Abstract
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that causes infections in the lungs of individuals with the genetic disease cystic fibrosis. Density-dependent production of toxic factors regulated by the Pseudomonas quinolone signal (2-heptyl-3-hydroxy-4-quinolone; PQS) have been proposed to be involved in P. aeruginosa virulence. PQS biosynthesis requires conversion of the central metabolite chorismate to anthranilate by anthranilate synthase. This reaction is also the first step in tryptophan biosynthesis. P. aeruginosa possesses two functional anthranilate synthases, TrpEG and PhnAB, and these enzymes are not functionally redundant, as trpEG mutants are tryptophan auxotrophs but produce PQS while mutants in phnAB are tryptophan prototrophs but do not produce PQS in minimal media. The goal of the work described in this paper was to determine the mechanism for this lack of functional complementation of TrpEG and PhnAB. Our results reveal that overexpression of either enzyme compensates for tryptophan auxotrophy and PQS production in the trpEG and phnAB mutants respectively, leading to the hypothesis that differential regulation of these genes is responsible for the lack of functional complementation. In support of this hypothesis, trpEG was shown to be expressed primarily during low-density growth while phnAB was expressed primarily at high density. Furthermore, dysregulation of phnAB expression eliminated tryptophan auxotrophy in the P. aeruginosa trpEG mutant. Based on these data, we propose a model for anthranilate sequestration by differential transcriptional regulation of the two P. aeruginosa anthranilate synthase enzymes.
Introduction
The Gram-negative, opportunistic pathogen Pseudomonas aeruginosa is a primary constituent of chronic, polymicrobial infections in the lungs of individuals with the genetic disease cystic fibrosis (CF) (Delhaes et al., 2012; Harrison, 2007). Because it is intrinsically resistant to many conventional antimicrobial therapies, P. aeruginosa is generally the most challenging pathogen to eradicate from these infections, and is the leading cause of morbidity and mortality for individuals with CF (Emerson et al., 2002; Hoiby et al., 1977). P. aeruginosa utilizes numerous virulence factors to colonize and persist in the CF lung, and many of those virulence factors are regulated in a density-dependent manner through a process called quorum sensing (QS) (Déziel et al., 2005; Rumbaugh et al., 1999; Schuster & Greenberg, 2006; Whiteley et al., 1999). In a canonical Gram-negative QS system, a small molecule called an autoinducer is synthesized constitutively, and the concentration of autoinducer increases as cell density increases. Upon reaching a threshold concentration, autoinducers interact with transcriptional regulators to alter gene expression. P. aeruginosa has three known QS systems: two require the production of acylhomoserine lactone autoinducers and one utilizes quinolone signals (Pesci et al., 1999; Schuster & Greenberg, 2006). The most potent quinolone signal is the Pseudomonas quinolone signal (2-heptyl-3-hydroxoy-4-quinlone; PQS), which regulates several genes involved in virulence (Déziel et al., 2005; Gallagher & Manoil, 2001). PQS signalling is likely to be relevant in the CF lung, as P. aeruginosa produces five times more PQS when grown in CF lung fluids (sputum) compared with when it is grown in a standard laboratory medium (Palmer et al., 2005). Using a defined synthetic CF sputum medium that mimics the nutritional contents of sputum, research in our laboratory has determined that phenylalanine and tyrosine are responsible for enhanced PQS production in CF sputum (Palmer et al., 2007). Subsequent work has demonstrated that aromatic amino acid induction of PQS production is not due to co-regulation of phenylalanine/tyrosine catabolism and PQS biosynthesis (Palmer et al., 2010).
Our favoured model for enhanced PQS production in the presence of phenylalanine and tyrosine involves flux of the central metabolite chorismate, a shared metabolic precursor for aromatic amino acid and PQS biosynthesis (Palmer et al., 2010, 2007). Chorismate is converted to anthranilate by an anthranilate synthase in the first step of PQS biosynthesis, however, degradation of tryptophan via the products of the kynABU operon has also been reported to be a significant source of anthranilate (Farrow & Pesci, 2007). Our measurements of CF sputum have indicated extremely low tryptophan levels (<10 µM) in the CF lung, thus chorismate-mediated synthesis of anthranilate for PQS production is likely to be the relevant biosynthetic scheme in the CF lung (Palmer et al., 2007).
It was discovered in 1990 that P. aeruginosa possesses two functional anthranilate synthases, each comprised of a large and small subunit encoded by the products of the trpE and trpG or phnA and phnB genes, respectively (Essar et al., 1990a, b). Interestingly, early evidence indicated that these enzymes are not functionally redundant, as growth experiments using a marked trpE mutant demonstrated tryptophan auxotrophy while a similar phnA mutant did not (Essar et al., 1990a, b). The authors also observed that the phnA mutant was deficient in production of the virulence factor pyocyanin while the trpE mutant was not (Essar et al., 1990a, b). At the time it was believed that PhnAB generated anthranilate for pyocyanin production, but later work determined that anthranilate is instead a precursor for PQS production, which is required to induce genes for pyocyanin biosynthesis (Calfee et al., 2001; Gallagher et al., 2002; Mavrodi et al., 2001). These observations led to the hypothesis that TrpEG generates anthranilate for tryptophan biosynthesis while PhnAB generates anthranilate for PQS biosynthesis; however, a mechanism explaining the lack of crosstalk between these pathways has not been determined.
Here, we investigated the two P. aeruginosa anthranilate synthases, TrpEG and PhnAB, in order to better characterize their roles in physiology and pathogenesis and determine why anthranilate does not appear to be shared between the tryptophan and PQS biosynthetic pathways. We provide evidence that differential expression of trpEG and phnAB is the mechanism for their lack of functional redundancy.
Methods
Bacterial strains and media.
Strains and plasmids used in this work are listed in Table 1. Luria–Bertani (LB) broth (Fisher) was used for growth of Escherichia coli and P. aeruginosa for molecular cloning and genetic manipulations. For growth and physiology experiments, P. aeruginosa cultures were grown in a MOPS minimal medium, a MOPS-buffered salts base (50 mM MOPS, pH 7.2, 93 mM NH4Cl, 43 mM NaCl, 3.7 mM KH2PO4, 1 mM MgSO4, 3.5 µM FeSO4 heptahydrate) supplemented with 20 mM succinate as a sole source of carbon and energy. When necessary for growth of tryptophan auxotrophic strains (see Table 1), MOPS minimal medium was additionally supplemented with 200 µM tryptophan unless otherwise stated. MOPS buffer was used to wash and starve cells and comprises MOPS minimal medium without a carbon source. Tetracycline concentrations for E. coli and P. aeruginosa were 10 and 50 µg ml−1 for selection and 5 and 25 µg ml−1 for maintenance, respectively. Gentamicin concentrations for P. aeruginosa containing pJN105-derived plasmids were 50 µg ml−1 for selection and 25 µg ml−1 for maintenance. Growth conditions were 37 °C with shaking at 250 r.p.m.
