Abstract
SVCT2 is the major transporter mediating vitamin C uptake in most organs. Its expression is driven by two promoters (CpG-poor exon 1a promoter and CpG-rich exon 1b promoter). In this work we mapped discrete elements within the proximal CpG-poor promoter responsible for the exon 1a transcription. We identified two E boxes for USF binding and one Y box for NF-Y binding. We further show that the formation of an NFY/USF complex on the exon 1a promoter amplifies each other's ability to bind to the promoter in a cooperativity-dependent manner and is absolutely required for the full activity of the exon 1a promoter. The analysis of the CpG site located at the upstream USF binding site in the promoter showed a strong correlation between expression and demethylation. It was also shown that the exon 1a transcription was induced in cell culture treated with demethylating agent decitabine. The specific methylation of this CpG site impaired both the binding of USF and the formation of the functional NF-Y/USF complex as well as promoter activity, suggesting its importance for the cell-specific transcription. Thus CpG methylation at the upstream USF binding site functions in establishing and maintaining cell-specific transcription from the CpG-poor SVCT2 exon 1a promoter.
Keywords: gene expression, NF-Y, USF, methylation, CpG
Introduction
Since most mammalian cells and all human cells are unable to synthesize vitamin C, or ascorbic acid, they are dependent upon uptake of the vitamin from their surroundings. This uptake is mediated primarily by one of two sodium-and energy-dependent vitamin C transporters, termed SVCT1(slc23a1) and SVCT2 (slc23a2) [1]. They belong to a family of nucleobase transporters and lack structural homology with any other mammalian membrane transporter. Although the SVCT1 and SVCT2 have a high sequence homology, they have distinct cellular distributions. The SVCT1 is located primarily in intestinal epithelium and renal proximal tubule cells, where it mediates ascorbate absorption and reabsorption, respectively. The SVCT2, on the other hand, has a more generalized tissue distribution in most major organs, with highest expression noted in brain and neuroendocrine tissues, such as pituitary and adrenal gland.
The presence of the SVCT2 is crucial for life, since its targeted deletion in the mouse fails to produce viable pups [2]. Although SVCT2-deficient embryos typically survive until birth, they die shortly thereafter, failing to take a first breath and inflate the lungs. The cause of death seems to relate to damage in the brain due to capillary hemorrhage. This is most evident in the cortex, but also occurs in areas of the lower brain crucial for control of body functions, including respiration.
Despite its importance for maintaining intracellular ascorbate, little is known about transcriptional regulation of the SVCT2. Both SVCT isoforms are predicted to cross the plasma membrane 12 times and contain potential intracellular protein kinase A and C phosphorylation sites [1]. In nucleated cells a variety of agents enhance SVCT2 expression at the level of mRNA, protein, and function. In some instances this accompanies cell differentiation, such as with zinc [3], calcium/phosphate ions [4] and phorbol ester stimulated human monocyte differentiation [5]. In others it is not related to cell differentiation, such as when induced by glucocorticoids [6], epidermal growth factor [7], or hydrogen peroxide [8]. Whereas these results show transcriptional regulation of the SVCT2, they do not define the molecular mechanism by which this occurs.
Gene regulation of the human SVCT2 is unusual in that it is driven by the interaction of two promoters [9]. Both SVCT2 promoters (P1 and P2) are located immediately upstream of the first two exons (named exon1a and exon1b). P1 and P2 originate two 5' untranslated region (5' UTR) variants, since the putative translation start site is located in exon 3 [9]. By computer analysis of the promoter sequence, only a small number of putative cis-acting elements are located in the proximal region of P1 including XBP/USF, NFY, HIF, FAST-1, SMAD3 and a gut-enriched Krueppel-like factor, while the proximal region of P2 appears to have multiple putative cis-acting elements for zinc finger transcription factor sites (4 sites, ZBT-89), Sp1 sites (3 sites), EGR-1 sites (2 sites), an AP2 site, a metal transcription factor site (MRE) and a Myc-associated zinc finger protein (MAZ) [9].
In this report, we map the discrete elements responsible for the activity of the P1/exon 1a promoter. The proximal binding sites for the transcription factors Upstream Stimulating Factor (USF) and Nuclear Factor-Y (NF-Y) mediate transcriptional regulation of the SVCT2 exon 1a. We show that NF-Y protein that binds to the Y box complexes and cooperates with USF1/2 bound to the upstream E box, activating the transcription of the exon 1a. These results prompted us to investigate the involvement of an epigenetic mechanism in the regulation of the cell-specific transcription of the exon 1a. Our results suggest that CpG methylation at the upstream USF binding site in the exon 1a promoter is important for its cell-specific expression and is the primary mechanism for the silencing of the exon 1a gene.
