Abstract
Uropathogenic Escherichia coli (UPEC) causes more than 90 % of all human urinary tract infections through type 1 piliated UPEC cells binding to bladder epithelial cells. The FimB and FimE site-specific recombinases orient the fimS element containing the fimA structural gene promoter. Regulation of fimB and fimE depends on environmental pH and osmolality. The EnvZ/OmpR two-component system affects osmoregulation in E. coli. To ascertain if OmpR directly regulated the fimB gene promoters, gel mobility shift and DNase I footprinting experiments were performed using OmpR or phosphorylated OmpR (OmpR-P) mixed with the fimB promoter regions of UPEC strain NU149. Both OmpR-P and OmpR bound weakly to one fimB promoter. Because there was weak binding to one fimB promoter, strain NU149 was grown in different pH and osmolality environments, and total RNAs were extracted from each population and converted to cDNAs. Quantitative reverse-transcriptase PCR showed no differences in ompR transcription among the different growth conditions. Conversely, Western blots showed a significant increase in OmpR protein in UPEC cells grown in a combined low pH/high osmolality environment versus a neutral pH/high osmolality environment. In a high osmolality environment, the ompR mutant expressed more fimB transcripts and Phase-ON positioning of the fimS element as well as higher type 1 pili levels than wild-type cells. Together these results suggest that OmpR may be post-transcriptionally regulated in UPEC cells growing in a low pH/high osmolality environment, which regulates fimB in UPEC.
Introduction
More than 90 % of the 7 million human urinary tract infections occurring in the United States annually (Foxman et al., 2000; Hooton & Stamm, 1997) are caused by uropathogenic Escherichia coli (UPEC) at a cost of $1.6 billion per year (Foxman, 2003; Hooton & Stamm, 1997). The ability of UPEC to bind to bladder epithelial cells is an important step in pathogenesis (Keith et al., 1986; van der Bosch et al., 1980; Virkola et al., 1988). Once the UPEC attach, they are able to grow in human urine with extreme fluctuations in both pH (5.0–8.0) and osmolality (50–1400 mOsm kg−1) (Kunin & Chambers, 1989; Ross & Neely, 1983).
Type 1 pili are the major adherence factor for UPEC in the lower urinary tract (Abraham et al., 1985a; Hultgren et al., 1985; Klemm, 1986; Ofek & Beachey, 1978; Schaeffer et al., 1979, 1987; van der Bosch et al., 1980), and expression is affected by phase variation that allows switching from a fully piliated state (Phase-ON) to a non-piliated state (Phase-OFF). This process involves the inversion of a 314 bp region of DNA (called fimS), containing the promoter for the main structural subunit fimA (Abraham et al., 1985b).
The fimB and fimE genes located upstream of the fimA gene code for site-specific recombinases that position the fimS invertible element, controlling type 1 pili phase variation (Blomfield et al., 1991; Freitag et al., 1985; Klemm, 1986; McClain et al., 1991, 1993; Pallesen et al., 1989). The FimB protein flips the invertible element in both the Phase-ON and the Phase-OFF orientations (McClain et al., 1991), whereas the FimE protein predominantly rearranges the fimS region in the Phase-OFF orientation (Pallesen et al., 1989; Stentebjerg-Olesen et al., 2000). One promoter has been identified for fimE (Olsen & Klemm, 1994), whereas 1–3 fimB promoters have been identified (Donato et al., 1997; El-Labany et al., 2003; Olsen & Klemm, 1994; Schwan et al., 1994).
Previously, high osmolality and low pH were shown to reduce type 1 fimbriae expression in UPEC in part by repressing fimB expression and flipping the fimS region to a Phase-OFF orientation (Schwan et al., 2002). An ompR mutant strain derepressed fimB expression in an acidic/high osmolality environment, suggesting that OmpR may be directly regulating fimB transcription in this environment.
OmpR is part of the EnvZ–OmpR two-component regulatory system that is affected by changes in osmolalities. Two-component systems have a histidine kinase (e.g. EnvZ) and a response regulator (e.g. OmpR). The EnvZ sensor protein monitors environmental osmotic changes. Under hypoosmotic conditions, EnvZ is autophosphorylated using ATP as its phosphoryl donor at the highly conserved histidine residue 243 located in the bacterial cytoplasm (Forst et al., 1989; Igo & Silhavy, 1988; Kenney et al., 1995). The OmpR response regulator is an autokinase that now uses the phosphorylated EnvZ histidine kinase as its phosphoryl donor to transfer the phosphate to aspartate residue 55 on the cytoplasmic OmpR protein (OmpR-P). OmpR-P then binds to a promoter region to either activate or repress transcription (Delgado et al., 1993; Forst et al., 1989; Mattison & Kenney, 2002; Rampersaud et al., 1994). In high osmotic stress environments, phosphorylation of OmpR by acetyl phosphate (Liu & Ferenci, 2001; Matsubara & Mizuno, 1999; McCleary & Stock, 1994; Prohinar et al., 2002; Wolfe, 2005) or alternative histidine kinase donors (Forst et al., 1988; Matsubara & Mizuno, 1999) may occur. OmpR regulates approximately 125 E. coli genes (Oshima et al., 2002).
To date, no studies have been performed, to our knowledge, to investigate whether OmpR or OmpR-P protein binds to the fimB gene promoters of UPEC. In this study, gel mobility shift and DNase I footprinting assays have shown unphosphorylated OmpR binding to one of the fimB promoter regions. Western blots demonstrated OmpR protein concentration changes in a clinical strain of UPEC when grown in different environments. The presence of more OmpR protein may influence fimB regulation when UPEC cells are growing in an acidic/high osmolality environment.
Methods
Bacterial strains, plasmids and growth conditions.
E. coli NU149, a UPEC clinical isolate (Schaeffer et al., 1987), was used in the PCR, quantitative real-time reverse transcriptase PCR (qRT-PCR), Western blot analyses, haemagglutination (HA) assays and electron microscopy. The NU149 OmpR1 strain with an ompR deletion has been described before (Schwan, 2009). Strains MC4100 (wild-type), MH1160 (ompR) and JMS6210 (envZ) were a kind gift from L. Kenney, University of Illinois-Chicago, and used to screen lacZ fusions. The E. coli strains were grown with shaking at 37 °C in media described previously (Schwan et al., 2002) for β-galactosidase assays as well as RNA and protein extractions. Plasmid pWRS2-10 (Schwan et al., 1992) containing the fimB and fimE genes from UPEC strain J96 was used as the template DNA for gel mobility shift and DNase I footprinting. Plasmid pJB5A (fimB : : lacZ) has been described previously (Schwan et al., 2002). For the ompR complementation experiments, plasmids pFR29* (Russo & Silhavy, 1991) containing the full-length unmutated ompR gene and pD55A (Schwan, 2009) containing a mutated ompR gene that does not allow phosphorylation, provided by L. Kenney, were used. The organisation of the fimB ORF is shown in Fig. S1, available with the online version of this paper.
β-Galactosidase assays.
The β-galactosidase activity of mid-exponential-phase bacteria permeabilized with SDS and CHCl3 was determined by using the method of Miller (1972). Assays were performed at least three times on different days, and the data were expressed as means±sd based on the values obtained.
Gel mobility shift assays.
The gel mobility shift assays were done as previously described (Schwan et al., 1994) using end-labelled fimB 445 bp FIMB2/PEXFIMB (Table S1) or end-labelled fimE 340 bp FIMEPE4/PEXFIME2 (Table S1) PCR products. Purified OmpR protein, a gift from L. Kenney, was mixed with both end-labelled DNA PCR products.
Table 1. Effects of ompR and envZ mutations on fimB expression compared with expression in wild-type, ompR, envZ and ompR complemented strains.
