Abstract
HU is a non-sequence-specific DNA-binding protein and one of the most abundant nucleoid-associated proteins in the bacterial cell. Like Escherichia coli, the genome of Porphyromonas gingivalis is predicted to encode both the HUα (PG1258) and the HUβ (PG0121) subunit. We have previously reported that PG0121 encodes a non-specific DNA-binding protein and that PG0121 is co-transcribed with the K-antigen capsule synthesis operon. We also reported that deletion of PG0121 resulted in downregulation of capsule operon expression and produced a P. gingivalis strain that is phenotypically deficient in surface polysaccharide production. Here, we show through complementation experiments in an E. coli MG1655 hupAB double mutant strain that PG0121 encodes a functional HU homologue. Microarray and quantitative RT-PCR analysis were used to further investigate global transcriptional regulation by HUβ using comparative expression profiling of the PG0121 (HUβ) mutant strain to the parent strain, W83. Our analysis determined that expression of genes encoding proteins involved in a variety of biological functions, including iron acquisition, cell division and translation, as well as a number of predicted nucleoid associated proteins were altered in the PG0121 mutant. Phenotypic and quantitative real-time-PCR (qRT-PCR) analyses determined that under iron-limiting growth conditions, cell division and viability were defective in the PG0121 mutant. Collectively, our studies show that PG0121 does indeed encode a functional HU homologue, and HUβ has global regulatory functions in P. gingivalis; it affects not only production of capsular polysaccharides but also expression of genes involved in basic functions, such as cell wall synthesis, cell division and iron uptake.
Introduction
Porphyromonas gingivalis is a Gram-negative obligate anaerobe belonging to the family Bacteroidaceae that persists as a natural member of the human oral microbiota. A shift in the microbial community leading to outgrowth of this anaerobe is directly linked to periodontitis, a chronic inflammatory disease that leads to destruction of the tissues supporting the gums and ultimately, exfoliation of the teeth (Choil et al., 1990; Dzink et al., 1988; Grossi et al., 1994; Lamont & Jenkinson, 2000; Moore et al., 1991). This commensal can colonize, invade and multiply within gingival epithelial cells, as well as penetrate into deeper epithelial cell layers, potentially releasing the whole organism and/or virulence factors into the bloodstream (reviewed by Yilmaz, 2008). In addition to its ability to cause disease in the oral cavity, there are data indicating a role in systemic disease, including its ability to invade vascular endothelial cells (Dorn et al., 2000, 2002; Jandik et al., 2008) and to cause aggregation of platelets (Pham et al., 2002). For many pathogenic bacteria, surface polysaccharides play a key role in immune modulation and evasion (Comstock & Kasper, 2006). It has recently been shown that the K-antigen capsule of P. gingivalis aids in immune evasion, promoting survival of the bacterium within host cells and increasing virulence (Singh et al., 2011). Previously, we have shown that expression of PG0121 affects synthesis of K-antigen capsule (Alberti-Segui et al., 2010). PG0121 is predicted to encode the nucleoid-associated DNA-binding protein HU.
The prokaryotic nucleoid is a defined, yet dynamic, structure, organized by complex interactions with nucleoid-associated proteins (NAPs) (Dillon & Dorman, 2010). HU protein is one of the most abundant NAPs in the bacterial cell. It exists in one of its homodimeric forms (HUα2 or HUβ2) or its heterodimeric form (HUαβ) in virtually all bacterial species (Grove, 2011). In Escherichia coli, HU proteins are differentially expressed during different phases of growth (Ali Azam et al., 1999) and recently it has been determined that this is also true for the bacterium Legionella pneumophila (Morash et al., 2009). With regard to function, HU has been shown to compact and prevent denaturation of DNA under detrimental conditions (Drlica & Rouviere-Yaniv, 1987; Oberto et al., 2009; Swinger & Rice, 2004) and to be involved in changing DNA topology (Swinger & Rice, 2004), specifically introducing negative supercoiling into relaxed DNA in the presence of topoisomerase I, in vitro (Bensaid et al., 1996). HU typically acts as an accessory protein in virtually all types of nucleoprotein-mediated processes (reviewed by Drlica & Rouviere-Yaniv, 1987; Grove, 2011); such as acting as a cofactor to either stimulate or repress transcription (Aki et al., 1996; Morales et al., 2002) or aiding in translation (Balandina et al., 2001). HU proteins bind with low affinity and without sequence specificity to both double-stranded DNA and RNA. However, these proteins have high affinity for cruciform DNA structures or DNA molecules with a nick or a gap (Bonnefoy et al., 1994; Pinson et al., 1999; Pontiggia et al., 1993), i.e. molecules that are either in a bend conformation or readily bendable. Furthermore, HU binds to nicked or gapped DNA–RNA hybrids and to small non-coding RNA molecules (Balandina et al., 2002).
Loss of HU function in E. coli results in a mild phenotype in laboratory strains (Grove, 2011), while HU appears to be essential in Bacillus subtilis, a result shared with all Gram-positive organisms tested to date (Fernández et al., 1997; Köhler & Marahiel, 1998; Swinger & Rice, 2004), with the exception of Mycobacterium smegmatis (Whiteford et al., 2011). This difference is, in part, due to a lack of functional redundancy in Gram-positive bacteria, while E. coli expresses a wide variety of NAPs, including H-NS, Fis, Dps and IHF (Thanbichler et al., 2005). Yet, even with a wide variety of functional NAP homologues, E. coli cells lacking HU do exhibit a variety of growth defects. Strains with mutations in both the α and β-subunits have increased sensitivity to UV and ionizing radiation (Boubrik & Rouviere-Yaniv, 1995; Li & Waters, 1998; Miyabe et al., 2000), have a lethal phenotype following cold or heat shock (Giangrossi et al., 2002; Wada et al., 1988), are defective in cell division (Dri et al., 1991) and are altered in outer-membrane protein composition (Painbeni et al., 1997). Recently, the HU regulon was determined in E. coli using transcription profiling of strains deficient in the alpha, beta, or both HU subunits (Oberto et al., 2009). This study discovered that HU affects expression of 8 % of the E. coli genome.
Similar to E. coli, the genome of P. gingivalis is predicted to encode both the HUα (PG1258) and HUβ (PG0121) subunits. Interestingly, there are 12 genes in the P. gingivalis W83 genome (PG0222, PG0330, PG0853, PG1276, PG2040, PG2152, PG0555, PG0566, PG1389, PG1205, PG1497, as well as an additional gene pgin_c_1_6688 annotated by BROP) that are annotated as histone-like DNA-binding proteins that are related to but longer (~160 amino acids) than HU (~90 amino acids), with the only exception being pgin_c_1_6688, which is 89 amino acids. These proteins have distinct domain architecture when compared not only with HU but also with other members of the DNABII family of proteins, such as IHF. Therefore, these proteins have been classified within a separate superfamily (TIGR01201), with the only other identified member found in the closely related gut bacterium, Bacteroides fragilis.
Here, we have established that PG0121 encodes a functional homologue of the E. coli HU protein. Additionally, we show that PG0121 is differentially expressed at the level of transcription during different phases of growth, and we provide further findings regarding the effect of a PG0121 (HUβ) deletion on global gene expression. Our studies show that deletion of PG0121 affects transcript levels of a diverse array of genes when the cells are in mid-exponential growth phase; including those predicted to encode proteins involved in surface polysaccharide synthesis, cell-division, iron uptake, translation and DNA binding. In addition, we provide evidence that HUβ plays an important role in cell survival under iron-limiting conditions. Given functional and sequence similarity, our working hypothesis is that PG0121 encodes HUβ, and this NAP functions as an accessory protein to stimulate or repress transcription. Collectively, our studies have established that expression of the HUβ subunit of P. gingivalis affects global gene expression, and NAPs may be a key mechanism by which this bacterium controls the switch from life as a quiescent commensal to a destructive pathogen. Global regulation by NAPs and the potential functional redundancy of NAPs in P. gingivalis are discussed.
Methods
Bacterial strains, media and primers.