Table 1. Strains and plasmids used in this study.
All mutant strains and plasmids were generated in this study unless otherwise referenced.
| Strain or plasmid | Description |
| Strains | |
| E. coli | |
| DH5α | Wild-type strain for molecular cloning (Sambrook & Russell, 2001) |
| SM10 | Conjugation strain for deletion mutant generation (de Lorenzo & Timmis, 1994) |
| P. aeruginosa | |
| PA14 | Wild-type Pseudomonas aeruginosa strain (Liberati et al., 2006) |
| ΔtrpE | trpE clean deletion in PA14 background |
| ΔkynA | kynA clean deletion in PA14 background |
| ΔphnAB | phnAB clean deletion in PA14 background |
| ΔtrpE ΔkynA | trpE and kynA double clean deletions in PA14 background |
| ΔtrpE ΔphnAB | trpE and phnAB double clean deletions in PA14 background |
| ΔtrpE ΔpqsA | trpE and pqsA double clean deletions in PA14 background |
| Plasmids | |
| pEX18Tc | Gene replacement vector, TcR (Hoang et al., 1998) |
| pJN105 | araC-pBAD expression vector, GmR (Newman & Fuqua, 1999) |
| pGPKO-trpE | pEX18-derived trpE deletion vector, TcR |
| pGPKO-kynA | pEX18-derived kynA deletion vector, TcR |
| pGPKO-phnAB | pEX18-derived phnAB deletion vector, TcR |
| pGPKO-pqsA | pEX18-derived pqsA deletion vector, TcR |
| pGP-trpEG | pJN105-derived trpEG overexpression vector, GmR |
| pKP-phnAB | pJN105-derived phnAB overexpression vector, GmR |
DNA manipulations.
Standard methods were utilized for handling DNA fragments and plasmids (Ausubel, 2002). Restriction endonucleases and other DNA-modifying enzymes were purchased from New England Biolabs. Genomic DNA preparations were performed using a DNeasy Tissue kit (Qiagen). Plasmid DNA preparations were performed using a GeneJet Plasmid Miniprep kit (Fermentas), and purification of DNA fragments was done using a GeneJet PCR Purification kit (Fermentas). PCR was performed using the Expand Long Template PCR System (Roche). DNA sequencing to confirm newly generated constructs was performed at the University of Texas at Austin Institute for Cell and Molecular Biology DNA core facility.
Anthranilate synthase phylogenetic analysis.
The amino acid sequences for P. aeruginosa PA14 TrpE and PhnA were obtained from http://www.pseudomonas.com and used to probe the database of non-redundant protein sequences at http://blast.ncbi.nlm.nih.gov with blast. Sequences for the top 30 hits from unique species were aligned using the mega 5 muscle alignment feature and phylogenetic trees were generated using the mega 5 maximum-likelihood tree generator with 100 bootstrap replicates (Tamura et al., 2011).
Generation of deletion mutants.
Unmarked deletion mutants were constructed by allelic exchange as described previously with some modifications (Hoang et al., 1998). To generate deletion constructs, ~1000 bp regions upstream and downstream of the desired locus to be deleted were PCR amplified using P. aeruginosa PA14 DNA as template and primers listed in Table 2. Amplicons were concatenated by overlap extension PCR, digested with EcoRI and XbaI or KpnI and XbaI (see restriction sites in Table 2) and ligated into pEX18Tc to generate the deletion vectors listed in Table 1. Deletion constructs were conjugated into PA14, or a previously generated mutant strain for double mutants, using the conjugation-competent E. coli strain SM10. Mutants were selected by growth on LB plates containing 50 µg tetracycline ml−1 and 25 µg nalidixic acid ml−1 for merodiploid transconjugants. Subsequent growth on LB plates containing 10 % sucrose was used to select for excision of the integrated plasmid with the desired deletion locus. Deletions were confirmed by PCR using confirmation primers outside the amplified flanking regions (Table 2) as well as phenotypic characterization for tryptophan auxotrophy (ΔtrpE, ΔtrpE ΔphnAB, ΔtrpE ΔpqsA), PQS production (ΔphnAB, ΔtrpE ΔphnAB, ΔtrpE ΔpqsA) and ability to grow on tryptophan (ΔkynA, ΔtrpE ΔkynA).
Table 2. Primers used in this study.
Underlined sequences represent recognition sites for restriction endonucleases.
| Description | Sequence |
| Deletion vector construction | |
| trpE flank 1 for | CTGAATTCGTCAACGTCAAGAACATCCGTGAG |
| trpE flank 1 rev | GTCCAGCCGAGCACAAAAAAGGCATCGGAGGAGAGAACGGTCAAC |
| trpE flank 2 for | GTTGACCGTTCTCTCCTCCGATGCCTTTTTTGTGCTCGGCTGGAC |
| trpE flank 2 rev | CTTCTAGAGCTGTACAAGGAAGTGGAGATGTC |
| kynA flank 1 for | CTGGTACCCGACTTTTTCCGTTCCCTGC |
| kynA flank 1 rev | GTACTACAGGTTGGAACGGAGCGTGAATCTCCTGAATTCGGC |
| kynA flank 2 for | GCCGAATTCAGGAGATTCACGCTCCGTTCCAACCTGTAGTAC |
| kynA flank 2 rev | CTTCTAGAGACATGACCGACGACATCGAC |
| phnAB flank 1 for | CTGAATTCGGTCAGCAACCTGGAAATCG |
| phnAB flank 1 rev | CGATGATGAACATGCCGTTGCCATCCCGAGTCGATTCTCAC |
| phnAB flank 2 for | GTGAGAATCGACTCGGGATGGCAACGGCATGTTCATCATCG |
| phnAB flank 2 rev | CTTCTAGACGTAAACCTGAGGAGGTGAAC |
| pqsA flank 1 for | CTGAATTCCCAGAATTGCCACCAAGACTC |
| pqsA flank 1 rev | GTCCACATTGGCCAACCTGACCCCTTTATCACGACAACCTTC |
| pqsA flank 2 for | GAAGGTTGTCGTGATAAAGGGGTCAGGTTGGCCAATGTGGAC |
| pqsA flank 2 rev | CTTCTAGACTATGGCAAGGTGCAACAATGG |
| Deletion confirmation | |
| trpE for | GGTACCCTCGACAAGTTGC |
| trpE rev | CATTGGTGCTGGAACCGCTG |
| kynA for | CGTACTGCGTTGGTGATGG |
| kynA rev | CCTCCATCGCATTACTCAGG |
| phnAB for | CGTGAACATGTTCCTCCAGG |
| phnAB rev | GGATCGTCTGGGCAACATG |
| pqsA for | CCAGGCTGAACTCGTTCTCG |
| pqsA rev | GGTTTCCAAACGCAGCAACC |
| Expression vectors | |
| phnAB for | GACTAGTGCGCGCTAGCGTCGCGCAGG |
| phnAB rev | GCTCTAGACCTGGCAACCGAGCATCGTCG |
| trpE for | GCCTGCAGCGTTTGCACCCTGTTGACC |
| trpE rev | GCAGAGCGTCGAGTAAGACGGAAATCAAGAGGTTACAGCC |
| trpG for | GGCTGTAACCTCTTGATTTCCGTCTTACTCGACGCTCTGC |
| trpG rev | GCTCTAGAGGTTGACGATGCGATTGAGG |
| RT-PCR | |
| phnA for | CGTTGAACGCCAATGGACG |
| phnA rev | CGGTACGATCTGGAACACG |
| trpE for | CGTAGTGGTATTCGACAACC |
| trpE rev | CGATCAGCATCAGGTGCTCG |
| rplU for | CGCAGTGATTGTTACCGGTG |
| rplU rev | AGGCCTGAATGCCGGTGATC |
Growth curves.