Values are mean±sd
| Strain | Plasmid | β-Galactosidase activity (Miller units) | |||
| pH 7† | pH 7+‡ | pH 5.5† | pH 5.5+‡ | ||
| MC4100 (wild-type) | pJB5A (fimB–lacZ) | 325±50 | 182±38 | 198±68 | 131±35 |
| MH1160 (ompR−) | pJB5A | 406±97 | 296±48 | 161±89 | 325±56 |
| JMS6210 (envZ−) | pJB5A | 302±98 | 234±36 | 227±29 | 144±19 |
| MC4100 | pJB5A/pFR29* | 222±15 | 180±35 | 199±49 | 157±39 |
| MH1160 | pJB5A/pFR29* | 185±21 | 147±29 | 218±56 | 136±27 |
| MC4100 | pJB5A/pD55A | 207±50 | 157±49 | 234±53 | 115±38 |
| MH1160 | pJB5A/pD55A | 225±32 | 147±26 | 221±47 | 87±23 |
LB growth (low osmolality) medium.
LB growth medium containing 800 mOsm NaCl (high osmolality).
DNase I footprinting.
Footprinting reactions were performed using pWRS2-10 DNA as a template for the PCR amplifications. A 445 bp fragment that contained fimB promoters 1 and 2 was generated by PCR using Platinum Taq (Invitrogen) and primer pair FIMB2/PEXFIMB (Table S1). The DNase I procedure followed Walthers et al. (2007). Images were acquired with a Molecular Dynamics Storm 860 Phosphorimager using the ImageQuant 5.3 software package.
Extraction of total RNA and conversion to cDNA.
Total RNA was extracted from E. coli NU149 cells grown with shaking to mid-exponential phase (OD600 0.5–0.8) at 37 °C in pH 5.5/low osmolality Luria–Bertani (LB) medium, pH 5.5/high osmolality (800 mOsm NaCl) LB medium, pH 7.0/low osmolality medium or pH 7.0/high osmolality (800 mOsm NaCl) LB medium using TRIzol Reagent (Invitrogen) with 50 µl lysozyme (10 mg ml−1).
For each RNA sample, 5 µg RNA was converted to cDNA using a SuperScript First-Strand Synthesis kit (Invitrogen) following the protocol recommended by the manufacturer.
Amplification and detection of ftsZ, ompR and fimB transcripts.
RT-PCRs were performed as previously described (Schwan, 2009). The ftsZ gene was used to standardize between the samples (Table S1; Schwan et al., 2002, 2007). For detection of ompR transcripts, a 319 bp ompR gene product was amplified using the primer pair OmpR3 and OmpR4 (Table S1). Detection of fimB transcripts was performed with the FimB5 and FimB6 (Table S1) primer pair, generating a 380 bp product. RT-PCR products were separated on 1.5 % agarose gels, stained with ethidium bromide and visualized with FOTO/Analyst PC Image Software (FOTODYNE Inc.).
qRT-PCR was performed using a Stratagene Mx3000P QPCR system (Agilent Technologies). One microlitre of a 1/10 dilution of cDNA was used in each qRT-PCR. A Power SYBR Green mastermix (Applied Biosystems) was used and products were detected with an FAM setting. The ftsZ gene was used to standardize between the samples (Table S1; Schwan et al., 2002, 2007). A 71 bp fragment of the ftsZ gene was amplified using the primer pair FtsZfwd and FtsZrev (Table S1). The 96 bp ompR gene segment was amplified using the primer pair OmpRfwd and OmpRrev (Table S1). A 380 bp fimB product was amplified using the primer pair FimB5 and FimB6 (Table S1). The real-time RT-PCR parameters were as follows: initial denaturation for 10 min at 95 °C followed by 38 cycles of 15 s at 95 °C and 1 min at 58 °C. Data were analysed by using the method (Livak & Schmittgen, 2001). Both the ompR and the fimB transcript levels were standardized to ftsZ transcript levels.
PCR for fimS orientation determination.
To determine the orientation of the fimS invertible element, previously described PCR techniques were used and products visualized with FOTO/Analyst PC Image Software (Schwan et al., 1992, 2007). To quantify the percentage of Phase-ON or Phase-OFF bacteria, a standard curve was prepared as described by Teng et al. (2005) using locked-ON (DH5α/pAON-1; Schwan et al., 2002) and locked-OFF bacteria (NU149 cells passaged five times on agar shown to be 100 % Phase-OFF; Schwan et al., 1992) as PCR templates and the ImageQuant 5.2 software.
Protein extraction and Western blotting.
SDS-PAGE was performed with 10–20 % Tris/HCl polyacrylamide gels (Bio-Rad), and 10 µg E. coli NU149 cell lysates from the same shaking exponential-phase cultures used for RNA extractions and processed as previously described (Schwan, 2009). For IPTG (New England Biolabs) induction, 1 mM of IPTG was added to the cultures. Proteins were electrotransferred to PVDF Immobilon membranes (Millipore) and processed as previously noted (Schwan, 2009). Rabbit polyclonal antibody to OmpR protein absorbed with NU149 OmpR1 cell lysates (1 : 1000 dilution; provided by L. Kenney) and goat anti-rabbit IgG fluorescein-conjugated secondary antibody (Millipore, 1 : 500 dilution) were used. Bound antibodies were detected using a Storm 860 Phosphorimager (Amersham Biosciences) and analysed with Image Quant 5.3 software (Amersham).
Haemagglutination assays.
The HA assays were performed with 1 % guinea pig erythrocytes (Hardy Diagnostics) as previously described (Schwan et al., 1992). The titres represent the average of three separate runs.
Electron microscopy.
Samples of NU149 and NU149 OmpR1 grown in pH 7.0 LB with 800 mOsm NaCl were attached to Formvar-coated grids and processed as described by Hultgren et al. (1985). Grids were observed using a JEOL 1200EXII transmission electron microscope. Images were gathered using a Gatan 791 digital camera running Digital Micrograph. A rank sharpening filter was applied globally to the images to help visualize the pili on the bacteria. Between 50 and 100 cells were examined for each growth condition.
Statistics.
A Student’s t-test was used to assess probabilities. P-values <0.05 were considered significant.
Results
Effects of growth medium and ompR/envZ mutations on type 1 pilus expression
Previous work suggested that OmpR repressed fimB expression in UPEC growing in acidic/high osmolality conditions (Schwan et al., 2002). To obtain further evidence that OmpR was involved in the fimB gene transcriptional changes, additional experiments were performed using the same fimB–lacZYA (pJB5A) fusion and growth environments as the previous study. Initially, fimB expression in wild-type cells was examined. Wild-type E. coli MC4100 had the highest level of fimB expression when grown in pH 7.0/low osmolality medium (LB) (Table 1). This expression decreased in response to high osmolality conditions (2-fold decrease in pH 7.0/800 mOsm NaCl medium), acidic conditions (1.6-fold decrease in pH 5.5 medium) and a combination of these two conditions (2.5-fold decrease in pH 5.5/800 mOsm NaCl medium). These results substantiated our previous study (Schwan et al., 2002) showing that fimB had its lowest expression in acidic/high osmolality medium.
The effects of envZ (JMS6210) and ompR (MH1160) mutations on fimB expression were then examined. First, an envZ mutation did not significantly alter the expression of fimB when compared with wild-type, suggesting that EnvZ does not play a role in regulating fimB transcription. However, an ompR mutation appeared to impact fimB transcription under certain growth conditions. Although no statistical difference in fimB transcription was observed when comparing the ompR mutant and wild-type strains grown in low osmolality media (regardless of pH), fimB expression was elevated in the ompR mutant strain when cells were grown in high osmolality media (Table 2). This derepression of fimB occurred under both pH conditions, specifically a 1.6-fold derepression in the pH 7.0/800 mOsm NaCl medium and a 2.5-fold derepression of fimB expression in the pH 5.5/800 mOsm NaCl medium (Table 1). These results confirmed the results from the previous study (Schwan et al., 2002) showing OmpR negatively regulated fimB transcription in cells grown in a high osmolality/acidic environment.