Bacterial strains and plasmids used in this study are shown in Table 1. P. gingivalis strains were maintained in a COY anaerobic chamber on trypticase soy agar plates (BAPHK), containing haemin (1 µg ml−1) and menadione (1 µg ml−1) and supplemented with 5 % defibrinated sheep blood (North-East Laboratory). For liquid culture the strains were grown in trypticase soy broth (TSBHK). Antibiotics were added at the following concentrations: E. coli, ampicillin (Amp) 100 µg ml−1, erythromycin (Erm) 200 µg ml−1, kanamycin (Km) 50 µg ml−1, chloramphenicol (Cm) 34 µg ml−1; and P. gingivalis, erythromycin (Erm) 10 µg ml−1, tetracycline (Tc) 0.5 µg ml−1. Unless otherwise stated, all chemicals were obtained from Sigma. Plasmids were constructed in E. coli DH5α or BL21DE3 and then electroporated into P. gingivalis strain W83 using previously described methods (Alberti-Segui et al., 2010; Davey & Duncan, 2006). P1 transduction, using hup alleles provided by Oberto et al. (2009), was performed as described by Thomason et al. (2007) to isolate the various hup mutants. E. coli strains were grown in Luria–Bertani broth (LB) and maintained on LB containing 1.5 % w/v agar (LB agar) at 37 °C. All primers used in this study are listed in Table S1 (available with the online version of this paper).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype | Source or reference |
| Strains | ||
| P. gingivalis | ||
| W83 (wild-type) | ATCC BAA-308 | |
| DEL0121 : : Erm insertion–deletion mutant (Emr) strain W83 | Alberti-Segui et al. (2010) | |
| PgTcKI_1 (Tcr); W83ΔPG0121 : : Erm complemented with PG0121 knock-in strain; Tcr | This study | |
| E. coli* | ||
| BL21-DE3+pET22b with PG0121 HU | Alberti-Segui et al. (2010) | |
| SG 632 | MG1655 wild-type | Dr Steven D. Finkel, USC† |
| SG 790 | MG1655 hupB : : Km hupA : : Cm | This study |
| SG 919 | MG1655 hupB : : Km hupA : : Cm+pTrcHis2C | This study |
| SG 883 | MG1655 WT+pTrcHis2A | This study |
| SG 886 | MG1655 hupB : : Km hupA : : Cm+pTrcHis2A | This study |
| SG 918 | MG1655 hupAB double mutant+pTrcHis2C expressing tagless PG0121 HU coding region | This study |
| SG 903 | MG1655 hupAB double mutant+pTrcHis2A expressing tagless E. coli HUβ coding region | This study |
| Plasmids | ||
| pT-COW | Cbr, Tcr | Gardner et al. (1996) |
| pTrcHis2A and C | Ampr | Invitrogen Life Technologies |
| pET22b | Ampr | Novagen |
The SG number represents the internal reference number for the Goodman Lab.
USC, University of Southern California.
Construction of the complementation plasmids.
The coding region for the HUβ gene (PG0121) was amplified by PCR from P. gingivalis strain W83 chromosomal DNA using primers oSG708 and oSG738. E. coli HUβ was also amplified by PCR from E. coli chromosomal DNA using primer pair oSG743 and oSG744. To generate a tagless protein, each of these primer pairs includes a stop codon at the end of the genes’ coding sequence. The PCR products were amplified with Herculase II Fusion DNA polymerase (Agilent Technologies) before digestion with the restriction enzymes BspHI and HindIII (New England Biolabs) and subsequent purification with the QIAquick Gel Extraction kit (Qiagen). The amplicon encoding HUβ was then cloned into pTrcHis-2C, while E. coli HUβ was cloned into pTrcHis-2A (Invitrogen), at the NcoI and HindIII sites, and transformed into E. coli Top10 cells and selected for ampicillin resistance. After the plasmids were confirmed by sequencing, they were transformed into the various hup mutant strains.
Antibody production.
Antibodies were generated with the following peptide to the C terminus of HU PG0121: ISIPARKVVRFKPGSTLELK by Robert Sargeant (Ramona, CA), as previously described for generating antibodies to histone-like proteins (Ditto et al., 1994).
Acid challenge assay.
Overnight cultures were diluted in fresh LB pH 7.0 to an OD600 of ~0.01 and incubated at 37 °C with shaking until the cells reached mid-exponential phase (OD600 ~0.2). Each strain was normalized to 108 cells in 500 µl LB at pH 2.5 then serially diluted in LB at neutral pH at various times (0, 15, 30, 60, 120 and 180 min) and plated on LB agar plates with the appropriate antibiotics. After overnight incubation at 37 °C the c.f.u. were counted.
Western blot.
To determine the expression level of HU PG0121 in our acid challenge assays, 200 µl of cultures at mid-exponential phase (OD600 ~0.2) were pelleted and resuspended in 200 µl 2× Laemmeli buffer (0.005 % bromophenol blue, 4 % SDS, 125 mM Tris pH 7.4, 20 % glycerol, v/v), boiled at 100 °C for 4 min and separated on a 4–20 % Tris-glycine gel (Invitrogen) at 150 V for 1.5 h before being transferred to a nitrocellulose membrane. After overnight blocking, the membrane was incubated with rabbit anti-HU PG0121 antibody (1 : 5000), detected with secondary HRP-conjugated anti-rabbit IgG antibody (1 : 20,000) (Cell Signaling Technology) and developed with Super Signal ELISA Femto Maximum Sensitivity Substrate (Thermo Scientific) before being exposed to film. The positive control HU (PG0121) protein was obtained from the previous characterization of the capsule operon (Alberti-Segui et al., 2010).
Generation of PG0121 P. gingivalis knock-in strain.
Our previous study showed that PG0121 is expressed as a monocistronic message and transcriptionally linked to the capsule operon, and that these transcripts are differentially expressed during different phases of growth; hence complementation from a plasmid did not fully restore the parental phenotype. To address this issue we generated a chromosomal ‘knock-in’ strain and used this strain for phenotypic analyses. We produced this strain by replacing the erythromycin cassette on the chromosome of the DEL0121 : : Erm insertion–deletion mutant with the PG0121 coding region from the parent strain W83 linked to a tetracycline resistance cassette. The knock-in cassette was prepared by amplifying a DNA fragment ~1000 bp upstream of the PG0121 (468 bp downstream of the start codon for PG0120) to ~1000 bp downstream of PG0123 (the 3′ end of PG0125) using the F-KI121 and R-KI121 primer set. This 3080 bp fragment was cloned into pGEMT and the tetracycline resistance cassette (Tcr) was inserted into a unique BmgBI site located between PG0123 and PG0124 in the non-coding region. The insertion of the Tcr was oriented such that it does not affect expression of the surrounding genes. This cassette was purified and then transformed as a linear fragment into the DEL0121 : : Erm insertion–deletion mutant, creating strain PgTcKI_1.
Phenotypic analysis of the DEL0121 : : Erm mutant.
For growth analysis under iron-limiting conditions, cells were grown in TSB broth or TSB blood agar plates supplemented with 1 µg menadione ml−1. Unless stated otherwise, the growth medium was not supplemented with haemin or antibiotics. Cells were first cultured on TSB blood plates and incubated for 48 h in the anaerobic chamber. The cells were then subcultured to fresh TSB blood plates and allowed to grow for 24 h. These cultures were then used to inoculate 1.5 ml TSB broth; these cultures were incubated overnight. From this overnight culture, cells were diluted 1 : 50 into 2 ml fresh TSB broth to which 125 µM 2,2′-dipyridyl (DPD) (Sigma–Aldrich) had been added to chelate iron. For growth analysis OD600 was measured at regular intervals.
For viability testing (survival of cells from stationary phase), the overnight cultures were serial diluted (10-fold dilutions) in the anaerobic chamber in pre-reduced 96-well microtitre plates containing TSB supplemented with 1 µg haemin ml−1 and 1 µg menadione ml−1 and the dilution was done in quadruplicate. The microtitre plates were then incubated for five days and each well was scored for growth or no growth. The most probable number (MPN) technique was then used to determine the number of viable cells in the overnight culture, as described by Oblinger & Koburger (1975). This technique was found to be much more reproducible than plate counts for P. gingivalis cells.