To determine growth characteristics of PA14, ΔphnAB and ΔtrpE strains, cultures of each were grown overnight in MOPS minimal medium supplemented with 200 µM tryptophan. Overnight cultures were subcultured into fresh MOPS minimal medium supplemented with 200 µM tryptophan and grown to mid-exponential phase (OD600 0.05) at which point cells were washed twice, concentrated and starved for 2 h in MOPS buffer. Washed, starved, exponential-phase cells were used to inoculate 25 ml MOPS minimal medium with no tryptophan to OD600 0.01, and growth was measured by OD600 readings every 30 min upon the initiation of exponential growth. To demonstrate tryptophan-dependent growth of ΔtrpE, tryptophan was added to a final concentration of 200 µM three hours after inoculation. Growth was monitored for 8 h after inoculation, and the generation times for each culture were calculated. The growth curve was repeated three times.
PQS extractions.
Overnight cultures of indicated strains grown in MOPS minimal medium supplemented with 200 µM tryptophan when necessary were subcultured into fresh MOPS minimal medium to OD600 0.05 and grown to mid-exponential phase (OD600 0.5). Mid-exponential-phase cells were washed twice, concentrated and starved for 2 h in MOPS buffer. Washed, starved, exponential-phase cells were used to inoculate 12.5 ml MOPS minimal medium containing 20 mM succinate supplemented with 5 mM of the stated amino acid or anthranilate. Cultures were grown for 24 h (final OD600 values were consistently within a range of 1.5–2.0) and 10 ml were removed and extracted twice with an equal volume of ethyl acetate (Fisher) acidified with 150 µl glacial acetic acid l−1 (Fisher). Ethyl acetate extracts were dried completely under a constant stream of N2 gas and resuspended in methanol (Fisher). PQS in extracts was resolved by separation on thin layer chromatography plates (EMD) as described previously (Palmer et al., 2007, 2010, 2011). PQS spots were imaged with excitation by long-wavelength UV light, and spots were quantified by densitometry using GeneTools software compared with a standard curve of synthetic PQS standards spotted on the same plate. For TLC images, 250 ng synthetic PQS or anthranilate were spotted on the TLC plate as migration standards.
Overexpression of TrpEG and PhnAB.
Overexpression constructs were generated by PCR amplification of the trpE and trpG genes and the phnAB operon using PA14 genomic DNA as template and primers listed in Table 2. Because the trpE and trpG genes are not adjacent on the P. aeruginosa chromosome (Essar et al., 1990b), overlap extension PCR was used to generate a fused construct. This was not necessary for phnAB, which are present in an operon (Cao et al., 2001; Essar et al., 1990a). The trpEG and phnAB PCR products were digested with PstI and XbaI or SpeI and XbaI (see restriction sites in Table 2), respectively, and ligated into the arabinose-inducible vector pJN105 to generate pGP-trpEG and pKP-phnAB (Table 1). Expression constructs and empty pJN105 were introduced into relevant P. aeruginosa strains by MgCl2 transformation as described previously (Whiteley et al., 2000).
To determine whether overexpression of PhnAB could restore growth to PA14 ΔtrpE, washed, starved, exponential-phase ΔtrpE cells containing pKP-phnAB or empty pJN105 were prepared as described above and used to inoculate 3 ml MOPS minimal medium with 0.5 % arabinose. After overnight growth (~16 h), a photograph was taken to demonstrate final growth yield. To determine whether overexpression of TrpEG restores PQS production in a minimal medium, washed, starved, exponential-phase PA14 and ΔphnAB cells containing either pGP-trpEG or empty pJN105 were prepared as described above and used to inoculate 12.5 ml MOPS minimal medium with 0.5 % arabinose. PQS was extracted and quantified as described above.
Tryptophan molar growth yield and semiquantitative RT-PCR.
To determine the tryptophan molar growth yield for ΔtrpE, washed, starved, exponential-phase ΔtrpE, ΔtrpE ΔphnAB or ΔtrpE ΔpqsA cells were prepared as described above and used to inoculate 100 µl MOPS minimal medium supplemented with increasing concentrations of tryptophan from 0 to 200 µM in the wells of a Nunc 96-well plate. Each well contained a borosilicate glass bead (Fisher) to improve aeration. Plates were grown overnight (~16 h) after which glass beads were removed and the volume in each well was increased to 200 µl by addition of MOPS buffer. Final growth yield for each well was determined by measuring OD600 using a BioTek SynergyMx 96-well plate reader with Gen5 software. Growth yields were averaged from five wells for each tryptophan concentration, and each plate set containing the entire range of tryptophan concentrations was repeated three times with unique biological replicates each time. For experiments with exogenous PQS, the same protocol and media were used with the addition of 20 µM PQS throughout growth and starvation. Reported OD600 readings are corrected for dilution in MOPS buffer and spectrophotometric path length.