Table 2. Measurement of haemagglutination (HA) titre for UPEC strains NU149, NU149 OmpR1 and NU149/OmpR1/pFR29+* grown in buffered LB at pH 5.5 and pH 7 with and without 800 mOsm NaCl.
Values are the mean of three runs.
| Strain | HA titre | |||
| pH 7 | pH 7+† | pH 5.5 | pH 5.5+† | |
| NU149 | 512 | 64 | 128 | 4 |
| NU149 OmpR1 | 512 | 256 | 16 | 32 |
| NU149 OmpR1/pFR29* | 512 | 64 | 128 | 8 |
Addition of 800 mOsm NaCl.
To confirm that the loss of OmpR caused the derepression of fimB expression, plasmids containing either the wild-type ompR gene (pFR29*) or a mutated ompR gene (pD55A) were used to complement the ompR mutant. Complementation of the ompR mutation occurred with the pFR29* (full-length unaltered OmpR) plasmid (Table 2). Surprisingly, the pD55A plasmid that contained a D55A substitution which changed the aspartate at position 55 to an alanine and generated an unphosphorylated OmpR protein, also complemented the ompR mutation, implying that unphosphorylated OmpR could repress fimB transcription. In addition, greater repression of fimB transcription was observed in the cell populations complemented with either pFR29* or pD55A because of a multicopy effect that allowed more OmpR protein expression.
Unphosphorylated and phosphorylated OmpR bind weakly to fimB promoter region 2
From the results in Table 1, we hypothesized that unphosphorylated OmpR bound to one or more of the fimB promoters to repress fimB transcription in E. coli cells grown in an acidic/high osmolality environment mirroring conditions in the human urinary tract. To determine if purified unphosphorylated OmpR bound to the fimB promoter region, a gel mobility shift assay was performed. Purified unphosphorylated OmpR shifted the mobility of the fimB promoter region in a concentration-dependent manner (Fig. 1a), suggesting unphosphorylated OmpR may be directly repressing fimB expression. When OmpR phosphorylated with phosphoramidate was used, the fimB promoter region also shifted (Fig. 1b). Purified D55A OmpR protein also caused a gel shift of the fimB DNA fragment, paralleling the unphosphorylated OmpR result (Fig. 1c). Another gel mobility shift with the radiolabelled fimE promoter region DNA showed no gel shift with unphosphorylated OmpR, demonstrating that the unphosphorylated OmpR protein did not bind non-specifically to DNA fragments (Fig. 1d). These results suggested that unphosphorylated OmpR was sufficient to repress fimB transcription.
Fig. 1.
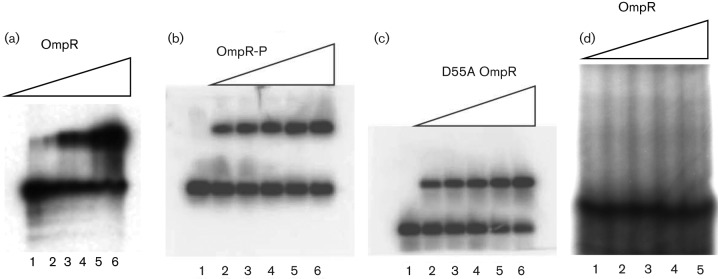
Gel mobility shift assays that use OmpR protein. (a) A gel shift with the 445 bp 32P-labelled fimB promoter region and OmpR. (b) A gel shift with the 445 bp 32P-labelled fimB promoter region and OmpR-P. (c) A gel shift with the 445 bp 32P-labelled fimB promoter region and D55A OmpR. (d) A gel shift with the 340 bp 32P-labelled fimE promoter region. Lanes were loaded as follows: (1) DNA alone, (2) DNA plus 0.5 µM OmpR, (3) DNA plus 1 µM OmpR, (4) DNA plus 2 µM OmpR, (5) DNA plus 5 µM Omp and (6) DNA plus 8 µM OmpR.
To determine more precisely where OmpR bound, DNase I footprinting assays were performed with DNA that was amplified by PCR with primers specific for the fimB promoter region 2. A sequencing ladder of the fimB sequence was used to determine exactly where OmpR/OmpR-P bound on the fimB promoter region 2 (data not shown). Unphosphorylated OmpR weakly bound to a 20 bp region (+2 to −18) (Fig. 2) and protected three bases of the −10 box, which included a hypersensitive site at the A at position −7 (Figs 2 and 3). In addition, a 24 bp region (+2 to −22) of the fimB promoter region 2 was protected by OmpR phosphorylated with phosphoramidate (Figs 2 and S1), spanning the entire −10 box plus 17 bases downstream of the −10 box.
Fig. 2.
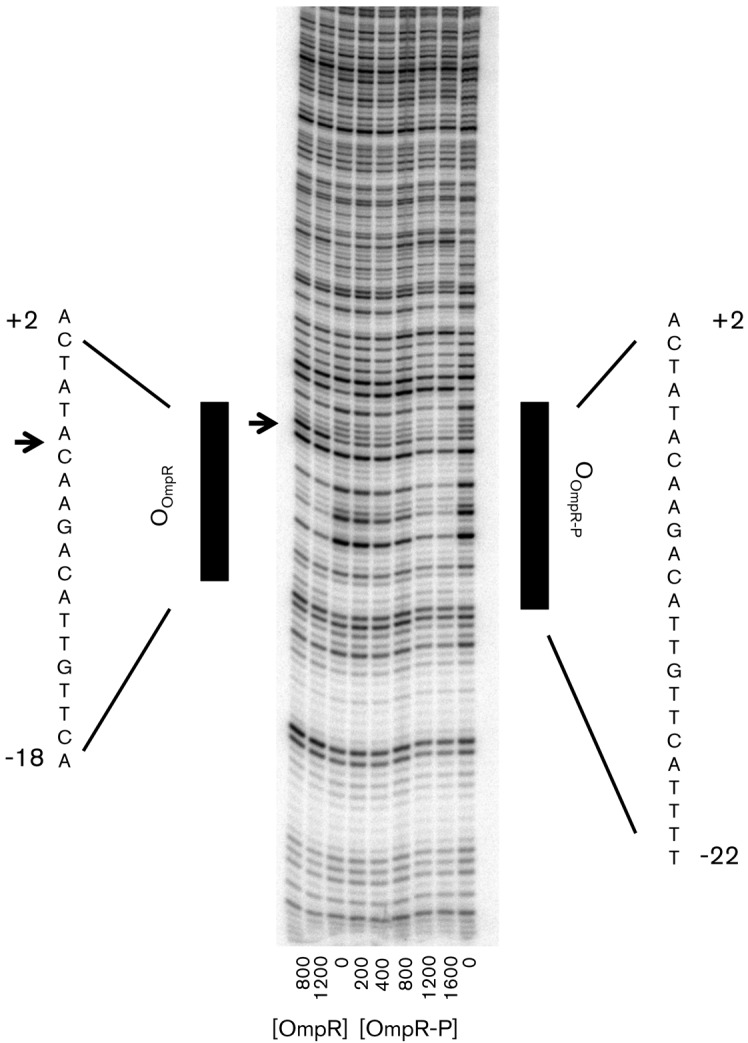
DNase I footprinting analysis of unphosphorylated (OmpR) and phosphorylated OmpR (OmpR-P) at the fimB promoter region 2. The concentration (nM) of OmpR and OmpR-P used in each lane is indicated at the bottom. The lanes with ‘0’ indicate the cleavage pattern by DNase I in the absence of OmpR/OmpR-P. The coordinates indicate positions relative to the fimB promoter 2 site. Regions of protection are indicated by black bars for OmpR (+2 to −18) and OmpR-P (+2 to −22) and a hypersensitive site is indicated by an arrow.
Fig. 3.