Dark field microscopy.
The cells were imaged with a Nikon Eclipse 80i microscope using NIS-Elements Imaging Software. Images were then analysed by using ImageJ software.
Isolation of total RNA.
Total RNA was isolated from cells grown to mid-exponential (OD600 0.5) growth phase. RNA was isolated using the MasterPure RNA purification kit (Epicentre) according to the manufacturer’s instructions, as previously described (Alberti-Segui et al., 2010).
Northern analysis.
Northern blot analysis was performed as previously described (Alberti-Segui et al., 2010), in brief total RNA was mixed with NorthernMax formaldehyde loading dye (Ambion), heat denatured and run on a 1 % denaturing agarose gel. After electrophoresis the RNA was transferred to a positively charged nylon membrane (Hybond-N+, Amersham) and then cross-linked with a UV Stratalinker (Stratagene). The probe for detection of transcripts encoding PG0121 was a 137 bp fragment amplified from strain W83 DNA using TIGR_F121 and TIGR_R121 primer set. The probe for detection of PG0106 transcript was a 233 bp PCR amplicon generated from W83 DNA using the TIGR_F106 and TIGR_R106 primer set. The primers are listed in Table S1. The labelling of the probe, hybridization of the blot and detection of the signal were performed using the AlkPhos Direct labelling and detection kit (Amersham) according to the manufacturer’s instructions.
Microarray analysis.
For comparative transcriptional profiling, we employed spotted array slides containing 70mer oligonucleotides based on ORFs annotated in the W83 genome sequence [manufactured and supplied by The Institute for Genomic Research/Pathogen Functional Genomics Resource Center (TIGR/PFGRC) of the J. Craig Venter Institute (JCVI)]. To perform these analyses, RNA was obtained from P. gingivalis strain W83 cells grown anaerobically to mid-exponential growth phase (OD600 0.5). For each Cy-labelling reaction, total RNA (15 µg) was reverse transcribed to cDNA using random hexamer primers and Superscript II (Invitrogen), and amino allyl-dUTP was incorporated for labelling with Cy-5 or Cy-3. For the coupling reaction, the cDNA was dissolved in 2× coupling buffer (0.2 M NaHCO3 pH 9.0) and mixed with 5 µl FluoroLink Cy-5 or Cy-3 monofunctional dye (Amersham) dissolved in DMSO. The reaction was incubated overnight at room temperature, in the dark, and then purified with a Qiagen PCR purification kit. The Cy-coupled cDNA samples from the wild-type and mutant strain (opposite labelling) were combined in a microcentrifuge tube along with 20 µg denatured salmon testes DNA (which facilitates precipitation) and precipitated with ethanol. After the ethanol has evaporated, the labelled cDNA from the wild-type and the mutant strains were each suspended in 25 µl hybridization buffer (50 % formamide, 5× SSC, 0.5 % SDS, in ultrapure water) and then combined into one tube. To denature the cDNA the mixture was boiled for 10 min and then placed on ice. The sample was then applied to the already prepared microarray slide and allowed to incubate overnight at 42 °C. Microarray slides were pre-hybridized and washed, as described previously (Nishikawa et al., 2004). Microarrays were scanned using the GenePix 4000B scanner (Axon).
Microarray data analysis.
Data analysis was performed using GenePix Pro 6.1 (Axon), as well as the Significance Analysis for Oral Pathogen Microarray Data (SAOPMD), an online microarray data analysis tool based on the LIMMA (Linear Models for Microarray Data) package for statistical inference. This analysis tool combines all the repeats within and between arrays and generates diagnostics plots and statistics. The analysis software is available at the Bioinformatic Resources for Oral Pathogens (BROP)(Chen et al., 2005). In our previous report on the PG0121 mutant (Alberti-Segui et al., 2010), we determined [using quantitative real-time PCR (qRT-PCR)] that subtle changes in expression (between 1.3- and 2-fold downregulation) of the capsule synthesis operon corresponded to a phenotypic decrease in the amount of capsular polysaccharide. Based on these previous experimental findings, we initiated our study by considering genes with ≥1.3-fold changes in expression and P-values of ≤0.05. We then focused our validation efforts on genes encoded in operons where there was a similar change in expression pattern throughout the locus, as well as genes predicted to encode histone-like proteins.
Quantitative real-time PCR (qRT-PCR) analysis.
To verify the fold change in expression of the identified genes, candidate genes from microarray analysis (35 genes in TSB and 48 genes under iron-limited growth conditions) were then analysed by qRT-PCR, as previously described (Christopher et al., 2010) with the following modifications. In brief, total RNA was confirmed to be free of genomic contamination by conventional PCR using the PG0521 (groES) primer and the RNA as a template. For cDNA generation, 2.5 µg of the purified RNA was used as a template using the SuperScript III (Invitrogen) kit and random hexamer primers (Invitrogen). The kit was used according to the manufacturer’s instructions with the recommended increase in the reaction temperature to 55 °C for difficult templates or templates with high secondary structure. cDNA was diluted 1 : 5-fold and 1 µl was used in 25 µl reactions. qPCR was carried out with a Roche LightCycler 480 using SYBR Fast Mastermix (Kapa Biosystems). The qPCR consisted of an initial denaturation step at 95 °C for 10 min followed by 40 cycles with a temperature profile of 30 s at 95 °C, 30 s at 55 °C and 30 s at 72 °C, followed by a melting curve. Three technical replicates were analysed for three biological samples. Data were analysed using the LightCycler software. Fold changes in gene expression were calculated in the following manner: crossing point (Cp) values were generated using the LightCycler 480 software and fitted to standard curves, which were generated for each primer set. Cycle numbers were normalized to the reference gene and the copy number was generated by fitting the normalized Cp to the standard curve for the corresponding primer set. Values were then divided by those for wild-type reference controls for each biological sample to generate the ratio, the three normalized values were then averaged and standard deviations were generated. TSB datasets were normalized to PG0881 (recA) and PG1853 (dnaN) for the DPD–TSB dataset, as neither gene was found to change in the respective microarray analyses.
Results
HU PG0121 protein complements the E. coli MG1655 hupAB mutant strain
Although PG0121 shares 48.8 % identity and high similarity (67.4 %) to the HUβ subunit of E. coli, its functional similarity to HU has not been determined. Therefore, we performed experiments to determine whether PG0121 expressed in trans complements an E. coli hupAB mutant strain. It has been shown that deficiencies in either HU subunit (α or β) result in minor if any phenotypic effects in E. coli (Drlica & Rouviere-Yaniv, 1987); however, there are significant phenotypic effects when both subunits are absent. Here we constructed an expression vector that expressed the coding sequence of PG0121, without an affinity tag (SG 918). We found that even in the absence of appropriate transcriptional inducer (IPTG) there was ample PG0121 protein expressed as shown by Western analysis (Fig. 1a, lane 5).
Fig. 1.
