Development (AusAID), Government of the People’s Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (SIDA), and the Department for International Development, UK (DFID). C. C. thanks the staff at the Office of Communicable Disease Control, Region 6, Khon Kaen, for their help with specimen collection.
Abbreviations:
- AMP
ampicillin
- CL
classical
- CTX
cholera toxin
- E
erythromycin
- ET
El Tor
- MAMA
mismatch amplification mutation assay
- MDR
multidrug resistant
- SXT
trimethoprim/sulfamethoxazole
- TE
tetracycline
- TLC
toxin-linked cryptic plasmid
References
- Alam M., Sultana M., Nair G. B., Siddique A. K., Hasan N. A., Sack R. B., Sack D. A., Ahmed K. U., Sadique A. & other authors (2007). Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci U S A 104, 17801–17806 10.1073/pnas.0705599104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Nusrin S., Islam A., Bhuiyan N. A., Rahim N., Delgado G., Morales R., Mendez J. L., Navarro A. & other authors (2010). Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol 48, 3666–3674 10.1128/JCM.00866-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang G. Y., Yu C. Y., Balqis K., Elina H. T., Azura H., Hani M. H., Yean C. Y. (2010). Molecular evidence of cholera outbreak caused by a toxigenic Vibrio cholerae O1 El Tor variant strain in Kelantan, Malaysia. J Clin Microbiol 48, 3963–3969 10.1128/JCM.01086-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaruzzaman M., Bhuiyan N. A., Nair B. G., Sack D. A., Lucas M., Deen J. L., Ampuero J., Chaignat C. L., Mozambique Cholera Vaccine Demonstration Project Coordination Group (2004). Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis 10, 2057–2059 10.3201/eid1011.040682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaruzzaman M., Bhuiyan N. A., Safa A., Sultana M., McUamule A., Mondlane C., Wang X. Y., Deen J. L., von Seidlein L., Clemens J. (2007). Genetic diversity of El Tor strains of Vibrio cholerae O1 with hybrid traits isolated from Bangladesh and Mozambique. Int J Med Microbiol 297, 443–449 10.1016/j.ijmm.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45, 493–496 [PubMed] [Google Scholar]
- Beltrán P., Delgado G., Navarro A., Trujillo F., Selander R. K., Cravioto A. (1999). Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol 37, 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake P. A. (1994). Endemic cholera in Australia and United States. In Vibrio cholerae and Cholera: Molecular to Global Perspectives, pp. 309–319 Edited by Wachsmuth I. K., Blake P. A., Olsvik O. Washington, DC: American Society for Microbiology [Google Scholar]
- Bureau of Epidemiology (2010). Annual epidemiological surveillance report. Nonthaburi, Thailand: Department of Disease Control, Ministry of Public Health
- Cameron D. N., Khambaty F. M., Wachsmuth I. K., Tauxe R. V., Barrett T. J. (1994). Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol 32, 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Sorenson J., Harris J. B., Robins W. P., Charles R. C., Jean-Charles R. R., Bullard J., Webster D. R., Kasarskis A. & other authors (2011). The origin of the Haitian cholera outbreak strain. N Engl J Med 364, 33–42 10.1056/NEJMoa1012928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Lee J. H., Jeon Y. S., Lee H. R., Kim E. J., Ansaruzzaman M., Bhuiyan N. A., Endtz H. P., Niyogi S. K. & other authors (2010). Multilocus variable-number tandem repeat analysis of Vibrio cholerae O1 El Tor strains harbouring classical toxin B. J Med Microbiol 59, 763–769 10.1099/jmm.0.017939-0 [DOI] [PubMed] [Google Scholar]
- Chow K. H., Ng T. K., Yuen K. Y., Yam W. C. (2001). Detection of RTX toxin gene in Vibrio cholerae by PCR. J Clin Microbiol 39, 2594–2597 10.1128/JCM.39.7.2594-2597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Grim C. J., Hasan N. A., Lee J. H., Choi S. Y., Haley B. J., Taviani E., Jeon Y. S., Kim D. W. & other authors (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106, 15442–15447 10.1073/pnas.0907787106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2010). Methods for Antimicrobial Dilution and Disc Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Approved Guideline, 2nd edn, document M45-A2 (ISBN 1-56238-732-4). Wayne, PA: Clinical and Laboratory Standards Institute.