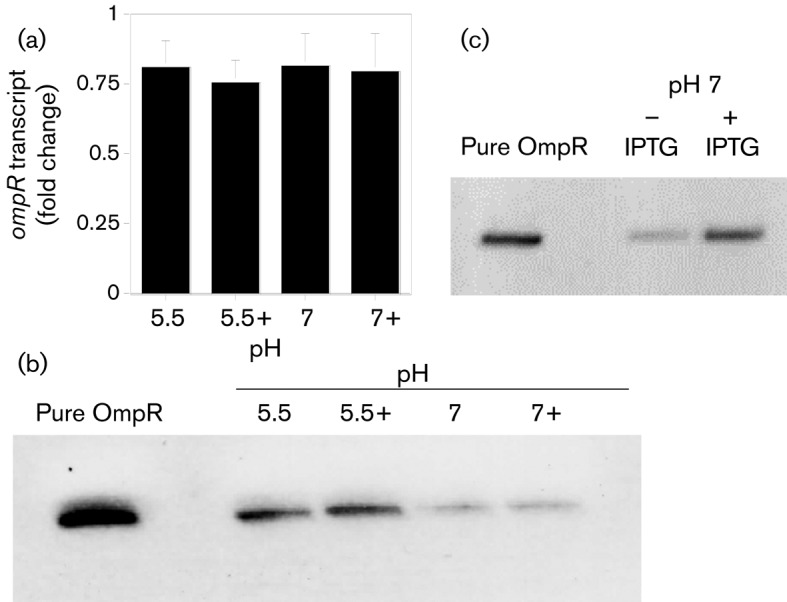
Quantitative determination of ompR transcript and OmpR expression levels in exponentially grown UPEC NU149 cells in different environments. (a) qRT-PCR analysis of UPEC NU149 cells grown to mid-exponential phase in pH 5.5, pH 5.5 plus 800 mOsm NaCl (5.5+), pH 7.0 and pH 7.0 plus 800 mOsm NaCl (7.0+) LB. The fold change in ompR transcript levels that were corrected using ftsZ are shown as the mean±sd from three separate runs. (b) Western blot analysis of OmpR protein expression. One hundred nanograms of purified OmpR protein or 10 µg of total protein isolated from NU149 grown in pH 5.5, 5.5+ (pH 5.5 LB with 800 mOsm NaCl), 7.0 and 7.0+ (pH 7.0 LB with 800 mOsm NaCl) LB. (c) One hundred nanograms of purified OmpR protein or 10 µg of total protein isolated from NU149 grown in pH 7.0 LB with (+) or without (−) IPTG. The blots were probed with absorbed polyclonal rabbit anti-OmpR antibody.
UPEC ompR transcription is constant regardless of growth conditions
As both OmpR and OmpR-P bound weakly to one of the fimB promoters, we hypothesized that a high OmpR protein concentration in a low pH/high osmolality environment could be responsible for the regulation of fimB. To test this idea, we first determined the level of ompR transcription using cDNAs converted from total RNAs extracted from UPEC NU149 grown in four different media [pH 5.5 (low osmolality), pH 5.5/800 mOsm NaCl (high osmolality), pH 7.0 (low osmolality) and pH 7.0/800 mOsm NaCl (high osmolality)]. We used the UPEC NU149 strain for the remainder of the experiments to have a more valid comparison. Using RT-PCR, no discernible difference in ompR transcription was observed for the UPEC grown in the four media (data not shown).
As PCR lacks sensitivity, qRT-PCR was subsequently performed to ascertain ompR transcription differences compared with an ftsZ housekeeping control. After applying the method (Livak & Schmittgen, 2001) to normalize the ompR transcripts to the ftsZ gene, no significant difference in ompR transcription was found in either logarithmic (Fig. 3a) or stationary phase UPEC cultures (data not shown) grown in the various media. Thus, the UPEC growth environment did not differentially regulate ompR transcription.
OmpR protein level increases in a combined low pH/high osmolality medium
Although ompR transcription did not differ in UPEC NU149 when grown in the four growth conditions, post-transcriptional regulation could affect OmpR protein levels and then fimB transcription. To determine whether there was a difference in OmpR protein levels in cultures from each growth condition, SDS-PAGE and Western blot analyses were performed on stationary phase UPEC NU149 cell lysates probed with absorbed polyclonal rabbit anti-OmpR antibody. One hundred nanograms of purified histidine-tagged OmpR protein reacted well with the antibody (Fig. 3b).
From the Western blot analysis, the steady-state levels of OmpR were higher in an acidic medium versus a neutral medium (Fig. 3b). A significant (fourfold) increase in OmpR protein expression occurred when NU149 cells grown in pH 5.5 (low osmolality) medium (29 715 pixels) were compared with the same strain grown in the pH 7.0 (low osmolality) medium (7443 pixels, P<0.0076). This result did not correlate with the results from Table 1 that showed no significant difference in fimB transcription under these conditions when comparing the wild-type and ompR strains. This discrepancy suggests that another global regulator may play a role under acidic conditions. Osmolality did not appear to have a major effect on steady-state OmpR levels. There was no significant difference in OmpR expression in cells grown in pH 5.5 medium (low osmolality, 29 715 pixels) versus pH 5.5/800 mOsm NaCl medium (high osmolality, 34 898 pixels) (P<0.266). A slightly significant effect was observed in cells grown in a pH 7.0 environment (low osmolality, 7443 pixels) compared with UPEC cells grown in pH 7.0/800 mOsm NaCl (10 896 pixels, 1.5-fold increase, P<0.02). These OmpR expression profiles did not entirely correlate with the fimB results from Table 1, again suggesting that another global regulator may be a co-factor. The difference in OmpR expression in strain NU149 was the highest (4.7-fold) when comparing cells grown in pH 5.5/800 mOsm NaCl medium with cells grown in pH 7.0 medium (P<0.0076). Cells grown to stationary phase also displayed significant differences in OmpR levels when comparing pH 5.5/800 mOsm with pH 7.0 grown cells (data not shown).
In addition, quantitative Western blots using different concentrations of purified OmpR protein confirmed the fold differences observed in Fig. 3(b) (data not shown). Collectively, these results demonstrated that UPEC grown in vitro in an acidic/high osmolality environment express more OmpR protein through a post-transcription change than cells grown in a neutral pH/low osmolality environment.
Because there was a low level of OmpR expression in cells grown in pH 7.0 medium, less OmpR would be available to repress fimB transcription and this could explain the high level of fimB transcription in a pH 7.0 environment shown in Table 1. We hypothesized that if OmpR levels were artificially increased in cells grown in pH 7.0 medium, this change would result in decreased fimB expression and decreased levels of HA of guinea pig erythrocytes, used as a relative measure of type 1 pili on the surface, due to decreased type 1 pili. To test this idea, the pFR29* plasmid, containing the wild-type ompR gene under IPTG control, was introduced into the clinical strain NU149. NU149/pFR29* cells were grown in pH 7.0 medium, divided into aliquots that were treated or untreated with IPTG, and whole-cell lysates prepared. Western blots performed on these lysates confirmed that addition of IPTG resulted in increased levels of OmpR. While the lysate derived from cells grown without IPTG displayed a level of OmpR expression (8516 pixels) consistent with the prior blot, the lysate derived from cells grown with IPTG had elevated OmpR (20 241 pixels) (Fig. 3c). Significantly, a qRT-PCR analysis of both populations showed 2.6-fold less fimB expression in the IPTG-induced population compared with the population grown in standard pH 7.0 LB. When both cultures were tested for their HA, the HA titre of the population without IPTG was 524, whereas the HA titre for the population with IPTG was 128, consistent with decreased levels of type 1 pili. These results demonstrate that overexpression of OmpR under pH 7.0 growth conditions decreases fimB expression and overrides post-transcriptional regulation of OmpR.