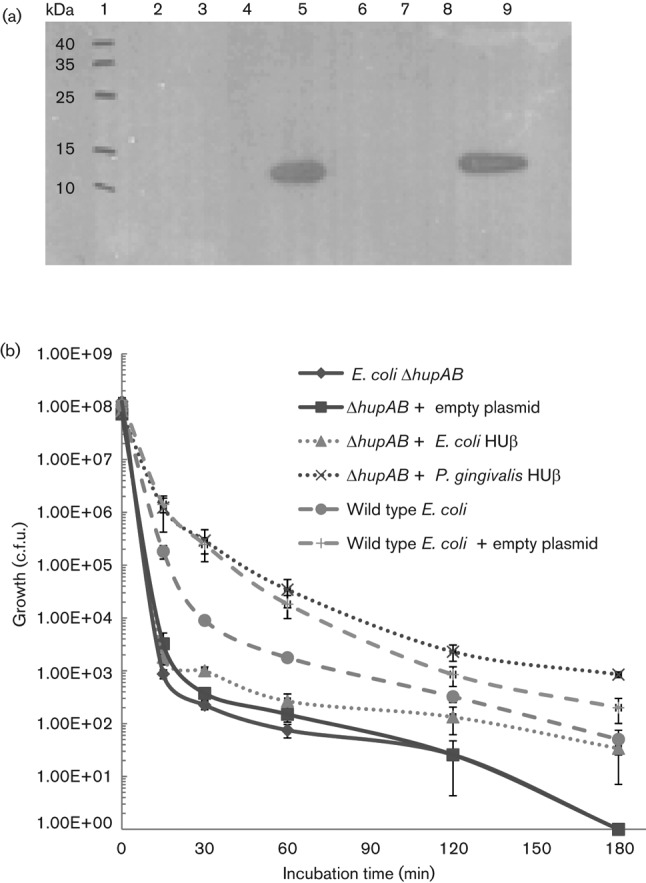
Western analysis of PG0121-HU expression in E. coli MG1655 hupAB double mutant and complementation of low pH sensitivity. (a) Western analysis of PG0121-HU (9 kDa) expression without addition of the transcriptional inducer IPTG using PG0121 antiserum. Lanes: (1) Molecular mass ladder; (2) SG 790, E. coli MG1655 hupAB double mutant; (3) SG 886, E. coli MG1655 hupAB double mutant + emptypTrcHis2A ; (4) SG 903, E. coli MG1655 hupAB double mutant + plasmid pTrcHis2A expressing E. coli HUβ coding region; (5) SG 918, E. coli MG1655 hupAB double mutant + plasmid pTrcHis2C expressing PG0121 HU coding region; (6) SG 919, E. coli MG1655 hupAB double mutant + empty pTrcHis2C; (7) SG 632, E. coli MG1655 wild-type; (8) SG 883, E. coli MG1655 WT + empty pTrcHis2A ; (9) 250 ng pure HU0121 control. (b) E. coli strains were incubated in LB broth at pH 2.5 for the time indicated. The cells were then diluted and plated and c.f.u. were determined. The data represent the mean of four biological replicates with sem.
It has been shown previously that an HU deficiency makes E. coli vulnerable to acidic conditions, resulting in a loss of viability (Bi et al., 2009; Oberto et al., 2009); therefore in our first complementation test, the HU-deficient HU (both HUα and HUβ absent) was tested for low-pH sensitivity. As shown in Fig. 1(b), it is clear that while the double mutant is more susceptible to low pH than the wild-type parent strain, the PG0121-complemented strain restores acid resistance. It has also been shown that an HU deficiency in E. coli results in a defect in cell division (Dri et al., 1991; Jaffe et al., 1997). As shown in Fig. 2, while the cells of the E. coli double mutant are elongated (Fig. 2b), indicative of a cell-division defect, the PG0121-complemented strain has a similar cell shape and size to the parental strain (Fig. 2e, a, respectively). Given these results, there is sufficient evidence that PG0121 adequately replaces one or both of the native E. coli HU subunits for some biological functions.
Fig. 2.
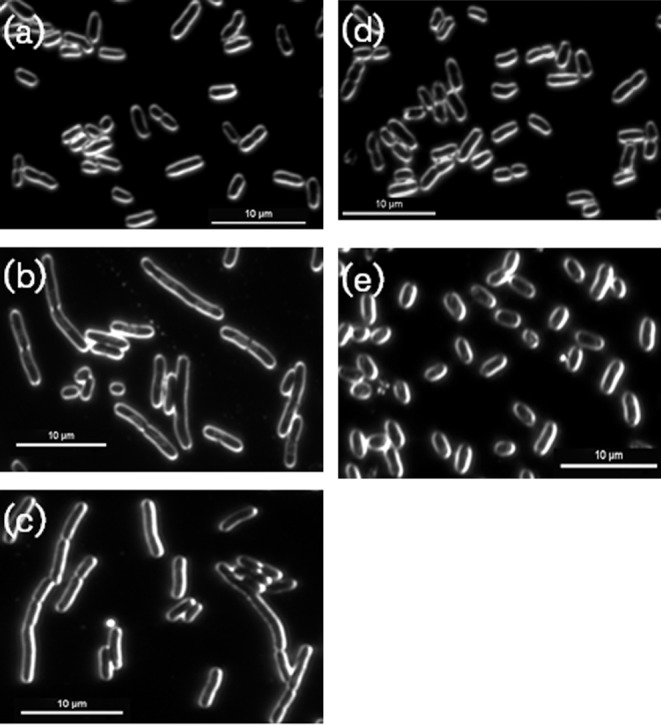
Complementation of cell-division defect of E. coli MG1655 hupAB double mutant with PG0121 HU. E. coli strains were grown to mid-exponential growth phase in LB broth and then imaged at 100× magnification with dark field microscopy. (a) E. coli MG1655 parent strain. (b) SG 790; E. coli MG1655 hupAB double mutant. (c) SG 886; E. coli MG1655 hupAB double mutant + empty pTrcHis2A. (d) SG 903; E. coli MG1655 hupAB double mutant + pTrcHis2A expressing E. coli HU-β coding region. (e) SG 918; E. coli MG1655 hupAB double mutant + pTrcHis2C expressing PG0121 HU coding region. The images shown are representative of three independent experiments. Bars, 10 µm.
PG0121 is transcribed from multiple promoters and differential expression is growth-phase-dependent
To investigate transcription of PG0121, we performed a Northern analysis using RNA obtained from cells in early exponential, mid-exponential and stationary phase growth. Two blots were prepared. One was hybridized with the 233 bp probe for PG0106 to monitor capsule operon expression, and the other blot was hybridized with the 137 bp probe to HUβ (PG0121). As shown in Fig. 3(b), expression of PG0121 switches from a predominantly monocistronic message (~300 bp) in early exponential growth (early) to being encoded in the same transcripts as the K-antigen capsule synthesis genes in mid-exponential and late stationary phase growth, as indicated by the hybridization pattern of the PG0106 probe (Fig. 3a). Thus, PG0121 is expressed in all phases of growth, but is encoded in different transcripts. Furthermore, the data indicate that when HU is expressed from its own promoter, expression of the capsule operon is either off or at very low levels.
Fig. 3.
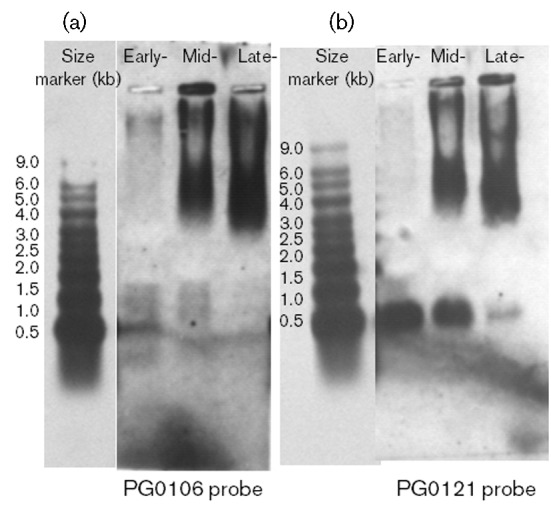
Northern blot analysis of PG0106 and PG121 expression during different phases of growth. Total RNA was isolated from the wild-type strain W83 during early (OD600 0.3), mid- (OD600 0.6) and late- (OD600 1.2) exponential growth. The blot on the left shows the transcripts that hybridize to PG0106 during the three different phases of growth. The blot on the right shows the transcripts that hybridized to a probe for PG0121 (HU) during the three different phases of growth. See text for description.