- Dalsgaard A., Skov M. N., Serichantalergs O., Echeverria P., Meza R., Taylor D. N. (1997). Molecular evolution of Vibrio cholerae O1 strains isolated in Lima, Peru, from 1991 to 1995. J Clin Microbiol 35, 1151–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M., Balon E., Boyd D., Fraser C. M., Heidelberg J. F., Mekalanos J. J. (2002). Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A 99, 1556–1561 10.1073/pnas.042667999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque S. M., Tam V. C., Chowdhury N., Diraphat P., Dziejman M., Heidelberg J. F., Clemens J. D., Mekalanos J. J., Nair G. B. (2007). Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci U S A 104, 5151–5156 10.1073/pnas.0700365104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R. I., Huq I., Alim A. R. M. A., Yunus M. (1980). Emergence of multiply antibiotic-resistant Vibrio cholerae in Bangladesh. J Infect Dis 142, 939–942 10.1093/infdis/142.6.939 [DOI] [PubMed] [Google Scholar]
- Goel A. K., Jain M., Kumar P., Sarguna P., Bai M., Ghosh N., Gopalan N. (2011). Molecular characterization reveals involvement of altered El Tor biotype Vibrio cholerae O1 strains in cholera outbreak at Hyderabad, India. J Microbiol 49, 280–284 10.1007/s12275-011-0317-9 [DOI] [PubMed] [Google Scholar]
- Greenough W. B., III, Gordon R. S., Jr, Rosenberg I. S., Davies B. I., Benenson A. S. (1964). Tetracycline in the treatment of cholera. Lancet 1, 355–357 10.1016/S0140-6736(64)92099-9 [DOI] [PubMed] [Google Scholar]
- Hasan N. A., Choi S. Y., Eppinger M., Clark P. W., Chen A., Alam M., Haley B. J., Taviani E., Hine E. & other authors (2012). Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109, E2010–E2017 10.1073/pnas.1207359109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K., Yamasaki S., Mukhopadhyay A. K., Chakraborty S., Basu A., Bhattacharya S. K., Nair G. B., Shimada T., Takeda Y. (1998). Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol 20, 201–207 10.1111/j.1574-695X.1998.tb01128.x [DOI] [PubMed] [Google Scholar]
- Ibarra J. O., Alvarado D. E. (2007). Antimicrobial resistance of clinical and environmental strains of Vibrio cholerae isolated in Lima-Peru during epidemics of 1991 and 1998. Braz J Infect Dis 11, 100–105 10.1590/S1413-86702007000100022 [DOI] [PubMed] [Google Scholar]
- Jain M., Goel A. K., Bhattacharya P., Ghatole M., Kamboj D. V. (2011). Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Trop 117, 152–156 10.1016/j.actatropica.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Kaper J. B., Morris J. G., Jr, Levine M. M. (1995). Cholera. Clin Microbiol Rev 8, 48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis D. K. R., Somara S., Maneval D. R., Jr, Johnson J. A., Kaper J. B. (1999). A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399, 375–379 10.1038/20715 [DOI] [PubMed] [Google Scholar]
- Kumar P., Wilson P. A., Bhai R., Thomas S. (2010a). Characterization of an SXT variant Vibrio cholerae O1 Ogawa isolated from a patient in Trivandrum, India. FEMS Microbiol Lett 303, 132–136 10.1111/j.1574-6968.2009.01868.x [DOI] [PubMed] [Google Scholar]
- Kumar P., Peter W. A., Thomas S. (2010b). Rapid detection of virulence-associated genes in environmental strains of Vibrio cholerae by multiplex PCR. Curr Microbiol 60, 199–202 10.1007/s00284-009-9524-6 [DOI] [PubMed] [Google Scholar]
- Lindenbaum J., Greenough W. B., Islam M. R. (1967). Antibiotic therapy of cholera. Bull World Health Organ 36, 871–883 [PMC free article] [PubMed] [Google Scholar]
- Mandomando I., Espasa M., Vallès X., Sacarlal J., Sigaúque B., Ruiz J., Alonso P. (2007). Antimicrobial resistance of Vibrio cholerae O1 serotype Ogawa isolated in Manhiça District Hospital, southern Mozambique. J Antimicrob Chemother 60, 662–664 10.1093/jac/dkm257 [DOI] [PubMed] [Google Scholar]
- Mhalu F. S., Mmari P. W., Ijumba J. (1979). Rapid emergence of El Tor Vibrio cholerae resistant to antimicrobial agents during first six months of fourth cholera epidemic in Tanzania. Lancet 1, 345–347 10.1016/S0140-6736(79)92889-7 [DOI] [PubMed] [Google Scholar]
- Morita M., Ohnishi M., Arakawa E., Bhuiyan N. A., Nusrin S., Alam M., Siddique A. K., Qadri F., Izumiya H. & other authors (2008). Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol 52, 314–317 10.1111/j.1348-0421.2008.00041.x [DOI] [PubMed] [Google Scholar]
- Morita M., Ohnishi M., Arakawa E., Yamamoto S., Nair G. B., Matsushita S., Yokoyama K., Kai A., Seto K. & other authors (2010). Emergence and genetic diversity of El Tor Vibrio cholerae O1 that possess classical biotype ctxB among travel-associated cases of cholera in Japan. J Med Microbiol 59, 708–712 10.1099/jmm.0.017624-0 [DOI] [PubMed] [Google Scholar]
- Mwansa J. C. L., Mwaba J., Lukwesa C., Bhuiyan N. A., Ansaruzzaman M., Ramamurthy T., Alam M., Balakrish Nair G. (2007). Multiply antibiotic-resistant Vibrio cholerae O1 biotype El Tor strains emerge during cholera outbreaks in Zambia. Epidemiol Infect 135, 847–853 10.1017/S0950268806007254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na-Ubol M., Srimanote P., Chongsa-Nguan M., Indrawattana N., Sookrung N., Tapchaisri P., Yamazaki S., Bodhidatta L., Eampokalap B. & other authors (2011). Hybrid & El Tor variant biotypes of Vibrio cholerae O1 in Thailand. Indian J Med Res 133, 387–394 [PMC free article] [PubMed] [Google Scholar]