Materials and methods
Reagents
The antibodies against USF1 (C-20), USF2 (C-20), TFII-I (H-58), and NF-YA (C-18) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Biotin end-labeled or unlabeled oligonucleotides and other chemicals were from Sigma Chemical Co.
Cell Culture
Human cell lines HeLa (cervical cancer), U2OS (osteosarcoma), HepG2 (liver carcinoma), SHS-Y5Y (neuroblastoma), HT1080 (fibrosarcoma), HEK293 (embryonic kidney), PMC42 (breast carcinoma), HCA-7 (colonic adenocarcinoma), HCT116 (colorectal carcinoma) and Caco-2 (epithelial colorectal adenocarcinoma) were maintained in DMEM with 10% FBS. Human umbilical vein endothelial cells (HUVEC) were cultured in Endothelial Cell Medium (ScienCell, CA). EA.hy926 cells derived from fusion of HUVEC with A549 (lung adenocarcinoma epithelial cell line) were cultured in DMEM that contained 10% FBS and HAT media supplement. THP-1 human myeloid leukemia cells were maintained in RPMI medium with 10% FBS, 0.05 mM 2-mercaptoethanol.
Plasmid constructs
The reporter constructs for the exon 1a promoter and the various exon 1a mutants as indicated were prepared by PCR with −1983/+266-luc [9] as the template. Digested PCR products were inserted into pGL3-basic vector (pCpGL-basic vector for data in Fig. 9B and C) and verified by sequencing. The expression vectors for NF-Y subunits, dominant-negative mutant NF-YA13m29, USF1/2, dominant-negative mutant A-USF, and c-Myc were all previously described [10–13]. The pCpGL-basic luciferase reporter vector which completely lacks CpG dinucleotides was kindly provided by Dr. Rehli [14].
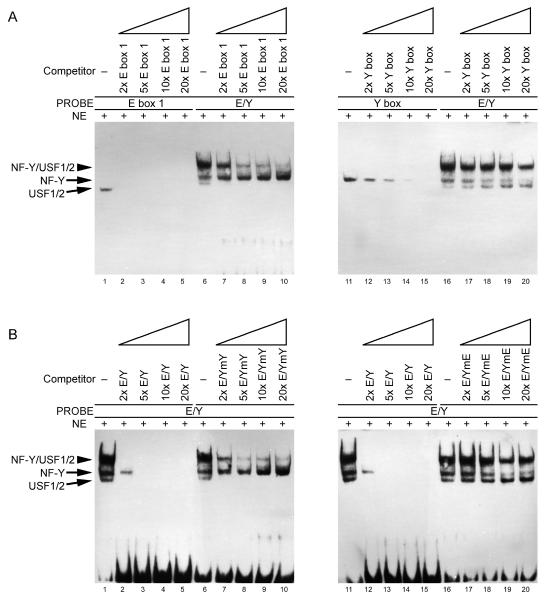
Figure 9.
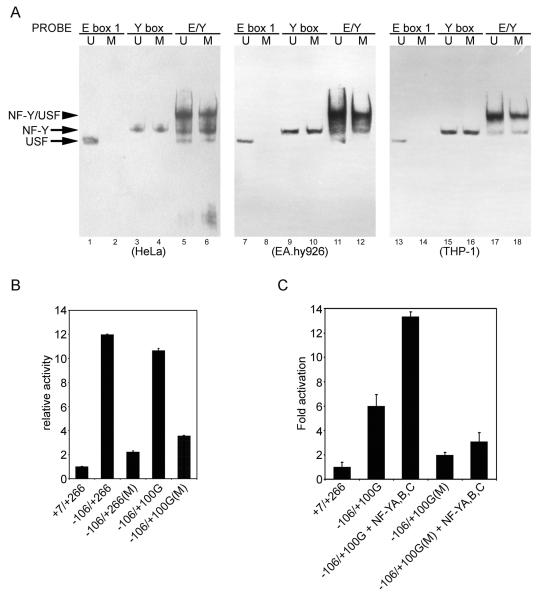
(A) CpG methylation at the upstream USF binding site impedes USF binding and inhibits the formation of NF-Y/USF complex. HeLa, EA.hy926 or THP-1 cell nuclear extract was incubated with methylated or mock-methylated E box 1, Y box, and a probe containing both the E and Y boxes (E/Y) for 20 min. Binding of added DNA and endogenous proteins in nuclear extracts was then assessed by EMSAs. (B) The methylated or mock-methylated promoter region was cloned into a pCpGL-basic vector and 500 ng of each was transiently transfected into HeLa cells. (C) Cells were transfected with 100 ng of methylated or mock-methylated exon 1a promoter reporter plasmid along with 400 ng of NF-YA/B/C plasmids (equimolar mixtures). Luciferase activity was measured 24 h after transfection. pRL-CMV was added as an internal control and relative luciferase activity is shown as the means of duplicate measurements from 1 of 3 similar experiments and the error bars show range of values, based on the activity of +7/+266. U, unmethylation; M, methylation.