To determine whether phnAB expression correlates with abolishment of ΔtrpE tryptophan auxotrophy, washed, starved, exponential-phase ΔtrpE cells were inoculated into 96-well plates as described above except that all wells contained MOPS minimal medium supplemented with 200 µM tryptophan. The RNA-stabilizing agent RNALater (Ambion) was added 1 : 1 (v/v) to wells containing cultures grown to OD600 0.07, 0.2 and 1.0. To determine trpE expression levels, washed, starved, exponential-phase PA14 cells were inoculated into 25 ml MOPS minimal medium and RNALater was added 1 : 1 (v/v) to cultures grown to OD600 0.15, 0.30 and 1.8. For all RT-PCR experiments, total RNA was extracted using RNA bee (Tel-test) as per the manufacturer’s instructions. DNA contaminants were removed by DNase treatment (Promega) and confirmed by PCR amplification of the rplU gene using primers listed in Table 2 as previously described (Palmer et al., 2010, 2007; Schuster et al., 2003). RNA integrity was confirmed by gel electrophoresis, and cDNA was synthesized using a SuperScript II First-Strand cDNA synthesis kit (Invitrogen) with random primers as described previously (Palmer et al., 2010, 2007; Schuster et al., 2003). cDNA was purified and used as the template for semiquantitative RT-PCR using phnA or trpE primers listed in Table 2 to PCR amplify phnA from 25 ng or trpE from 20 ng cDNA, and rplU primers listed in Table 2 to PCR amplify rplU from 5 ng cDNA as a constitutively expressed loading control. RNA alone and genomic DNA served as negative and positive controls, respectively.
Results and discussion
Evidence that phnAB was acquired by horizontal gene transfer
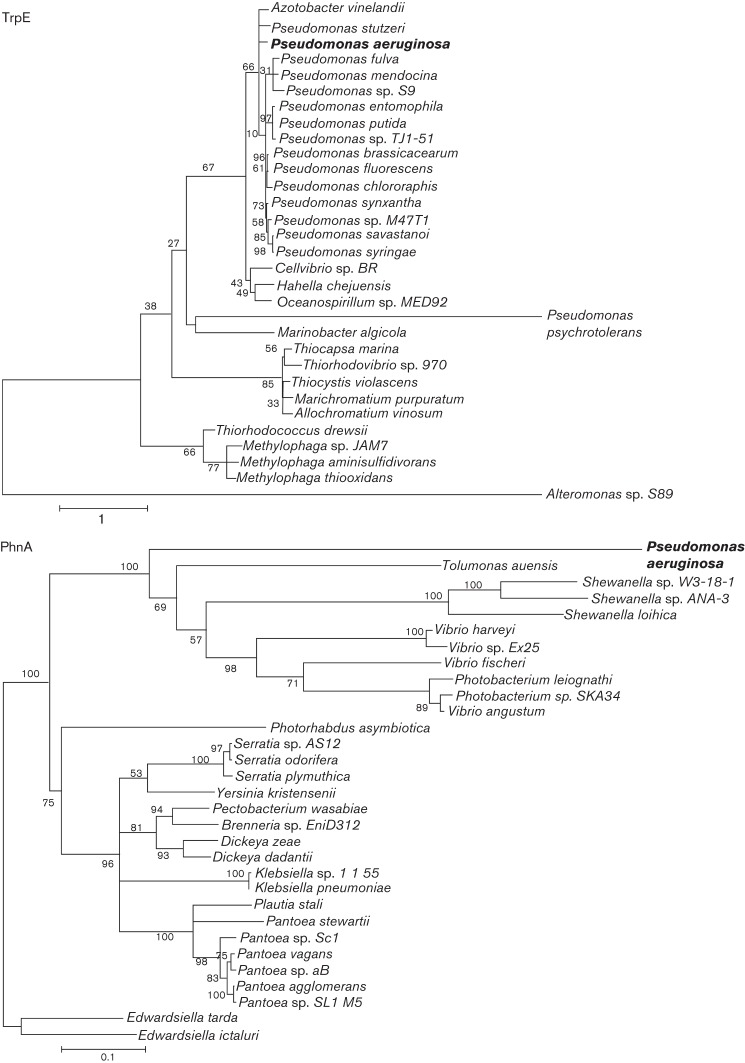
P. aeruginosa appears to be rare in that it possesses two functional anthranilate synthases, and it is probably not a coincidence that this bacterium also produces over 50 unique quinolone compounds that require anthranilate for biosynthesis (Essar et al., 1990a, b; Lépine et al., 2004). A complete quinolone biosynthesis operon has been identified in Burkholderia pseudomallei, Burkholderia thailandensis and Burkholderia ambifaria, though these organisms produce fewer quinolones than P. aeruginosa (Diggle et al., 2006; Lépine et al., 2004; Vial et al., 2008). Pseudomonas putida also contains quinolone biosynthesis genes throughout its genome (Diggle et al., 2006). Among the quinolones reportedly produced by these organisms is the immediate precursor to PQS, 2-heptyl-4-quinoline (HHQ) (Diggle et al., 2006); however, a blast analysis using the P. aeruginosa PA14 TrpEG and PhnAB sequences performed for this work indicated that these organisms only possess homologues of TrpEG. As a first step toward determining how P. aeruginosa came to possess two anthranilate synthases, we used blast analysis of the TrpE and PhnA protein sequences to identify the most closely related anthranilate synthase homologues. Identified sequences were aligned and used to generate phylogenetic trees. Our results demonstrate that TrpE shares significant homology with the tryptophan biosynthesis enzymes of closely related species including many pseudomonads (Fig. 1). By contrast, the protein sequence of PhnA was most closely related to multiple divergent species (Fig. 1). The GC content of phnA (68 %) is similar to that of the rest of the P. aeruginosa genome (66 %). By including many new genomes which have been sequenced since 2004, these results extend the findings of a study by Xie and colleagues indicating that P. aeruginosa acquired the phnAB operon from the tryptophan biosynthetic machinery of another bacterium via horizontal gene transfer and that this event probably occurred after the diversification of fluorescent pseudomonads, as no close relatives of P. aeruginosa possess a phnAB-like operon (Merino et al., 2008; Xie et al., 2004).
Fig. 1.
Anthranilate synthase phylogenetic trees. The top 30 homologues identified by blast analysis using the P. aeruginosa TrpE and PhnA sequences reveal the evolutionary relationships of each anthranilate synthase enzyme to those of other species. TrpEG are most closely related to anthranilate synthases from other members of the fluorescent pseudomonad family, while PhnAB are most closely related to anthranilate synthases from more distantly related organisms. The absence of a phnAB-like operon in other pseudomonads is evidence that PhnAB acquisition occurred after the family’s diversification. Bootstrap values at tree nodes indicate likelihood the node represents a genuine phylogenetic relationship.
TrpE is required for growth without tryptophan
It has previously been reported that a mutation in trpE results in tryptophan auxotrophy in P. aeruginosa, while phnA and phnB mutants retain the ability to grow in the absence of tryptophan (Essar et al., 1990a, b). To determine whether unmarked deletion mutants also displayed these phenotypes, ΔtrpE and ΔphnAB mutants were generated. P. aeruginosa PA14, ΔtrpE and ΔphnAB were grown to exponential phase, washed, starved and inoculated into MOPS minimal medium with no tryptophan, and growth was monitored over 8 h. P. aeruginosa ΔphnAB grew at the same rate without tryptophan, however, only upon addition of tryptophan to a final concentration of 200 µM after 3 h of non-growth was ΔtrpE able to grow at the same rate as wild-type P. aeruginosa (Fig. 2). These results demonstrated that ΔtrpE is a tryptophan auxotroph and that endogenous phnAB is unable to complement tryptophan auxotrophy in ΔtrpE.