Expression of genes involved in synthesis of surface polysaccharides, cell division, iron acquisition and DNA binding are altered in a PG0121 HU mutant during mid-exponential growth phase
It was recently determined that HU protein is a master regulator in E. coli; controlling genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction (Oberto et al., 2009). Our previous studies determined that PG0121 (HUβ) expression affects production of K-antigen capsule in P. gingivalis (Alberti-Segui et al., 2010). In order to further investigate the global regulatory role of HUβ protein in P. gingivalis, we performed whole-genome comparative transcription profiling of the PG0121 mutant to the parent strain W83 using spotted array slides containing 70-mer oligonucleotides based on ORFs annotated in the W83 genome sequence. The data has been submitted to NCBI Gene Expression Omnibus (GEO) functional genomics data repository; http://www.ncbi.nlm.nih.gov/geo/. The GEO accession number is GSE34009. The results are derived from four independent experiments, using RNA extracted from cells grown to mid-exponential phase. It should be noted that RNA extracted from cells in early exponential growth (two biological replicates) was also interrogated and no differential expression was detected (data not shown). qRT-PCR was used to further validate differential expression of select genes, these data are presented in Table S2.
As a starting point, we considered genes with at least a 1.3-fold change in expression with a P-value of ≤0.05 in our analysis and all of these data are presented in Table S3. Our working hypothesis was that other NAPs compensate for the lack of PG0121 HU, so we considered subtle changes in gene expression. In fact, many of the differentially expressed genes were statistically significantly, yet not substantially changed (less than twofold). Subsets of the identified genes are encoded within predicted operons and the operons showed a similar expression pattern throughout the entire locus, so we focused on these loci. Moreover, we focused on genes with basic functions and those whose expression has been shown to be regulated by HU protein in other organisms. We performed qRT-PCR analysis on representative genes to support or refute subtle changes in expression. This validation was done with three biological replicates. In particular, we focused on the genes encoding histone-like proteins that were differentially expressed. Here, we report a subset of our findings, specifically genes belonging to the following five categories inferred by gene description: cell division; capsular polysaccharide and LPS synthesis; DNA binding and DNA replication; iron acquisition; and translation.
The PG0121 HU mutant demonstrates a cell-division defect under iron-depleted growth conditions and cell-division genes are concurrently underexpressed
As shown in Fig. S1, the growth rates of the parent P. gingivalis strain and the PG0121 HU mutant were similar when grown under iron-limiting (no haemin added) or depleted conditions. However, when the cultures treated with DPD (iron-depleted conditions) were examined microscopically, as shown in Fig. 4, the PG0121 HU mutant cells were elongated, while the complemented strain PgTcKI_1 had a similar cell shape and size to the parent strain. This elongation phenotype was not observed under any other growth condition tested, as shown in Figure S2. These data indicate that like E. coli HU, PG0121 plays a role in coordinating cell division in P. gingivalis. In addition there was at least a four-log decrease in viability of stationary phase cells when grown in the DPD-treated medium (most probable number ≥2.4×107 for the wild type strain in TSB and DPD-treated TSB, 2.4×105 for the PG0121 mutant in TSB and 2.4×103 for the mutant in DPD-treated medium); thus the PG0121 mutant is not able to survive as well as the wild-type strain when iron is depleted. To further investigate a role for PG0121 when iron (haemin) is depleted, qRT-PCR analysis was used to examine the expression of the genes that we identified as differentially expressed in the PG0121 mutant, in rich medium (TSB). As shown in Fig. S3, gene clusters differentially expressed in TSB were also altered under iron-depleted conditions. Of particular interest are the cell division genes that were expressed at significantly lower levels in the mutant; as well as the iron acquisition genes which were also expressed at much lower levels relative to the parent strain W83. In addition, the differential expression of a variety of DNA-binding proteins, including putative transcriptional repressors, sigma factors and NAPs was also of significance. These findings suggest that a complex regulatory network controls the ability of this ‘microbial vampire’ (O’Brien-Simpson et al., 2003) to acquire iron. Overall, our expression analysis studies show that HU plays a role in regulating a number of basic functions in P. gingivalis, such as growth, cell wall synthesis and iron acquisition under various growth conditions.
Fig. 4.
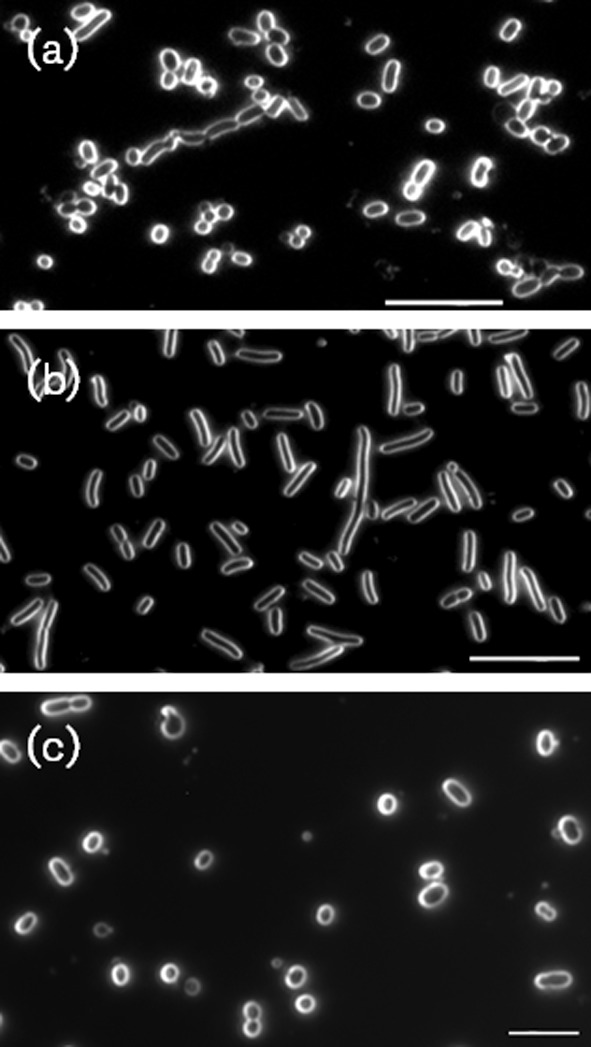
The PG0121 HU mutant demonstrates an altered cell-division phenotype under iron-limiting growth conditions. P. gingivalis strains were grown to mid-exponential growth phase in TSB broth that was treated with, 2′-dipyridyl to chelate the iron and then imaged at 100× magnification with dark field microscopy. (a) Parent strain P. gingivalis W83. (b) PG0121 HU mutant strain. (c) Knock-in complemented P. gingivalis strain PgTcKI_1. The images shown are representative of three independent experiments. Bar, 10 µm.
Discussion
Loss of HU function in E. coli results in a subtle phenotype, yet HU is essential in most Gram-positive organisms studied to date. This difference has been attributed, in part, to a lack of functional redundancy in Gram-positive bacteria. Unlike E. coli, which has a variety of NAPs, including H-NS, Fis, Dps and IHF, Gram-positive bacteria have a more limited repertoire (Dillon & Dorman, 2010; Grove, 2011). The P. gingivalis genome is predicted to encode 13 genes that are annotated as histone-like DNA-binding proteins, related to but longer than HU. Although these genes bear striking sequence similarity to the DNABII family, their ability to act as nucleoproteins has yet to be determined. Nonetheless, our working hypothesis has been that these proteins represent a potential for functional redundancy in P. gingivalis; therefore we assessed their expression in the PG0121 mutant. Of these putative NAPs, two (PG0222 and PG0330) were found to be overexpressed during mid-exponential growth phase in rich medium, potentially reflecting an adjustment strategy to compensate for the lack of HUβ. We propose that this adjustment results in more subtle transcriptional and phenotypic changes. Our studies did show that two other NAPs [one of the histone-like proteins (PG1497) and Dps (PG0090), which plays a role in protection from oxidative stress in P. gingivalis (Ueshima et al., 2003)] are underexpressed in the PG0121 HU mutant; showing that expression of these genes is also adjusted when the levels of HUβ change. In summary, our data indicate that there is the potential for dynamic changes of the levels of NAPs within the cell and that NAPs as a whole may be an important mechanism for gene regulation in this organism.