Levels of fimB transcripts change in the ompR strain grown in a high osmolality environment
The β-galactosidase assays suggested that in an ompR strain, expression of fimB increased compared with the wild-type cells when grown in a high osmolality environment. To verify these results, qRT-PCR was performed on total RNAs isolated from NU149 and NU149 OmpR1 cells grown in the same media as described above. Data points were standardized to ftsZ expression. The pH 7.0 medium with no added salt was used as a baseline as there was little difference between the NU149 and NU149 OmpR1 data (Fig. 4, P<0.493). When the osmolality was increased to 800 mOsm at pH 7.0, fimB levels dropped approximately 4.2-fold in the wild-type lane versus a 1.4-fold drop in the ompR lane (P<0.024). In the pH 5.5 LB, fimB transcript levels dropped about 2.6-fold in the wild-type lane versus pH 7.0 (P<0.032), but there was an 8.7-fold drop in fimB transcript levels in the ompR lane under the same growth condition as compared with pH 7.0 (P<0.019). The fimB levels dropped further (8-fold decline compared with pH 7.0, P<0.013) in wild-type cells grown in the pH 5.5 LB with 800 mOsm medium, but fimB transcript levels declined less (2.8-fold, P<0.06) in the ompR strain when compared with pH 7.0. The results paralleled the β-galactosidase assay findings, demonstrating that fimB transcript levels are higher in the ompR versus wild-type strain when both were grown in a high osmolality environment.
Fig. 4.
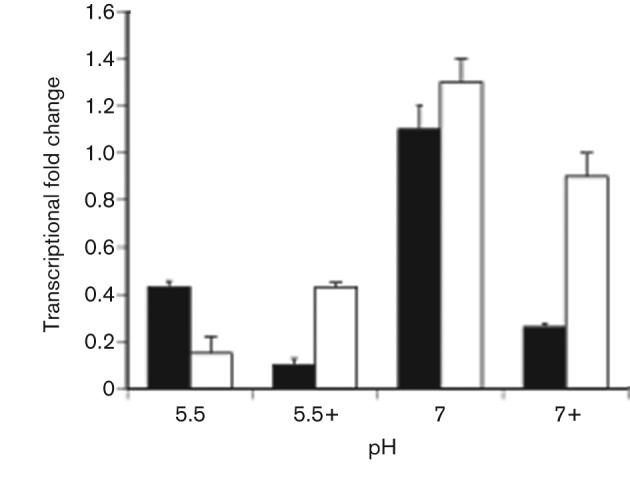
Quantitative determination of fimB transcript expression by qRT-PCR. UPEC NU149 (solid black bars) and NU149 OmpR1 (open white bars) cells were grown to mid-exponential phase in pH 5.5, pH 5.5 plus 800 mOsm NaCl (5.5+), pH 7.0 and 7.0 plus 800 mOsm NaCl (7.0+) LB. The fold change in fimB transcript levels that were corrected using ftsZ are shown as the mean±sd from two separate runs.
Orientation of the fimS element changes in the ompR strain grown under high osmolality conditions
Previously, we have demonstrated that the orientation of the 314 bp fimS element was affected by pH and osmolality (Schwan et al., 1992, 2002). Because the fimB transcript level changed in the ompR mutant strain when grown under high osmolality conditions, we investigated whether there was also an effect on the positioning of the fimS element. A PCR-based approach (Schwan et al., 1992, 2007; Fig. S2) was used combined with a quantitative comparison of fimS orientation (Teng et al., 2005) to measure the prevalence of Phase-ON and Phase-OFF DNA in NU149, NU149 OmpR1 and NU149 OmpR1/pFR29* populations when grown in LB with varying pH and osmolality. This analysis showed that ompR mutant cells grown at pH 7.0 (ratio ON/OFF = 83 : 17 %) had the invertible element positioned approximately equal to the wild-type cells (ratio ON/OFF = 81 : 19 %) and the complemented mutant (ratio ON/OFF = 78 : 22 %, Fig. 5). Under pH 7.0 with high salt growth conditions, the wild-type cells shifted significantly towards a Phase-OFF orientation (ratio ON/OFF = 48 : 52 %), but the ompR mutant appeared still to position the fimS element to favour the Phase-ON orientation (ratio ON/OFF = 70 : 30 %) compared with wild-type cells (ratio ON/OFF = 48 : 52 %). In pH 5.5 LB, the ompR mutant had more cells in the Phase-OFF orientation (ratio ON/OFF = 18 : 82 %) than the wild-type (ratio ON/OFF = 62 : 38 %), consistent with the observed effect on fimB transcription. However, more Phase-ON oriented cells were present in the ompR mutant in pH 5.5 LB with 800 mOsm NaCl (ratio ON/OFF = 51 : 49 %) versus wild-type cells (ratio ON/OFF = 20 : 80 %). Together, these results indicated that the derepression of fimB in the ompR mutant led to more Phase-ON oriented cells when grown in a high osmolality environment. In a low osmolality/low pH environment, less fimB was transcribed in the ompR strain compared with wild-type, suggesting another regulatory system such as an acid tolerance system regulator may be involved in regulating fimB in the absence of OmpR.
Fig. 5.
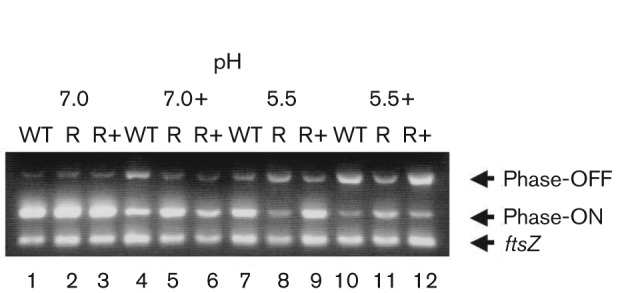
Determination of the fimS invertible element orientation by PCR on chromosomal DNAs isolated from NU149, NU149 OmpR1 and NU149 OmpR1/pFR29* cells grown in pH 5.5 or 7.0 LB with or without added NaCl (+, added NaCl). Cells were harvested during mid-exponential phase following aerobic growth with agitation at 250 r.p.m. at 37 °C in pH 7.0 LB media with either no added NaCl (low salt) or 800 mOsm added NaCl (high salt) or in pH 5.5 LB medium with either no added NaCl (low salt) or 800 mM NaCl (high salt). Multiplex PCRs were set up with INV and FIMA primers to amplify Phase-ON-oriented DNA (ON, 450 bp product) (Schwan et al., 1992), FIME and INV primers to amplify Phase-OFF-oriented DNA (OFF, 750 bp product) (Schwan et al., 1992), and EcFtsZ 1 and 2 primers to amplify the ftsZ gene (302 bp product) (Schwan et al., 2007). Each multiplex was run at least three separate times. WT, wild-type NU149; R, ompR strain; R+, ompR strain complemented with pFR29*. The lanes were loaded onto a 1.5 % agarose gel as follows: lane 1, NU149 (pH 7.0, low salt); lane 2, NU149 OmpR1 (pH 7.0, low salt); lane 3, NU149 OmpR1/pFR29* (pH 7.0, low salt); lane 4, NU149 (pH 7.0, high salt); lane 5, NU149 OmpR1 (pH 7.0, high salt); lane 6, NU149 OmpR1/pFR29* (pH 7.0, high salt); lane 7, NU149 (pH 5.5, low salt); lane 8, NU149 OmpR1 (pH 5.5, low salt); lane 9, NU149 OmpR1/pFR29* (pH 5.5, low salt); lane 10, NU149 (pH 5.5, high salt); lane 11, NU149 OmpR1 (pH 5.5, high salt); and lane 12, NU149 OmpR1/pFR29* (pH 5.5, high salt). For each lane, the intensities of the OFF and ON states were quantified using ImageQuant software (Molecular Dynamics) and corrected to the intensity of the ftsZ band. The corrected values for both states were standardized to the respective wild-type band (lane 1).