Fig. 2.
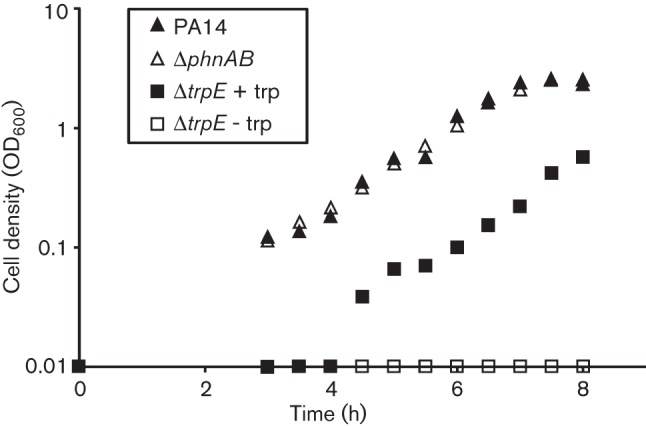
Determination of ΔtrpE tryptophan auxotrophy. Washed, starved, exponential-phase PA14, ΔphnAB and ΔtrpE strains were grown in MOPS minimal medium with no tryptophan. Wild-type growth was observed for ΔphnAB; however, ΔtrpE did not grow until the addition of tryptophan to a final concentration of 200 µM after three hours of non-growth. ΔtrpE did not grow in the absence of tryptophan.
PhnAB supplies anthranilate for PQS production
P. aeruginosa enhances production of PQS in the presence of aromatic amino acids (Farrow & Pesci, 2007; Palmer et al., 2007, 2005). For tryptophan, catabolism via the products of the kynABU genes generates anthranilate for PQS production (Farrow & Pesci, 2007); however the role of PhnAB during growth with tryptophan was not known. Examination of PQS production in the presence of tryptophan revealed that wild-type P. aeruginosa and ΔphnAB produced ~6 and ~3.5 µM PQS, respectively (Fig. 3a). The decreased levels of PQS in ΔphnAB suggest that PhnAB generates anthranilate for PQS biosynthesis even in the presence of high levels of tryptophan, indicating that degradation of tryptophan via KynABU is not the only source of anthranilate when the amino acid is in excess (Farrow & Pesci, 2007). This was confirmed as ΔkynA, which is unable to degrade tryptophan and ΔtrpE ΔkynA produced low levels (~1 µM) of PQS in the presence of 5 mM tryptophan (Fig. 3a). These experiments were performed using fresh tryptophan stock solutions to minimize possible confounding effects of spontaneous tryptophan degradation in solution. Because PhnAB is the only known source of anthranilate in ΔtrpE ΔkynA, it is likely that PhnAB is a source of anthranilate for quinolone biosynthesis in the presence of tryptophan; however, construction of a trpE kynA phnAB quadruple mutant would be necessary to confirm this.
Fig. 3.
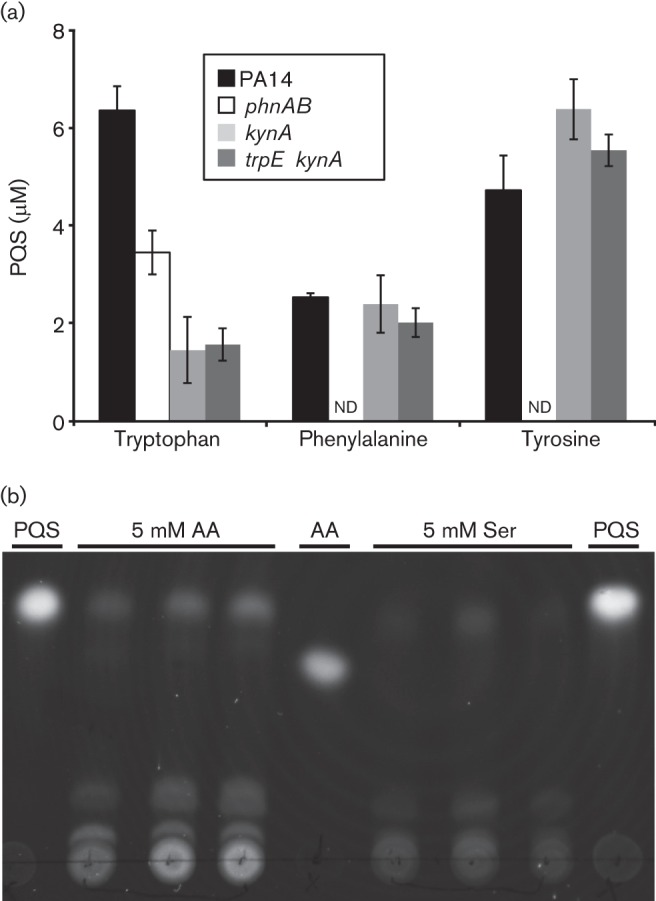
PQS production in the presence of aromatic amino acids and anthranilate. (a) PQS was extracted and quantified from whole cultures of PA14, ΔphnAB, ΔkynA and ΔtrpE ΔkynA grown for 24 h in MOPS minimal medium supplemented with 5 mM of the indicated aromatic amino acids. ND indicates no detectable levels of PQS and error bars represent sem. (b) PQS was extracted from whole cultures of ΔphnAB grown for 24 h in MOPS minimal medium supplemented with 5 mM anthranilate or serine. Extracts were separated by TLC and imaged under long-wave UV light. Extractions from three independently grown cultures are shown for anthranilate and serine addition. 250 ng synthetic PQS (PQS) or anthranilate (AA) were included as TLC migration standards.