It should be noted that in other organisms HUα will compensate for the lack of HUβ. The predicted HUα protein in P. gingivalis is PG1258 and results from our previous studies (Alberti-Segui et al., 2010) as well as studies by others (Klein et al., 2012) indicate that it is essential in the organism. We are currently developing an inducible promoter system for P. gingivalis in order to confirm this essentiality and to more thoroughly investigate HUα function. Surprisingly, our microarray and qRT-PCR data indicate that transcription of PG1258 is not significantly altered in the PG0121 HU mutant, indicating that HUα may not play a compensatory role in this organism. On the other hand, post-transcriptional control of HU protein levels has been shown in E. coli (Bonnefoy et al., 1989); therefore further studies at the protein level are required to evaluate the role of PG1258.
Another fundamentally important area of differential expression was observed in genes involved in iron acquisition. The primary form of iron that is utilized by P. gingivalis is haemin (iron protoporphyrin IX), which is acquired from haemoglobin via the enzymic activity of R- and K-specific gingipains. The molecular mechanisms underlying iron uptake have been extensively studied (Olczak et al., 2005). Genes involved in iron acquisition include the hmuYRSTUV locus (PG1551–PG1556), which encodes a novel hybrid haemin-uptake system (Brunner et al., 2010; Lewis et al., 2006; Olczak et al., 2005, 2008; Simpson et al., 2000, 2004; Smalley et al., 2011); the hbp35 gene (PG0616), which encodes a haemin-binding protein that also exhibits thioredoxin activity (Hiratsuka et al., 2010; Shoji et al., 2010); the surface protein HusA (PG2227), which acts as a haemophore with very high affinity (Gao et al., 2010); the tlr (PG0644) iron transport locus (Dashper et al., 2000; Slakeski et al., 2000); the haemin-binding protein FetB (PG0669) and the surrounding genes in that locus; and PG0465, the ferric uptake regulator (fur) (Olczak et al., 2005). In addition, regulatory mechanisms controlling expression of iron acquisition genes have been identified, including the transcriptional activator encoded by PG1237, which has been shown to control expression of the hmu locus (Wu et al., 2009). It has also been shown that haemin uptake is significantly reduced in the absence of Ltp1 (PG1641), a tyrosine phosphatase with regulatory function (Maeda et al., 2008). There are also data indicating that expression of luxS (PG0498) is linked to changes in expression of iron acquisition genes (Chung et al., 2001; James et al., 2006). Interestingly, strain comparisons have shown that unlike strain W83, strain 33277 grows poorly under iron-limiting conditions; indicating that there is strain-to-strain variability in efficiency of iron acquisition (Grenier et al., 2001). In our present study with strain W83, we found that a subset of iron acquisition genes are differentially expressed when HUβ is absent; specifically the hmu locus and the high-affinity haemophore PG2227 are significantly underexpressed in the mutant during mid-exponential-phase growth in rich medium. In addition, PG1326 which is predicted to encode a haemagglutinin with a cleaved adhesion domain was significantly underexpressed. In contrast, PG0127, which is predicted to encode a ferrochelatase (hemH), was overexpressed when HUβ is absent. Although the function of this gene has not been verified, it has been suggested that this ferrochelatase could function in the removal of iron from exogenously imported haem (Roper et al., 2000).
Changes in expression of iron acquisition genes in rich medium (TSB) led us to investigate growth and gene expression under iron-depleted conditions. These experiments determined that PG0121 HU expression affects cell division and viability when iron is limiting. Cell division has been well characterized in a number of bacteria. It is initiated by the formation of a ring-like structure by the tubulin homologue, FtsZ (Dewar & Dorazi, 2000). This ring serves as a scaffold for the assembly of more than 15 proteins leading to formation of a septal ring or divisiome (Errington et al., 2003). FtsA is recruited early to the Z-ring that is critical to the stability of the ring. It also plays a role in tethering the Z-ring to the inner membrane. Once the Z-ring is established, late proteins like FtsK, FtsQ, FtsL, FtsW, FtsI and FtsN are assembled in a linear-dependent fashion. It has been shown that E. coli strains lacking both subunits of HU protein have cell division defects, demonstrating a filamentation phenotype (Wada et al., 1988). Although the precise mechanism causing cell filamentation has not been determined, this defect is likely to be due to changes in the expression of fts genes. Downregulation of a number of cell division, genes was observed in the P. gingivalis PG0121 HU single mutant. Both microarray and qRT-PCR revealed that transcript levels of FtsZ (PG0584), the tubulin homologue which initiates cell division, were downregulated along with those of FtsA (PG0583), the protein required for Z-ring stabilization in E. coli. Apart from these, FtsW (PG0579) and penicillin-binding protein (PG0575), two proteins essential for septal peptidoglycan synthesis, were also expressed at low levels. Moreover, phenotypic analysis determined that the PG0121 HU mutant is defective in cell division under iron-depleted conditions. Expression of genes encoding enzymes involved in peptidoglycan precursor synthesis (PG0576, PG0577, PG0578, PG0580 and PG0581) was also down, indicating that the P. gingivalis PG0121 HU mutant may have cell wall defects.
HU is also known to play a key role in DNA replication initiation. In E. coli, it is required for efficient initiation of oriC, the primary origin of replication (Bahloul et al., 2001) and proper chromosome partitioning along with MukB (Jaffe et al., 1997). Our results indicate that genes located at the origin of replication (PG0001, PG0002, PG2226 and PG2227) are downregulated in the P. gingivalis PG0121 HU mutant, which includes the chromosomal replication initiator protein DnaA (PG0001). Interestingly, the expression of a suite of genes encoding ribosomal proteins was upregulated in the PG0121 mutant, yet growth rate was not increased. However, a linkage to expression of ribosomal proteins is consistent with previous studies showing a physical association of HU to ribosomes in E. coli (Oberto et al., 1996) and Mycobacterium (Matsumoto et al., 1999).
Taken together, our data provide the first insight, to our knowledge, into the effects of HU protein on global gene expression in P. gingivalis. Our studies show that HUβ controls basic functions, such as cell wall synthesis, growth and the surface properties of the cell. Our data also indicate that the transcript levels of other predicted NAPs are altered when the cells lack HUβ. Collectively, our studies show that NAPs as a whole may play a central role in adjusting gene expression in this bacterium. It has been proposed that nucleoid structural reorganization by HU could serve as an efficient mechanism to synchronize a genetic response to changes in environmental parameters (Kar et al., 2005). Given our current understanding of HU’s function as an accessory protein in transcriptional regulation, future studies on mechanisms controlling expression of the loci identified in this study may need to consider a model consisting of a DNA–multiprotein complex, such as that described for the gal operon repressosome in E. coli (Kar & Adhya, 2001). Further characterization of all the NAPs encoded in the genome of P. gingivalis is warranted and these studies are ongoing in our laboratories.
Acknowledgements
The authors wish to thank Jennifer Downey for her technical expertise and advice. This work was supported by a grant from the National Institute of Dental and Craniofacial Research NIDCR (DE-019117) to M. E. D.
Abbreviations:
- Cp
crossing point
- NAP
nucleoid-associated protein
- qRT-PCR
quantitative real time-PCR
Footnotes
Three supplementary tables and three supplementary figures are available with the online version of this paper.