Production of type 1 pili is altered in the ompR cells versus wild-type cells
Both fimB transcript levels and positioning of the fimS invertible element changed in the ompR strain grown under high osmotic stress conditions. To determine if the level of type 1 pili also changed in the ompR mutant strain, HA assays were done using guinea pig erythrocytes. The results showed that NU149, NU149 OmpR1 (ompR) and NU149 OmpR1/pFR29* cells had the same HA assay titres when grown at pH 7.0 (Table 2). However, when the osmolality increased by 800 mOsm at pH 7.0, both the wild-type and the complemented mutant populations had an eightfold decline in the HA titre, whereas the ompR population had only a twofold drop. Furthermore, transmission electron microscopy demonstrated the ompR mutant strain exhibited more type 1 pili (Fig. 6a) compared with the wild-type strain (Fig. 6b).
Fig. 6.
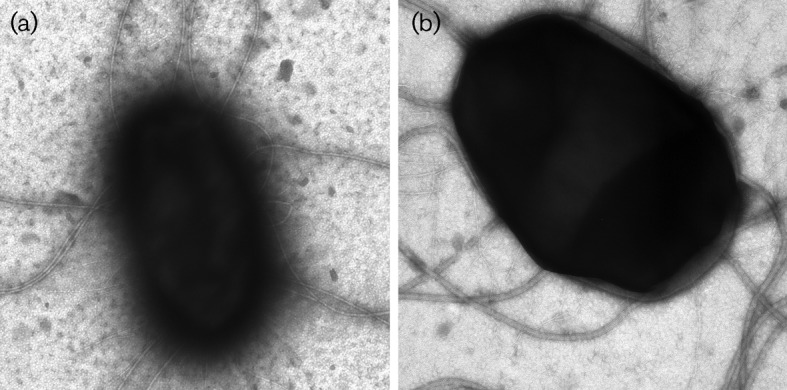
Electron micrographs of NU149 OmpR1 (a) and NU149 (b) cells grown in pH 7.0 LB with 800 mOsm NaCl. Magnification, ca. ×100 000.
In an acidic growth environment (pH 5.5), the wild-type and complemented mutant strains showed a fourfold drop in the HA titre versus the pH 7.0 baseline. The ompR strain showed an additional eightfold drop over the HA titre displayed by the wild-type strain. On the other hand, when the osmolality was increased by 800 mOsm in the same pH 5.5 environment, wild-type and complemented mutant strains had further declines in the HA titre to 4, but the ompR strain had a rebound in the HA titre up to 32. These results demonstrate that OmpR lowers type 1 pili expression under high osmotic stress conditions.
Discussion
In the human urinary tract, UPEC survive in the differing pH/osmotically stressed environments by utilizing the EnvZ/OmpR system. Previously, we showed that the osmolality of the growth environment affects UPEC type 1 pilus expression and our results suggested that OmpR was regulating several fim genes (Schwan et al., 2002). Our working hypothesis was that in an acidic/high osmolality environment, OmpR repressed fimB transcription in UPEC by binding to a fimB promoter. Using gel mobility shift and DNase footprinting assays, we have demonstrated that OmpR binds to a fimB promoter region. In addition, genetic and phenotypic data indicated that under high osmolality conditions, OmpR represses fimB transcription, leading to an increased Phase-OFF position for the fimS switch and a corresponding decrease in expression of type 1 pili.
OmpR regulates other E. coli genes (Oshima et al., 2002). Several studies have demonstrated OmpR/OmpR-P binding upstream of the −35 and −10 boxes of the ompF and ompC promoter regions (Head et al., 1998; Maeda et al., 1988; Tsung et al., 1989). OmpR binding to a 20 bp region in the ompF promoter region has been previously observed (Harlocker et al., 1995; Huang & Igo, 1996), and a loose consensus sequence containing an AC at positions 1,2 and 11,12 with a central GXXXC sequence was proposed for OmpR binding (Head et al., 1998). A more recent study further defined the OmpR binding site as having a core sequence comprising a central GTXTCA (5′–3′ with the X being either an A or a T) (Rhee et al., 2008).
Our results showed that OmpR and OmpR-P shifted the mobility of a DNA sequence spanning the fimB promoter 1 and 2 region. Furthermore, DNase footprinting experiments demonstrated that OmpR and OmpR-P protected 20 and 24 bp sites, respectively, at the fimB promoter 2. Binding of the 20 bp site by OmpR may be due to binding of an unphosphorylated OmpR monomer. The expanded 24 bp protected area might be due to another phosphorylated OmpR monomer attaching to the first OmpR monomer bound to the DNA, resulting in a dimer. Rhee et al. (2008) have proposed such a model to explain some of the OmpR-mediated gene regulation. In our study, both of the OmpR protected regions spanned the −10 box of the fimB promoter 2. Within these protected regions, a putative OmpR binding motif sequence, 5′-ACTTGTTACAGAAC-3′, with an AC at positions 1, 2 as well as an internal GTTACA sequence, was found (Fig. 3). Although the internal sequence for the fimB promoter 2 does not precisely match the core GTXTCA OmpR binding sequence (Rhee et al., 2008), it only differs by one base, with an A rather than a T at the fourth position. This one base mismatch may explain the relatively low affinity of OmpR/OmpR-P binding for the fimB promoter 2 region as compared with the high-affinity sequence for ompF and ompC promoter regions. Low-affinity OmpR-P binding has been demonstrated for the flhDC promoter region, but no conserved core sequence was observed (Shin & Park, 1995).
A previous study by Cai & Inouye (2002) indicated that at low osmolality, phosphorylation of only 3.5 % (120 nM or 70 OmpR-P molecules per cell) of total OmpR protein (2024 molecules of OmpR per cell) would be necessary for promoting ompF transcription. However, in a high osmolality medium, phosphorylation of 10 % (580 nM or 350 OmpR-P molecules per cell) of total OmpR molecules in a cell (3500 molecules of OmpR per cell) would be enough to repress the ompF gene and stimulate expression of the ompC gene (Cai & Inouye, 2002). Our fimB DNase footprint assays showed protection by OmpR and OmpR-P at 800 nM (Fig. 5), equating to 466 OmpR molecules per cell, or 13.5 % of total OmpR protein. Our observed binding of OmpR to the fimB promoter 2 region occurs at levels well within the physiological range of OmpR protein that would be present in UPEC growing in a high osmolality environment. Whether OmpR binds to either of the two remaining fimB promoters remains unknown and will serve as an area of further exploration.
Since both OmpR and OmpR-P bound weakly to the fimB promoter region 2, we investigated whether OmpR protein itself might be influenced by pH or osmolality in UPEC cells, through either transcriptional or translational changes. We hypothesized that OmpR levels would be higher in an acidic/high osmolality environment, leading to low-affinity binding to the fimB promoter and direct regulation of fimB transcription. However, the RT-PCR and qRT-PCR analyses showed no difference in ompR transcript levels in strain NU149 grown in the four growth conditions used previously (Schwan et al., 2002). These results ruled out the possibility that transcriptional changes in ompR were responsible for the fimB regulation.
In E. coli, envZ and ompR are co-transcribed, but 35 times more OmpR protein is present compared with EnvZ, presumably because of differences in translational initiation (Cai & Inouye, 2002). Based on this previous study and the lack of change in our ompR transcriptional assays, we hypothesized post-transcriptional regulation of OmpR in response to low pH and/or high osmolality conditions. Western blots confirmed that OmpR protein levels increased (fourfold) in UPEC cells in response to acidic conditions, providing evidence for post-transcriptional regulation of OmpR. The post-transcriptional change could be tied to greater translational initiation of the ompR transcript in an acidic environment; greater binding by RNA-binding proteins (e.g. Hfq-dependent stimulation of translation) under stress conditions (Nogueira & Springer, 2000); or through interactions with small, non-coding RNAs that affect gene regulation (Valentin-Hansen et al., 2007). Indeed, Hfq-binding sRNA is linked to a number of target-mRNAs, including porin proteins, regulated at the post-transcriptional level within E. coli (Guillier & Gottesman, 2006). OmpR is regulated by at least two sRNAs, OmrA and OmrB, bound to Hfq (Guillier & Gottesman, 2008). As sRNAs derepress translation by smoothing out mRNA hairpin loops (Majdalani et al., 1998), the sRNAs bound to Hfq may be allowing better translation of ompR transcripts when UPEC cells are grown under acidic conditions.