- Nair G. B., Qadri F., Holmgren J., Svennerholm A. M., Safa A., Bhuiyan N. A., Ahmad Q. S., Faruque S. M., Faruque A. S. G. & other authors (2006). Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44, 4211–4213 10.1128/JCM.01304-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi B., Nandy R. K., Mukhopadhyay S., Nair G. B., Shimada T., Ghose A. C. (2000). Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol 38, 4145–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B. M., Lee J. H., Cuong N. T., Choi S. Y., Hien N. T., Anh D. D., Lee H. R., Ansaruzzaman M., Endtz H. P. & other authors (2009). Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol 47, 1568–1571 10.1128/JCM.02040-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrin S., Gil A. I., Bhuiyan N. A., Safa A., Asakura M., Lanata C. F., Hall E., Miranda H., Huapaya B. & other authors (2009). Peruvian Vibrio cholerae O1 El Tor strains possess a distinct region in the Vibrio seventh pandemic island-II that differentiates them from the prototype seventh pandemic El Tor strains. J Med Microbiol 58, 342–354 10.1099/jmm.0.005397-0 [DOI] [PubMed] [Google Scholar]
- O’Shea Y. A., Reen F. J., Quirke A. M., Boyd E. F. (2004). Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J Clin Microbiol 42, 4657–4671 10.1128/JCM.42.10.4657-4671.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Chantaroj S., Roobthaisong A., Hamada S., Sawanpanyalert P. (2010). A cholera outbreak of the Vibrio cholerae O1 El Tor variant carrying classical CtxB in northeastern Thailand in 2007. Am J Trop Med Hyg 82, 875–878 10.4269/ajtmh.2010.09-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Roobthaisong A., Nakagawa I., Hamada S., Chantaroj S. (2012). Genotypic and PFGE/MLVA analyses of Vibrio cholerae O1: geographical spread and temporal changes during the 2007–2010 cholera outbreaks in Thailand. PLoS ONE 7, e30863 10.1371/journal.pone.0030863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsvik O., Wahlberg J., Petterson B., Uhlén M., Popovic T., Wachsmuth I. K., Fields P. I. (1993). Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol 31, 22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic T., Bopp C., Olsvik O., Wachsmuth K. (1993). Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol 31, 2474–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilici M. L., Massenet D., Gake B., Bwalki B., Olson D. M. (2010). Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg Infect Dis 16, 1804–1805 10.3201/eid1611.100568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhuri A., Mukhopadhyay A. K., Ramamurthy T., Nandy R. K., Takeda Y., Nair G. B. (2008). Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J Med Res 128, 695–698 [PubMed] [Google Scholar]
- Reimer A. R., Van Domselaar G., Stroika S., Walker M., Kent H., Tarr C., Talkington D., Rowe L., Olsen-Rasmussen M. & other authors (2011). Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis 17, 2113–2121 10.3201/eid1711.110794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera I. N. G., Chun J., Huq A., Sack R. B., Colwell R. R. (2001). Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl Environ Microbiol 67, 2421–2429 10.1128/AEM.67.6.2421-2429.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa A., Nair G. B., Kong R. Y. C. (2010). Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18, 46–54 10.1016/j.tim.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Saha D., Karim M. M., Khan W. A., Ahmed S., Salam M. A., Bennish M. L. (2006). Single-dose azithromycin for the treatment of cholera in adults. N Engl J Med 354, 2452–2462 10.1056/NEJMoa054493 [DOI] [PubMed] [Google Scholar]
- Siddique A. K., Nair G. B., Alam M., Sack D. A., Huq A., Nizam A., Longini I. M., Jr, Qadri F., Faruque S. M. & other authors (2010). El Tor cholera with severe disease: a new threat to Asia and beyond. Epidemiol Infect 138, 347–352 10.1017/S0950268809990550 [DOI] [PubMed] [Google Scholar]
- Supawat K., Huttayananont S., Sawanpanyalert P., Aswapokee N., Mootsikapun P. (2009). Antimicrobial resistance surveillance of Vibrio cholerae in Thailand from 2000 to 2004. J Med Assoc Thai 92 (Suppl. 4), S82–S86 [PubMed] [Google Scholar]
- Tangkanakul W., Hanpanjakit C. (2007). Weekly Epidemiological Surveillance Report. Bangkok. Bangkok, Thailand: Bureau of Epidemiology, Department of Disease Control, Ministry of Public Health [Google Scholar]
- Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. (1987). Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84, 2833–2837 10.1073/pnas.84.9.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. D., Alam M., Trung N. V., Kinh N. V., Nguyen H. H., Pham V. C., Ansaruzzaman M., Rashed S. M., Bhuiyan N. A. & other authors (2012). Multi-drug resistant Vibrio cholerae O1 variant El Tor isolated in northern Vietnam between 2007 and 2010. J Med Microbiol 61, 431–437 10.1099/jmm.0.034744-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]