Our group has previously reported that in addition to tryptophan, phenylalanine and tyrosine also enhance PQS production (Palmer et al., 2007, 2005). However unlike tryptophan, this induction is not due to catabolism of these amino acids to anthranilate, but instead increased flux of shared precursors of anthranilate biosynthesis to PQS biosynthesis. Thus, we hypothesized that deletion of phnAB would completely eliminate PQS production in the presence of phenylalanine and tyrosine unless trpEG could complement this deletion. To test this, PQS was extracted and quantified from washed, starved, exponential-phase cells inoculated into MOPS minimal medium supplemented with 5 mM phenylalanine or tyrosine. P. aeruginosa PA14, ΔtrpE ΔkynA and ΔkynA produced ~2 µM PQS in the presence of 5 mM phenylalanine and ~5–6 µM PQS in the presence of 5 mM tyrosine (Fig. 3a). No detectable PQS was produced by ΔphnAB, indicating that PhnAB is the anthranilate synthase responsible for PQS production in the presence of phenylalanine and tyrosine (Fig. 3a). Consistent with Farrow and Pesci’s observations, ΔphnAB PQS production is restored upon growth with anthranilate (Fig. 3b) (Palmer et al., 2010; Farrow & Pesci, 2007; Palmer et al., 2007). Taken together, these experiments indicate that TrpEG-generated anthranilate is used for tryptophan biosynthesis while PhnAB-generated anthranilate is used for quinolone biosynthesis.
Overexpression of either anthranilate synthase complements the loss of the other
The inability of these seemingly redundant anthranilate synthases to compensate for each other in genetic experiments is an interesting conundrum. In order to determine whether it is possible for anthranilate to cross pathways, overexpression constructs were generated for TrpEG and PhnAB and their ability to cross complement each of the respective anthranilate synthase mutant phenotypes was tested (Fig. 4). Overexpression of PhnAB restored growth of ΔtrpE in the absence of tryptophan, while no growth was observed for ΔtrpE cells containing empty vector (Fig. 4a). To determine whether PQS production could be restored in ΔphnAB, PQS was extracted and quantified from ΔphnAB cells overexpressing TrpEG or containing empty vector. Expression of TrpEG in wild-type P. aeruginosa and ΔphnAB resulted in high levels of PQS production (Fig. 4b). This effect was dependent upon TrpEG, as ΔphnAB cells containing empty vector did not produce detectable levels of PQS (Fig. 4b). These experiments indicate that it is possible for metabolic crosstalk to occur between the tryptophan and quinolone biosynthesis pathways, however, why this does not occur with the endogenous enzymes remains unclear.
Fig. 4.
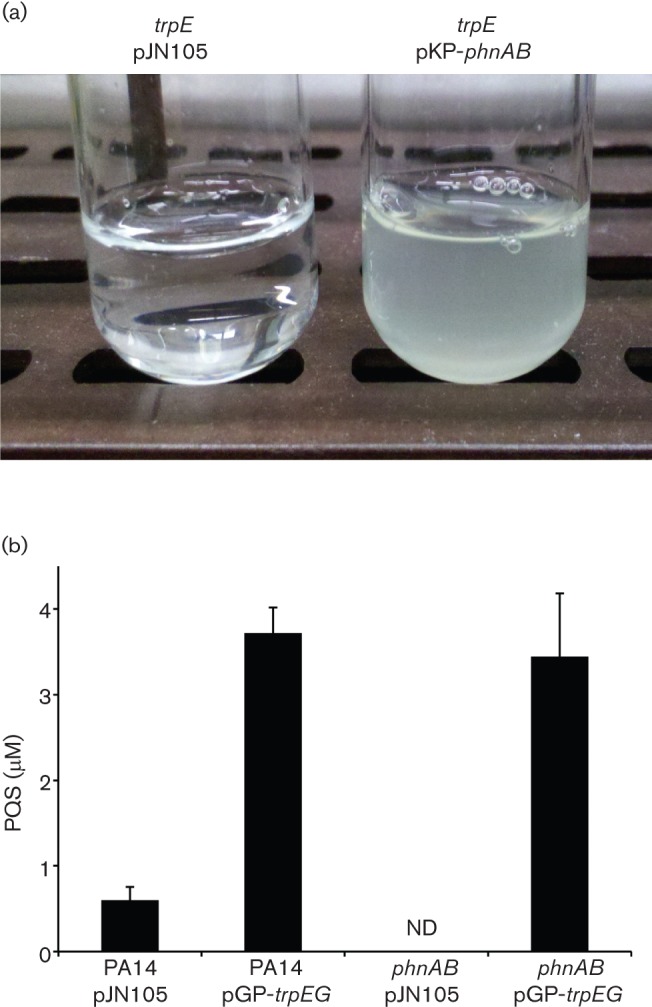
Overexpression of anthranilate synthase genes results in pathway crosstalk. (a) Overexpression of phnAB in ΔtrpE restored growth in MOPS minimal medium without tryptophan, while empty vector did not. (b) Overexpression of fused trpEG restored the ability of ΔphnAB to generate PQS in a minimal medium, while empty vector did not. ND indicates no detectable levels of PQS and error bars represent sem.
ΔtrpE tryptophan auxotrophy is dependent upon phnAB expression
One possible explanation for the lack of crosstalk of TrpEG- and PhnAB-generated anthranilate is that PhnAB is expressed at levels too low to compensate for loss of TrpEG. Expression of phnAB is cell-density dependent, showing increased expression at mid- to late-exponential phase (Cao et al., 2001); thus it is possible that sufficient PhnAB levels are not present at low cell densities to complement ΔtrpE-mediated tryptophan auxotrophy. However at high cell densities, we hypothesize that tryptophan auxotrophy would be eliminated in ΔtrpE due to increased phnAB expression. To test this hypothesis, washed, starved, exponential-phase ΔtrpE cells were grown overnight in MOPS minimal medium supplemented with tryptophan at concentrations ranging from 0 to 200 µM and final growth yields were determined. Results revealed that ΔtrpE final growth yields were dependent upon tryptophan concentrations until mid-exponential phase, at which point ΔtrpE reached maximal growth yields regardless of the concentration of tryptophan in the medium (Fig. 5a). When phnAB was deleted in ΔtrpE, growth yields remained dependent on the tryptophan concentration in the medium (Fig. 5a).
Fig. 5.