References
- Aki T., Choy H. E., Adhya S. (1996). Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells 1, 179–188. 10.1046/j.1365-2443.1996.d01-236.x [DOI] [PubMed] [Google Scholar]
- Alberti-Segui C., Arndt A., Cugini C., Priyadarshini R., Davey M. E. (2010). HU protein affects transcription of surface polysaccharide synthesis genes in Porphyromonas gingivalis. J Bacteriol 192, 6217–6229. 10.1128/JB.00106-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A. (1999). Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahloul A., Boubrik F., Rouviere-Yaniv J. (2001). Roles of Escherichia coli histone-like protein HU in DNA replication: HU-beta suppresses the thermosensitivity of dnaA46ts. Biochimie 83, 219–229. 10.1016/S0300-9084(01)01246-9 [DOI] [PubMed] [Google Scholar]
- Balandina A., Claret L., Hengge-Aronis R., Rouviere-Yaniv J. (2001). The Escherichia coli histone-like protein HU regulates rpoS translation. Mol Microbiol 39, 1069–1079. 10.1046/j.1365-2958.2001.02305.x [DOI] [PubMed] [Google Scholar]
- Balandina A., Kamashev D., Rouviere-Yaniv J. (2002). The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J Biol Chem 277, 27622–27628. 10.1074/jbc.M201978200 [DOI] [PubMed] [Google Scholar]
- Bensaid A., Almeida A., Drlica K., Rouviere-Yaniv J. (1996). Cross-talk between topoisomerase I and HU in Escherichia coli. J Mol Biol 256, 292–300. 10.1006/jmbi.1996.0086 [DOI] [PubMed] [Google Scholar]
- Bi H., Sun L., Fukamachi T., Saito H., Kobayashi H. (2009). HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Curr Microbiol 58, 443–448. 10.1007/s00284-008-9340-4 [DOI] [PubMed] [Google Scholar]
- Bonnefoy E., Almeida A., Rouviere-Yaniv J. (1989). Lon-dependent regulation of the DNA binding protein HU in Escherichia coli. Proc Natl Acad Sci U S A 86, 7691–7695. 10.1073/pnas.86.20.7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E., Takahashi M., Yaniv J. R. (1994). DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J Mol Biol 242, 116–129. 10.1006/jmbi.1994.1563 [DOI] [PubMed] [Google Scholar]
- Boubrik F., Rouviere-Yaniv J. (1995). Increased sensitivity to γ irradiation in bacteria lacking protein HU. Proc Natl Acad Sci U S A 92, 3958–3962. 10.1073/pnas.92.9.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner J., Scheres N., El Idrissi N. B., Deng D. M., Laine M. L., van Winkelhoff A. J., Crielaard W. (2010). The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol 10, 5. 10.1186/1471-2180-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Abbey K., Deng W. J., Cheng M. C. (2005). The bioinformatics resource for oral pathogens. Nucleic Acids Res 33 (Web Server issue), W734–W740. 10.1093/nar/gki361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choil J.-I., Nakagawa T., Yamada S., Takazoe I., Okuda K. (1990). Clinical, microbiological and immunological studies on recurrent periodontal disease. J Clin Periodontol 17, 426–434. 10.1111/j.1600-051X.1990.tb02341.x [DOI] [PubMed] [Google Scholar]
- Christopher A. B., Arndt A., Cugini C., Davey M. E. (2010). A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology 156, 3469–3477. 10.1099/mic.0.042671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. O., Park Y., Lamont R. J., McNab R., Barbieri B., Demuth D. R. (2001). Signaling system in Porphyromonas gingivalis based on a LuxS protein. J Bacteriol 183, 3903–3909. 10.1128/JB.183.13.3903-3909.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock L. E., Kasper D. L. (2006). Bacterial glycans: key mediators of diverse host immune responses. Cell 126, 847–850. 10.1016/j.cell.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Dashper S. G., Hendtlass A., Slakeski N., Jackson C., Cross K. J., Brownfield L., Hamilton R., Barr I., Reynolds E. C. (2000). Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol 182, 6456–6462. 10.1128/JB.182.22.6456-6462.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E., Duncan M. J. (2006). Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol 188, 5510–5523. 10.1128/JB.01685-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S. J., Dorazi R. (2000). Control of division gene expression in Escherichia coli. FEMS Microbiol Lett 187, 1–7. 10.1111/j.1574-6968.2000.tb09127.x [DOI] [PubMed] [Google Scholar]
- Dillon S. C., Dorman C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8, 185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- Ditto M. D., Roberts D., Weisberg R. A. (1994). Growth phase variation of integration host factor level in Escherichia coli. J Bacteriol 176, 3738–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn B. R., Burks J. N., Seifert K. N., Progulske-Fox A. (2000). Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol Lett 187, 139–144. 10.1111/j.1574-6968.2000.tb09150.x [DOI] [PubMed] [Google Scholar]
- Dorn B. R., Harris L. J., Wujick C. T., Vertucci F. J., Progulske-Fox A. (2002). Invasion of vascular cells in vitro by Porphyromonas endodontalis. Int Endod J 35, 366–371. 10.1046/j.0143-2885.2001.00489.x [DOI] [PubMed] [Google Scholar]
- Dri A. M., Rouviere-Yaniv J., Moreau P. L. (1991). Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J Bacteriol 173, 2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. (1987). Histonelike proteins of bacteria. Microbiol Rev 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzink J. L., Socransky S. S., Haffajee A. D. (1988). The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol 15, 316–323. 10.1111/j.1600-051X.1988.tb01590.x [DOI] [PubMed] [Google Scholar]
- Errington J., Daniel R. A., Scheffers D. J. (2003). Cytokinesis in bacteria. Microbiol Mol Biol Rev 67, 52–65. . 10.1128/MMBR.67.1.52-65.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández S., Rojo F., Alonso J. C. (1997). The Bacillus subtilis chromatin-associated protein Hbsu is involved in DNA repair and recombination. Mol Microbiol 23, 1169–1179. 10.1046/j.1365-2958.1997.3061670.x [DOI] [PubMed] [Google Scholar]
- Gao J. L., Nguyen K. A., Hunter N. (2010). Characterization of a hemophore-like protein from Porphyromonas gingivalis. J Biol Chem 285, 40028–40038. 10.1074/jbc.M110.163535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Russell J. B., Wilson D. B., Wang G. R., Shoemaker N. B. (1996). Use of a modified Bacteroides–Prevotella shuttle vector to transfer a reconstructed β-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl Environ Microbiol 62, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrossi M., Giuliodori A. M., Gualerzi C. O., Pon C. L. (2002). Selective expression of the β-subunit of nucleoid-associated protein HU during cold shock in Escherichia coli. Mol Microbiol 44, 205–216. 10.1046/j.1365-2958.2002.02868.x [DOI] [PubMed] [Google Scholar]
- Grenier D., Goulet V., Mayrand D. (2001). The capacity of Porphyromonas gingivalis to multiply under iron-limiting conditions correlates with its pathogenicity in an animal model. J Dent Res 80, 1678–1682. 10.1177/00220345010800071501 [DOI] [PubMed] [Google Scholar]
- Grossi S. G., Zambon J. J., Ho A. W., Koch G., Dunford R. G., Machtei E. E., Norderyd O. M., Genco R. J. (1994). Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol 65, 260–267. 10.1902/jop.1994.65.3.260 [DOI] [PubMed] [Google Scholar]
- Grove A. (2011). Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol 13, 1–12. [PubMed] [Google Scholar]
- Hiratsuka K., Kiyama-Kishikawa M., Abiko Y. (2010). Hemin-binding protein 35 (HBP35) plays an important role in bacteria–mammalian cells interactions in Porphyromonas gingivalis. Microb Pathog 48, 116–123. 10.1016/j.micpath.2010.01.001 [DOI] [PubMed] [Google Scholar]
- Jaffe A., Vinella D., D’Ari R. (1997). The Escherichia coli histone-like protein HU affects DNA initiation, chromosome partitioning via MukB, and cell division via MinCDE. J Bacteriol 179, 3494–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C. E., Hasegawa Y., Park Y., Yeung V., Tribble G. D., Kuboniwa M., Demuth D. R., Lamont R. J. (2006). LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun 74, 3834–3844. 10.1128/IAI.01768-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandik K. A., Bélanger M., Low S. L., Dorn B. R., Yang M. C., Progulske-Fox A. (2008). Invasive differences among Porphyromonas gingivalis strains from healthy and diseased periodontal sites. J Periodontal Res 43, 524–530. 10.1111/j.1600-0765.2007.01064.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S., Adhya S. (2001). Recruitment of HU by piggyback: a special role of GalR in repressosome assembly. Genes Dev 15, 2273–2281. 10.1101/gad.920301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S., Edgar R., Adhya S. (2005). Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc Natl Acad Sci U S A 102, 16397–16402. 10.1073/pnas.0508032102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. A., Tenorio E. L., Lazinski D. W., Camilli A., Duncan M. J., Hu L. T. (2012). Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13, 578. 10.1186/1471-2164-13-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler P., Marahiel M. A. (1998). Mutational analysis of the nucleoid-associated protein HBsu of Bacillus subtilis. Mol Gen Genet 260, 487–491. 10.1007/s004380050921 [DOI] [PubMed] [Google Scholar]