Transient transfection and luciferase assays
Cells were seeded in 24-well plates and grown to ~70% confluence. On the following day, the cells were co-transfected with 0.1 to 0.5 μg of reporter plasmid, 5 ng of Renilla plasmid pRL-CMV, 0.2 to 0.4 μg of plasmids expressing the genes of interest or empty vector plasmid to compensate for the amount of DNA. Fugene HD reagent (Roche, IN) was used for the delivery of plasmids into cells. At 24 h after transfection, cell lysates for measurement of firefly and Renilla luciferase activities were prepared using Passive Lysis Buffer (Promega) according to the manufacturer's manual.
Electrophoresis mobility shift assays (EMSA)
For in vitro binding reactions, 2 μl of NE was incubated with the biotin end-labeled probes at room temperature for 20 minutes in 10 mM Tris pH 7.5, 50 mM KCl, 50 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 5% glycerol, 0.1% NP-40, 5 mM MgCl2, 1 mg/ml BSA, and 50 ng/μl poly dI-dC. For competition or super-shift experiments, the NEs were treated with excessive amounts of unlabeled probes or 1 to 4 μg antibody for 30 minutes at room temperature prior to the addition of the biotin end-labeled probes. The reaction products were then loaded onto 4.5% polyacrylamide gel electrophoresis (PAGE) and run at 100 V in 0.5× TBE buffer for 1.5 hours following the detection according to the manufacturer's. The nucleotide sequences for each of the probes are depicted in Table 1.
Table 1.
Probes used in the current study
| Name | Sequence |
|---|---|
| E box 1 | 5-TCCACTTTCACCCACGTGAGCAGGCATCAT-3 |
| mE box 1 | 5-TCCACTTTCACCAGATCTAGCAGGCATCAT-3 |
| E box 2 | 5-GGCCTTTAGAATCACGTGAGCTAATTCCTG-3 |
| mE box 2 | 5-GGCCTTTAGAATAGATCTAGCTAATTCCTG-3 |
| XDH/XO | 5-CCGGGAGGCGTATCTTTCAAGTTGCAGGGCAGT-3 |
| Y box | 5-AGCAGGCATCATCCAATCCACTGTGGGTC-3 |
| mY box | 5-AGCAGGCATCATCCAGGCCACTGTGGGTC-3 |
| E/Y | 5-AAAAGTTCCACTTTCACCCACGTGAGCAG GCATCATCCAATCCACTGTGGGTCCAGACA-3 |
| E/YmE | 5-AAAAGTTCCACTTTCACCCATATGAGCAG GCATCATCCAATCCACTGTGGGTCCAGACA-3 |
| E/YmY | 5-AAAAGTTCCACTTTCACCCACGTGAGCAG GCATCATCCGTTCCACTGTGGGTCCAGACA-3 |
| INS1 | 5-ACTTTCACCCACGTGAGCAGGCATCATC CTGAGCAGGCATCATCCAATCCACTGTGG-3 |
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) assays were performed via a commercially chromatin immunoprecipitation kit (Cell Signaling, MA), using antibodies against either USF1, USF2, NF-YA or TFII-I. Briefly, cell contents were first cross-linked by adding formaldehyde. Cross-linked lysates were then digested by Micrococcal nuclease. After digestion, the samples were centrifuged and the supernatants were diluted 5-fold in ChIP buffer. Cross-linked chromatin was incubated overnight with USF1, USF2, NF-YA antibody or normal rabbit IgG at 4°C. Antibody-protein-DNA complexes were isolated by immunoprecipitation with 30 μl of protein G magnetic beads. After extensive washing, pellets were eluted and formaldehyde cross-linking was reversed by 2-h incubation at 65°C after addition of Proteinase K and NaCl. Samples were purified and used as a template for PCR. ChIP primers 5-GTTCCACTTTCACCCACGTGAGC-3 and 5-GAGAAGATGAATGGCCCTGCTCCA-3 were used to amplify a 119-base pair fragment corresponding to the core exon 1a promoter.