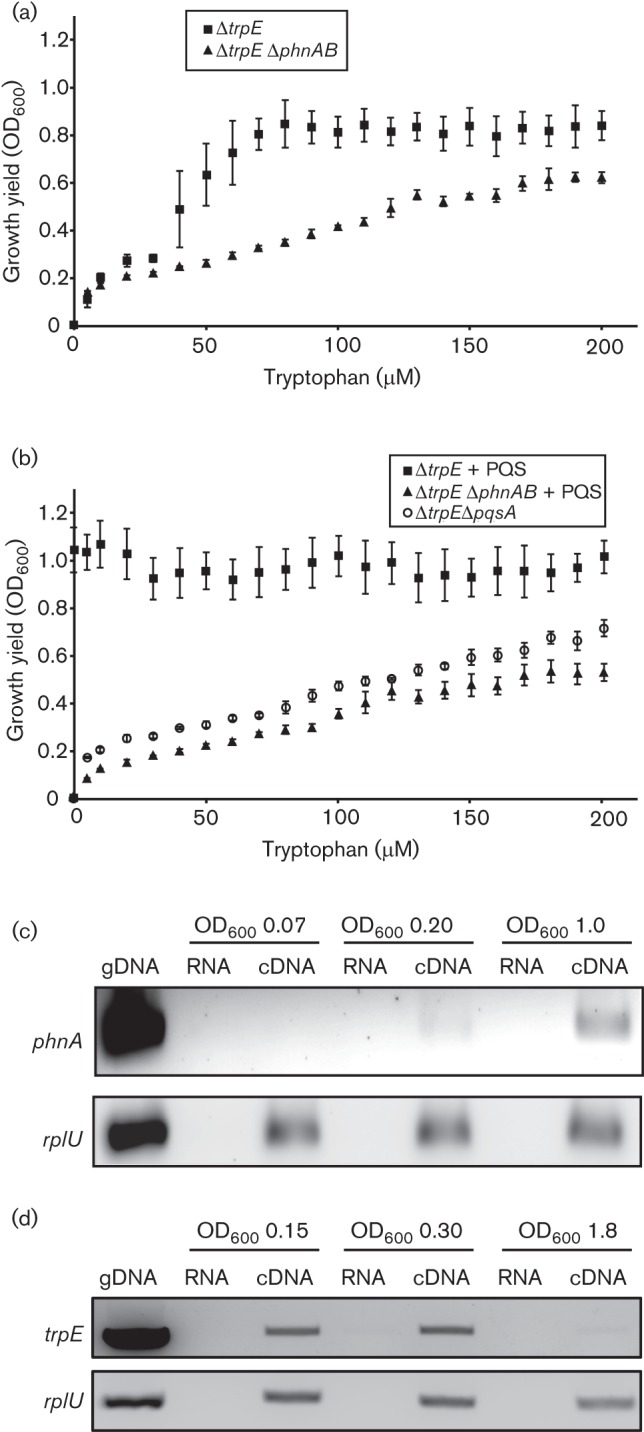
ΔtrpE tryptophan auxotrophy is dependent upon phnAB expression. (a) The final growth yields of ΔtrpE and ΔtrpE ΔphnAB on increasing concentrations of tryptophan were determined from OD600. ΔtrpE growth became tryptophan-independent upon reaching OD600 ~ 0.5. Growth of ΔtrpE ΔphnAB was tryptophan-dependent. (b) Exogenous addition of 20 µM PQS rescued ΔtrpE tryptophan auxotrophy, while ΔtrpE ΔphnAB remained auxotrophic for tryptophan in the presence of PQS. Growth yields were also tryptophan-dependent in ΔtrpE ΔpqsA, which is unable to generate PQS. (c) RT-PCR confirms that phnA expression levels correspond to the loss of ΔtrpE tryptophan auxotrophy. Expression of phnA was analysed at three stages of growth (OD600 0.07, 0.2 and 1.0) by PCR amplification from 25 ng cDNA, and levels were compared with constitutive expression of rplU, amplified from 5 ng cDNA. Genomic DNA (gDNA) and RNA served as positive and negative controls, respectively. Gel images are inverted for clarity. (d) RT-PCR confirms that trpE expression levels decrease late in growth when phnAB expression levels are increasing. Expression of trpE was analysed at OD600 0.15, 0.3 and 1.8 by PCR amplification from 20 ng cDNA, and levels are compared with constitutive expression of rplU, amplified from 5 ng cDNA. gDNA and RNA serve as positive and negative controls, respectively. Gel images are inverted for clarity.
The previous results suggest that PhnAB is a source of anthranilate for tryptophan biosynthesis at high cell densities. If this is the case, we hypothesized that addition of PQS to low-density cultures would eliminate the ΔtrpE auxotrophy since phnAB is induced by PQS. In support of this hypothesis, induction of phnAB expression with a physiologically relevant concentration of PQS (20 µM) resulted in complete rescue of tryptophan auxotrophy in ΔtrpE, as ΔtrpE reached maximal growth yields regardless of the concentration of tryptophan in the medium (Fig. 5b). To further confirm the ability of PQS-dependent PhnAB expression to mitigate ΔtrpE tryptophan auxotrophy, ΔtrpE ΔpqsA was generated. Because the pqsA gene product is required for PQS biosynthesis, its deletion renders this strain unable to generate PQS endogenously (Bredenbruch et al., 2005; Coleman et al., 2008; Mashburn & Whiteley, 2005). In the absence of endogenous PQS, ΔtrpE ΔpqsA growth yields were similar to those of ΔtrpE ΔphnAB, and thus, were entirely dependent on the concentration of tryptophan in the medium (Fig. 5b). As a final line of evidence for the correlation of ΔtrpE tryptophan auxotrophy and low levels of phnAB expression, RT-PCR was used to examine phnA expression levels during growth in the presence of tryptophan, compared with the constitutively expressed rplU gene. Consistent with other PQS-regulated genes (Cao et al., 2001; Xiao et al., 2006), phnA displayed low levels of expression in lag (OD600 0.07) and early exponential (OD600 0.20) phases of growth; however, expression increased substantially as density increased (OD600 1.0) (Fig. 5c). Importantly, as previously shown, trpE expression levels were high early in growth (OD600 0.15 and 0.30) and low upon reaching stationary phase (OD600 1.8) (Fig. 5d) (Essar et al., 1990a). Taken together these results are consistent with cell-density-dependent PhnAB expression rescuing ΔtrpE tryptophan auxotrophy.
Conclusions
P. aeruginosa has been known to possess two anthranilate synthases for over 20 years, and the unique phenotypes resulting from mutations in the anthranilate synthase genes suggested that TrpEG generates anthranilate exclusively for tryptophan biosynthesis while PhnAB generates anthranilate exclusively for quinolone biosynthesis (Essar et al., 1990a, b). Here we extend this work by presenting additional evidence that phnAB was acquired through horizontal gene transfer following the diversification of fluorescent pseudomonads (Merino et al., 2008; Xie et al., 2004). Based on its organization in an operon and quinolone-dependent regulation, PhnAB has probably evolved to generate anthranilate for quinolone biosynthesis (Essar et al., 1990a, b; Merino et al., 2008), and this is consistent with the ability of P. aeruginosa to produce more quinolones than its quinolone-producing relatives (Lépine et al., 2004; Vial et al., 2008). Furthermore, our data suggest that differential expression of trpEG and phnAB explains the observed anthranilate synthase mutant phenotypes. While our data do not rule out the physical sequestration of tryptophan and quinolone biosynthesis precursors via channelling or some other mechanism, the ability of exogenous anthranilate to restore PQS production in ΔphnAB suggests this is not the case (Fig. 3b) (Farrow & Pesci, 2007). Regardless, the importance of temporal expression of phnAB is clear, as phnAB expression levels are the primary determinant of ΔtrpE tryptophan auxotrophy (Fig. 5). Additionally, trpE and trpG expression levels are decreased at high cell densities (Fig. 5d) (Essar et al., 1990a), which leads to a model for P. aeruginosa anthranilate production in which density-dependent expression of PhnAB explains the apparent lack of redundancy between TrpEG and PhnAB. At low cell densities PhnAB expression is low and TrpEG is the primary anthranilate synthase enzyme present. As PQS-mediated phnAB expression is activated at higher cell densities, PhnAB becomes the primary anthranilate synthase enzyme present. The results presented here underscore the importance of studying basic microbial physiology and metabolism to understand bacterial signalling and pathogenesis.