Levels of fimB expression were highest in a neutral pH/low osmolality condition and lowest in a low pH/high osmolality medium. A switch from a neutral pH/low osmolality environment that favours fimB activation by SlyA (a proposed activator of fimB transcription, McVicker et al., 2011) or RcsB (another proposed activator of fimB transcription, Schwan et al., 2007) to a low pH/high osmolality environment found in the human urinary tract that favours OmpR could have relevance in regulating fimB (Schwan, 2011). OmpR could displace SlyA or RcsB on the fimB promoter site as the OmpR level increases in UPEC growing in an acidic/high osmolality environment. These results suggest a possible physiological role for OmpR in E. coli growing in acidic/high osmolality environments, such as the human kidney.
Prior studies have indicated that OmpR protein expression in Salmonella and enterohaemorrhagic E. coli increased after acid shock or growth in an acidic environment compared with a neutral pH environment (Bang et al., 2000; Huang et al., 2007). These previous studies provide evidence that the OmpR protein is important for survival in acidic environments, and our data showing that the OmpR protein is elevated in response to low pH is consistent with these prior works.
Previously, wild-type E. coli grown in a high osmolality environment (added sucrose) had a 1.7-fold increase in OmpR protein as compared with cells grown in a low osmolality medium (Cai & Inouye, 2002). Although our data indicated only subtle changes in OmpR protein levels in response to high osmolality, OmpR/OmpR-P ratios could be influenced by osmolality and may be a contributing factor in vivo. If OmpR/OmpR-P represses fimB expression, an ompR mutant strain should display greater levels of fimB transcripts and higher expression of type 1 pili in UPEC grown in a high osmolality growth environment. Indeed, fimB transcript levels were four- to eightfold higher in the ompR mutant compared with wild-type cells under high osmolality conditions, which in turn led to more Phase-ON cells and higher type 1 pili expression. Our model is that greater concentrations of OmpR and OmpR-P in UPEC growing in kidney urine (acidic/high osmolality) may allow OmpR-P to directly bind first to the higher affinity sites on the fimB promoter region followed by binding to lower affinity sites as the concentration of OmpR increased. This scenario would be similar to the binding affinity regulating the ompF gene (Huang & Igo, 1996; Qin et al., 2001; Rampersaud et al., 1994). OmpR and OmpR-P binding to the fimB promoter region would repress fimB transcription, thereby increasing the cellular ratio of FimE to FimB. In turn, FimE would bind to the invertible element, switching it to the Phase-OFF orientation and resulting in non-piliated bacteria in the kidneys observed in previous studies (Hultgren et al., 1985; Schaeffer et al., 1987). Non-piliated UPEC cells in the kidney would be less likely to be phagocytized by phagocytic cells targeting the type 1 pili (Perry et al., 1983; Silverblatt et al., 1979).
Acknowledgements
We thank Linda Kenney, Leslie Morgan and Don Walthers for their assistance with the DNase I footprinting, anti-OmpR antibody, purified OmpR protein and mutant strains used in this study. All of the DNase I footprints were performed in Linda Kenney’s laboratory. We also thank David Anderson and Matthew Zuberbuehler for technical assistance with the RNA extractions. This study was funded by a University of Wisconsin-La Crosse graduate student Research, Service and Educational Leadership grant to A. E. R. and a National Institutes of Health grant AI065432-01A2 to W. R. S.
Abbreviations:
- HA
haemagglutination
- OmpR-P
phosphorylated OmpR
- UPEC
uropathogenic Escherichia coli
Footnotes
One supplementary table and two supplementary figures are available with the online version of this paper.
References
- Abraham S. N., Babu J. P., Giampapa C. S., Hasty D. L., Simpson W. A., Beachey E. H. (1985a). Protection against Escherichia coli-induced urinary tract infections with hybridoma antibodies directed against type 1 fimbriae or complementary d-mannose receptors. Infect Immun 48, 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. (1985b). An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A 82, 5724–5727. 10.1073/pnas.82.17.5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang I. S., Kim B. H., Foster J. W., Park Y. K. (2000). OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol 182, 2245–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomfield I. C., McClain M. S., Princ J. A., Calie P. J., Eisenstein B. I. (1991). Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol 173, 5298–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. J., Inouye M. (2002). EnvZ–OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277, 24155–24161. 10.1074/jbc.M110715200 [DOI] [PubMed] [Google Scholar]
- Delgado J., Forst S., Harlocker S., Inouye M. (1993). Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol 10, 1037–1047. 10.1111/j.1365-2958.1993.tb00974.x [DOI] [PubMed] [Google Scholar]
- Donato G. M., Lelivelt M. J., Kawula T. H. (1997). Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol 179, 6618–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Labany S., Sohanpal B. K., Lahooti M., Akerman R., Blomfield I. C. (2003). Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol Microbiol 49, 1109–1118. 10.1046/j.1365-2958.2003.03624.x [DOI] [PubMed] [Google Scholar]
- Forst S., Delgado J., Ramakrishnan G., Inouye M. (1988). Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J Bacteriol 170, 5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Delgado J., Inouye M. (1989). Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86, 6052–6056. 10.1073/pnas.86.16.6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman B. (2003). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon 49, 53–70. 10.1016/S0011-5029(03)90000-9 [DOI] [PubMed] [Google Scholar]
- Foxman B., Barlow R., D’Arcy H., Gillespie B., Sobel J. D. (2000). Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol 10, 509–515. 10.1016/S1047-2797(00)00072-7 [DOI] [PubMed] [Google Scholar]
- Freitag C. S., Abraham J. M., Clements J. R., Eisenstein B. I. (1985). Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol 162, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M., Gottesman S. (2006). Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59, 231–247. 10.1111/j.1365-2958.2005.04929.x [DOI] [PubMed] [Google Scholar]
- Guillier M., Gottesman S. (2008). The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36, 6781–6794. 10.1093/nar/gkn742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlocker S. L., Bergstrom L., Inouye M. (1995). Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem 270, 26849–26856. [DOI] [PubMed] [Google Scholar]
- Head C. G., Tardy A., Kenney L. J. (1998). Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol 281, 857–870. 10.1006/jmbi.1998.1985 [DOI] [PubMed] [Google Scholar]
- Hooton T. M., Stamm W. E. (1997). Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin North Am 11, 551–581. 10.1016/S0891-5520(05)70373-1 [DOI] [PubMed] [Google Scholar]
- Huang K. J., Igo M. M. (1996). Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol 262, 615–628. 10.1006/jmbi.1996.0540 [DOI] [PubMed] [Google Scholar]
- Huang Y. J., Tsai T. Y., Pan T. M. (2007). Physiological response and protein expression under acid stress of Escherichia coli O157 : H7 TWC01 isolated from Taiwan. J Agric Food Chem 55, 7182–7191. 10.1021/jf071014s [DOI] [PubMed] [Google Scholar]
- Hultgren S. J., Porter T. N., Schaeffer A. J., Duncan J. L. (1985). Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect Immun 50, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Silhavy T. J. (1988). EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol 170, 5971–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B. R., Maurer L., Spears P. A., Orndorff P. E. (1986). Receptor-binding function of type 1 pili effects bladder colonization by a clinical isolate of Escherichia coli. Infect Immun 53, 693–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney L. J., Bauer M. D., Silhavy T. J. (1995). Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci U S A 92, 8866–8870. 10.1073/pnas.92.19.8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. (1986). Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J 5, 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin C. M., Chambers S. T. (1989). Osmoprotective properties for bacteria of renal papilla and urine: role of betaines as osmoprotectant molecules. In Host–arasite Interactions in Urinary Tract Infections, pp. 327–332. Edited by Kass E., Svanborg Eden C. Chicago: University of Chicago Press. [Google Scholar]