- Lamont R. J., Jenkinson H. F. (2000). Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol 15, 341–349. 10.1034/j.1399-302x.2000.150601.x [DOI] [PubMed] [Google Scholar]
- Lewis J. P., Plata K., Yu F., Rosato A., Anaya C. (2006). Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology 152, 3367–3382. 10.1099/mic.0.29011-0 [DOI] [PubMed] [Google Scholar]
- Li S., Waters R. (1998). Escherichia coli strains lacking protein HU are UV sensitive due to a role for HU in homologous recombination. J Bacteriol 180, 3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Tribble G. D., Tucker C. M., Anaya C., Shizukuishi S., Lewis J. P., Demuth D. R., Lamont R. J. (2008). A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol 69, 1153–1164. 10.1111/j.1365-2958.2008.06338.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S., Yukitake H., Furugen M., Matsuo T., Mineta T., Yamada T. (1999). Identification of a novel DNA-binding protein from Mycobacterium bovis bacillus Calmette–Guérin. Microbiol Immunol 43, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Miyabe I., Zhang Q. M., Kano Y., Yonei S. (2000). Histone-like protein HU is required for recA gene-dependent DNA repair and SOS induction pathways in UV-irradiated Escherichia coli. Int J Radiat Biol 76, 43–49. 10.1080/095530000138998 [DOI] [PubMed] [Google Scholar]
- Moore W. E., Moore L. H., Ranney R. R., Smibert R. M., Burmeister J. A., Schenkein H. A. (1991). The microflora of periodontal sites showing active destructive progression. J Clin Periodontol 18, 729–739. 10.1111/j.1600-051X.1991.tb00064.x [DOI] [PubMed] [Google Scholar]
- Morales P., Rouviere-Yaniv J., Dreyfus M. (2002). The histone-like protein HU does not obstruct movement of T7 RNA polymerase in Escherichia coli cells but stimulates its activity. J Bacteriol 184, 1565–1570. 10.1128/JB.184.6.1565-1570.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morash M. G., Brassinga A. K., Warthan M., Gourabathini P., Garduño R. A., Goodman S. D., Hoffman P. S. (2009). Reciprocal expression of integration host factor and HU in the developmental cycle and infectivity of Legionella pneumophila. Appl Environ Microbiol 75, 1826–1837. 10.1128/AEM.02756-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Yoshimura F., Duncan M. J. (2004). A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol 54, 546–560. 10.1111/j.1365-2958.2004.04291.x [DOI] [PubMed] [Google Scholar]
- O’Brien-Simpson N. M., Veith P. D., Dashper S. G., Reynolds E. C. (2003). Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci 4, 409–426. 10.2174/1389203033487009 [DOI] [PubMed] [Google Scholar]
- Oberto J., Bonnefoy E., Mouray E., Pellegrini O., Wikström P. M., Rouvière-Yaniv J. (1996). The Escherichia coli ribosomal protein S16 is an endonuclease. Mol Microbiol 19, 1319–1330. 10.1111/j.1365-2958.1996.tb02476.x [DOI] [PubMed] [Google Scholar]
- Oberto J., Nabti S., Jooste V., Mignot H., Rouviere-Yaniv J. (2009). The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS ONE 4, e4367. 10.1371/journal.pone.0004367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblinger J. L., Koburger J. A. (1975). Understanding and teaching the most probable number technique. J Milk Food Technol 38, 540–545. [Google Scholar]
- Olczak T., Simpson W., Liu X., Genco C. A. (2005). Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev 29, 119–144. 10.1016/j.femsre.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Olczak T., Sroka A., Potempa J., Olczak M. (2008). Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch Microbiol 189, 197–210. 10.1007/s00203-007-0309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painbeni E., Caroff M., Rouviere-Yaniv J. (1997). Alterations of the outer membrane composition in Escherichia coli lacking the histone-like protein HU. Proc Natl Acad Sci U S A 94, 6712–6717. 10.1073/pnas.94.13.6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham K., Feik D., Hammond B. F., Rams T. E., Whitaker E. J. (2002). Aggregation of human platelets by gingipain-R from Porphyromonas gingivalis cells and membrane vesicles. Platelets 13, 21–30. 10.1080/09537100120104863 [DOI] [PubMed] [Google Scholar]
- Pinson V., Takahashi M., Rouviere-Yaniv J. (1999). Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J Mol Biol 287, 485–497. 10.1006/jmbi.1999.2631 [DOI] [PubMed] [Google Scholar]
- Pontiggia A., Negri A., Beltrame M., Bianchi M. E. (1993). Protein HU binds specifically to kinked DNA. Mol Microbiol 7, 343–350. 10.1111/j.1365-2958.1993.tb01126.x [DOI] [PubMed] [Google Scholar]
- Roper J. M., Raux E., Brindley A. A., Schubert H. L., Gharbia S. E., Shah H. N., Warren M. J. (2000). The enigma of cobalamin (Vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J Biol Chem 275, 40316–40323. 10.1074/jbc.M007146200 [DOI] [PubMed] [Google Scholar]
- Shoji M., Shibata Y., Shiroza T., Yukitake H., Peng B., Chen Y. Y., Sato K., Naito M., Abiko Y. & other authors (2010). Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol 10, 152. 10.1186/1471-2180-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson W., Olczak T., Genco C. A. (2000). Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol 182, 5737–5748. 10.1128/JB.182.20.5737-5748.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson W., Olczak T., Genco C. A. (2004). Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim Pol 51, 253–262. [PubMed] [Google Scholar]
- Singh A., Wyant T., Anaya-Bergman C., Aduse-Opoku J., Brunner J., Laine M. L., Curtis M. A., Lewis J. P. (2011). The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun 79, 4533–4542. 10.1128/IAI.05016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slakeski N., Dashper S. G., Cook P., Poon C., Moore C., Reynolds E. C. (2000). A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol 15, 388–392. 10.1034/j.1399-302x.2000.150609.x [DOI] [PubMed] [Google Scholar]
- Smalley J. W., Byrne D. P., Birss A. J., Wojtowicz H., Sroka A., Potempa J., Olczak T. (2011). HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis. PLoS ONE 6, e17182. 10.1371/journal.pone.0017182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger K. K., Rice P. A. (2004). IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol 14, 28–35. 10.1016/j.sbi.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Wang S. C., Shapiro L. (2005). The bacterial nucleoid: a highly organized and dynamic structure. J Cell Biochem 96, 506–521. 10.1002/jcb.20519 [DOI] [PubMed] [Google Scholar]
- Thomason L. C., Costantino N., Court D. L. (2007). E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1, 17. [DOI] [PubMed] [Google Scholar]
- Ueshima J., Shoji M., Ratnayake D. B., Abe K., Yoshida S., Yamamoto K., Nakayama K. (2003). Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect Immun 71, 1170–1178. 10.1128/IAI.71.3.1170-1178.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kano Y., Ogawa T., Okazaki T., Imamoto F. (1988). Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol 204, 581–591. 10.1016/0022-2836(88)90357-9 [DOI] [PubMed] [Google Scholar]
- Whiteford D. C., Klingelhoets J. J., Bambenek M. H., Dahl J. L. (2011). Deletion of the histone-like protein (Hlp) from Mycobacterium smegmatis results in increased sensitivity to UV exposure, freezing and isoniazid. Microbiology 157, 327–335. 10.1099/mic.0.045518-0 [DOI] [PubMed] [Google Scholar]
- Wu J., Lin X., Xie H. (2009). Regulation of hemin binding proteins by a novel transcriptional activator in Porphyromonas gingivalis. J Bacteriol 191, 115–122. 10.1128/JB.00841-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O. (2008). The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology 154, 2897–2903. 10.1099/mic.0.2008/021220-0 [DOI] [PMC free article] [PubMed] [Google Scholar]