Acknowledgements
This work was supported by a grant from the National Institutes For Health (NIH) (5R01AI075068 to M. W.). M. W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease. We thank Kelli L. Palmer for generating the phnAB overexpression construct and Taejoon Kwon for assistance with bioinformatic analyses.
Abbreviations:
- CF
cystic fibrosis
- gDNA
genomic DNA
- PQS
Pseudomonas quinolone signal
- QS
quorum sensing
References
- Ausubel F. M. (2002). Short Protocols in Molecular Biology: a Compendium of Methods from Current Protocols in Molecular Biology, 5th edn New York: Wiley. [Google Scholar]
- Bredenbruch F., Nimtz M., Wray V., Morr M., Müller R., Häussler S. (2005). Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol 187, 3630–3635. 10.1128/JB.187.11.3630-3635.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee M. W., Coleman J. P., Pesci E. C. (2001). Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 98, 11633–11637. 10.1073/pnas.201328498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Krishnan G., Goumnerov B., Tsongalis J., Tompkins R., Rahme L. G. (2001). A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98, 14613–14618. 10.1073/pnas.251465298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. P., Hudson L. L., McKnight S. L., Farrow J. M., III, Calfee M. W., Lindsey C. A., Pesci E. C. (2008). Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol 190, 1247–1255. 10.1128/JB.01140-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Timmis K. N. (1994). Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235, 386–405. 10.1016/0076-6879(94)35157-0 [DOI] [PubMed] [Google Scholar]
- Delhaes L., Monchy S., Fréalle E., Hubans C., Salleron J., Leroy S., Prevotat A., Wallet F., Wallaert B., et al. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS ONE 7, e36313. 10.1371/journal.pone.0036313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E., Gopalan S., Tampakaki A. P., Lépine F., Padfield K. E., Saucier M., Xiao G., Rahme L. G. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol Microbiol 55, 998–1014. 10.1111/j.1365-2958.2004.04448.x [DOI] [PubMed] [Google Scholar]
- Diggle S. P., Lumjiaktase P., Dipilato F., Winzer K., Kunakorn M., Barrett D. A., Chhabra S. R., Cámara M., Williams P. (2006). Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol 13, 701–710. 10.1016/j.chembiol.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Emerson J., Rosenfeld M., McNamara S., Ramsey B., Gibson R. L. (2002). Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34, 91–100. 10.1002/ppul.10127 [DOI] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Hadero A., Crawford I. P. (1990a). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172, 884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Han C. Y., Crawford I. P. (1990b). DNA sequences and characterization of four early genes of the tryptophan pathway in Pseudomonas aeruginosa. J Bacteriol 172, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. M., III, Pesci E. C. (2007). Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. J Bacteriol 189, 3425–3433. 10.1128/JB.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Manoil C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol 183, 6207–6214. 10.1128/JB.183.21.6207-6214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., McKnight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184, 6472–6480. 10.1128/JB.184.23.6472-6480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison F. (2007). Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923. 10.1099/mic.0.2006/004077-0 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hoiby N., Flensborg E. W., Beck B., Friis B., Jacobsen S. V., Jacobsen L. (1977). Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of Pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. Scand J Respir Dis 58, 65–79. [PubMed] [Google Scholar]
- Lépine F., Milot S., Déziel E., He J., Rahme L. G. (2004). Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom 15, 862–869. 10.1016/j.jasms.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Liberati N. T., Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F. M. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103, 2833–2838. 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn L. M., Whiteley M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- Mavrodi D. V., Bonsall R. F., Delaney S. M., Soule M. J., Phillips G., Thomashow L. S. (2001). Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183, 6454–6465. 10.1128/JB.183.21.6454-6465.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E., Jensen R. A., Yanofsky C. (2008). Evolution of bacterial trp operons and their regulation. Curr Opin Microbiol 11, 78–86. 10.1016/j.mib.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. R., Fuqua C. (1999). Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227, 197–203. 10.1016/S0378-1119(98)00601-5 [DOI] [PubMed] [Google Scholar]
- Palmer K. L., Mashburn L. M., Singh P. K., Whiteley M. (2005). Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 187, 5267–5277. 10.1128/JB.187.15.5267-5277.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K. L., Aye L. M., Whiteley M. (2007). Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189, 8079–8087. 10.1128/JB.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G. C., Palmer K. L., Jorth P. A., Whiteley M. (2010). Characterization of the Pseudomonas aeruginosa transcriptional response to phenylalanine and tyrosine. J Bacteriol 192, 2722–2728. 10.1128/JB.00112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G. C., Schertzer J. W., Mashburn-Warren L., Whiteley M. (2011). Quantifying Pseudomonas aeruginosa quinolones and examining their interactions with lipids. Methods Mol Biol 692, 207–217. 10.1007/978-1-60761-971-0_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E. C., Milbank J. B., Pearson J. P., McKnight S., Kende A. S., Greenberg E. P., Iglewski B. H. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96, 11229–11234. 10.1073/pnas.96.20.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh K. P., Griswold J. A., Iglewski B. H., Hamood A. N. (1999). Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun 67, 5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schuster M., Greenberg E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296, 73–81. 10.1016/j.ijmm.2006.01.036 [DOI] [PubMed] [Google Scholar]
- Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185, 2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial L., Lépine F., Milot S., Groleau M. C., Dekimpe V., Woods D. E., Déziel E. (2008). Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol 190, 5339–5352. 10.1128/JB.00400-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Lee K. M., Greenberg E. P. (1999). Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96, 13904–13909. 10.1073/pnas.96.24.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Parsek M. R., Greenberg E. P. (2000). Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol 182, 4356–4360. 10.1128/JB.182.15.4356-4360.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., He J., Rahme L. G. (2006). Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152, 1679–1686. 10.1099/mic.0.28605-0 [DOI] [PubMed] [Google Scholar]
- Xie G., Bonner C. A., Song J., Keyhani N. O., Jensen R. A. (2004). Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy. BMC Biol 2, 15. 10.1186/1741-7007-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]