- Liu X., Ferenci T. (2001). An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147, 2981–2989. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maeda S., Ozawa Y., Mizuno T., Mizushima S. (1988). Stereospecific positioning of the cis-acting sequence with respect to the canonical promoter is required for activation of the ompC gene by a positive regulator, OmpR, in Escherichia coli. J Mol Biol 202, 433–441. 10.1016/0022-2836(88)90276-8 [DOI] [PubMed] [Google Scholar]
- Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S. (1998). DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 95, 12462–12467. 10.1073/pnas.95.21.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M., Mizuno T. (1999). EnvZ-independent phosphotransfer signaling pathway of the OmpR-mediated osmoregulatory expression of OmpC and OmpF in Escherichia coli. Biosci Biotechnol Biochem 63, 408–414. 10.1271/bbb.63.408 [DOI] [PubMed] [Google Scholar]
- Mattison K., Kenney L. J. (2002). Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J Biol Chem 277, 11143–11148. 10.1074/jbc.M111128200 [DOI] [PubMed] [Google Scholar]
- McClain M. S., Blomfield I. C., Eisenstein B. I. (1991). Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 173, 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M. S., Blomfield I. C., Eberhardt K. J., Eisenstein B. I. (1993). Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol 175, 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary W. R., Stock J. B. (1994). Acetyl phosphate and the activation of two-component response regulators. J Biol Chem 269, 31567–31572. [PubMed] [Google Scholar]
- McVicker G., Sun L., Sohanpal B. K., Gashi K., Williamson R. A., Plumbridge J., Blomfield I. C. (2011). SlyA protein activates fimB gene expression and type 1 fimbriation in Escherichia coli K-12. J Biol Chem 286, 32026–32035. 10.1074/jbc.M111.266619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Nogueira T., Springer M. (2000). Post-transcriptional control by global regulators of gene expression in bacteria. Curr Opin Microbiol 3, 154–158. 10.1016/S1369-5274(00)00068-0 [DOI] [PubMed] [Google Scholar]
- Ofek I., Beachey E. H. (1978). Mannose binding and epithelial cell adherence of Escherichia coli. Infect Immun 22, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P. B., Klemm P. (1994). Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol Lett 116, 95–100. 10.1111/j.1574-6968.1994.tb06681.x [DOI] [PubMed] [Google Scholar]
- Oshima T., Aiba H., Masuda Y., Kanaya S., Sugiura M., Wanner B. L., Mori H., Mizuno T. (2002). Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46, 281–291. 10.1046/j.1365-2958.2002.03170.x [DOI] [PubMed] [Google Scholar]
- Pallesen L., Madsen O., Klemm P. (1989). Regulation of the phase switch controlling expression of type 1 fimbriae in Escherichia coli. Mol Microbiol 3, 925–931. 10.1111/j.1365-2958.1989.tb00242.x [DOI] [PubMed] [Google Scholar]
- Perry A., Ofek I., Silverblatt F. J. (1983). Enhancement of mannose-mediated stimulation of human granulocytes by type 1 fimbriae aggregated with antibodies on Escherichia coli surfaces. Infect Immun 39, 1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohinar P., Forst S. A., Reed D., Mandic-Mulec I., Weiss J. (2002). OmpR-dependent and OmpR-independent responses of Escherichia coli to sublethal attack by the neutrophil bactericidal/permeability increasing protein. Mol Microbiol 43, 1493–1504. 10.1046/j.1365-2958.2002.02804.x [DOI] [PubMed] [Google Scholar]
- Qin L., Yoshida T., Inouye M. (2001). The critical role of DNA in the equilibrium between OmpR and phosphorylated OmpR mediated by EnvZ in Escherichia coli. Proc Natl Acad Sci U S A 98, 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud A., Harlocker S. L., Inouye M. (1994). The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem 269, 12559–12566. [PubMed] [Google Scholar]
- Rhee J. E., Sheng W., Morgan L. K., Nolet R., Liao X., Kenney L. J. (2008). Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem 283, 8664–8677. 10.1074/jbc.M705550200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. L., Neely A. E. (1983). Renal function. In Textbook of Urinalysis and Body Fluids, pp. 97–112. Norwalk, VA: Appleton Century Crofts. [Google Scholar]
- Russo F. D., Silhavy T. J. (1991). EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol 222, 567–580. 10.1016/0022-2836(91)90497-T [DOI] [PubMed] [Google Scholar]
- Schaeffer A. J., Amundsen S. K., Schmidt L. N. (1979). Adherence of Escherichia coli to human urinary tract epithelial cells. Infect Immun 24, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer A. J., Schwan W. R., Hultgren S. J., Duncan J. L. (1987). Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect Immun 55, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R. (2009). Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155, 1832–1839. 10.1099/mic.0.026187-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R. (2011). Regulation of fim genes in uropathogenic Escherichia coli. World J Clin Infect Dis 1, 17–25. 10.5495/wjcid.v1.i1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R., Seifert H. S., Duncan J. L. (1992). Growth conditions mediate differential transcription of fim genes involved in phase variation of type 1 pili. J Bacteriol 174, 2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R., Seifert H. S., Duncan J. L. (1994). Analysis of the fimB promoter region involved in type 1 pilus phase variation in Escherichia coli. Mol Gen Genet 242, 623–630. 10.1007/BF00285286 [DOI] [PubMed] [Google Scholar]
- Schwan W. R., Lee J. L., Lenard F. A., Matthews B. T., Beck M. T. (2002). Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun 70, 1391–1402. 10.1128/IAI.70.3.1391-1402.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan W. R., Shibata S., Aizawa S.-I., Wolfe A. J. (2007). The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J Bacteriol 189, 7159–7163. 10.1128/JB.00705-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Park C. (1995). Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177, 4696–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverblatt F. J., Dreyer J. S., Schauer S. (1979). Effect of pili on susceptibility of Escherichia coli to phagocytosis. Infect Immun 24, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentebjerg-Olesen B., Chakraborty T., Klemm P. (2000). FimE-catalyzed off-to-on inversion of the type 1 fimbrial phase switch and insertion sequence recruitment in an Escherichia coli K-12 fimB strain. FEMS Microbiol Lett 182, 319–325. 10.1111/j.1574-6968.2000.tb08915.x [DOI] [PubMed] [Google Scholar]
- Teng C. H., Cai M., Shin S., Xie Y., Kim K. J., Khan N. A., Di Cello F., Kim K. S. (2005). Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect Immun 73, 2923–2931. 10.1128/IAI.73.5.2923-2931.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsung K., Brissette R. E., Inouye M. (1989). Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J Biol Chem 264, 10104–10109. [PubMed] [Google Scholar]
- Valentin-Hansen P., Johansen J., Rasmussen A. A. (2007). Small RNAs controlling outer membrane porins. Curr Opin Microbiol 10, 152–155. 10.1016/j.mib.2007.03.001 [DOI] [PubMed] [Google Scholar]
- van der Bosch J. F., Verboom-Sohmer U., Postma P., de Graaff J., MacLaren D. M. (1980). Mannose-sensitive and mannose-resistant adherence to human uroepithelial cells and urinary virulence of Escherichia coli. Infect Immun 29, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virkola R., Westerlund B., Holthöfer H., Parkkinen J., Kekomäki M., Korhonen T. K. (1988). Binding characteristics of Escherichia coli adhesins in human urinary bladder. Infect Immun 56, 2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walthers D., Carroll R. K., Navarre W. W., Libby S. J., Fang F. C., Kenney L. J. (2007). The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol 65, 477–493. 10.1111/j.1365-2958.2007.05800.x [DOI] [PubMed] [Google Scholar]
- Wolfe A. J. (2005). The acetate switch. Microbiol Mol Biol Rev 69, 12–50. 10.1128/MMBR.69.1.12-50.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]