Bisulfite genomic sequencing to analyze methylation patterns
Genomic DNA was isolated from HeLa, HepG2, U2OS, THP-1, EA.hy926, CaCo-2, SHS-Y5Y, HCA-7 and HCT116 via a commercially Genomic DNA purification kit (Promega, WI). Bisulfite modification of genomic DNA was carried out using the CpGenome Fast DNA Modification Kit (Millipore, MA). The PCR primers for amplification of the exon 1a promoter containing the upstream E box were: forward primer 5-gggagctcTGGGTTAAGAGATGTTTAGATAGTTGGTAAAAT-3, and reverse primer 5-cggctagcCCCTACCTTTTTATTCTATCTAAACCCACAATAA-3. The digested PCR fragments were sub-cloned into the pGL3-basic vector for sequencing.
In vitro methylation of oligonucleotides and reporter plasmids
SssI methylase (NEB, MA) was used for the methylation of EMSA oligonucleotides and of the SVCT2 exon 1a promoter-luciferase constructs. Oligonucleotides and pCpGL-basic constructs were incubated with methylase according to the manufacturers' recommendations. In each case, half of the DNA was treated in the same manner in the absence of methylase as mock methylation.
RNA isolation and RT-PCR
Total RNA was isolated using TRIZOL reagent (GIBCO, Grand Island, NY), and 2 μg was reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad, Carlsbad, CA). Amplification was performed using Advantage 2 PCR kit (Clontech, Mountain View, CA). The PCR products were separated on 1% agarose gels. The parameters and primers for SVCT2 were described previously [5].
Results
The exon 1a promoter activity is dependent on the −92/−87-bp E box
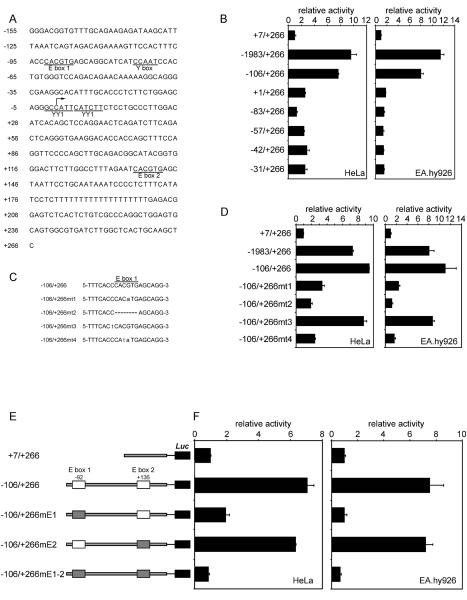
Most of the activity of the human exon 1a promoter in both HeLa and EA.hy926 cells is present in the 106 base pair region upstream from the transcription start site (Fig. 1A). This region of the promoter contains two highly conserved E box elements that are near-consensus matches for the optimal USF binding site. To determine whether one or both of these are important for the exon 1a promoter activity, reporter constructs with deleted or mutated E boxes were constructed. Deletion of the upstream E box beyond −83 bp completely eliminated the activity of the exon 1a promoter in both HeLa and EA.hy926 cells (Fig. 1B). That this decrease in activity was due to deletion of the E box was evident with mutations of the upstream E box (Fig. 1C and D), which also showed nearly complete loss of promoter activity in both cell types when elements of the E box were mutated (mt1, mt2 and mt4) [15–17], but not when a preceding cytosine was changed to a thymidine (mt3) [18]. Changing the E box sequence to that of the TFE3 binding site (CACATG), an alternative binding site for Myc/Max [19–21] but not for USF [16], dramatically impaired the exon 1a promoter activity (mt1). Furthermore, the T/A flanking residue modification preferred by USF [18] did not affect the exon 1a promoter activity, strongly suggesting the upstream E box is a USF-type (mt3). In contrast to the upstream E box, mutation of the downstream USF-type E box (−106/+266mE2) [22]did not significantly affect the basal transcriptional activity of the exon 1a promoter (Fig. 1E and F). These results illustrate the crucial importance of the upstream USF-type E box for the exon 1a promoter activity. As the control, the +7/+266 fragment showed no significant promoter activity compared with the pGL3-basic vector (data not shown).
Figure 1.
Reporter analysis of the human SVCT2 exon 1a gene. (A) Human SVCT2 exon 1a promoter sequence of −155 bp to +266 bp relative to the transcription start site. The potential transcription factor binding sites are underlined. (B) Transient transfection of 500 ng of serial deletion luciferase reporter constructs into HeLa and EA.hy926 cells. 5 ng of pRL-CMV was added as an internal control, and luciferase activities were measured 24 hours after transfection and normalized by Renilla reporter activities. (D) Effect of an E box 1 mutation on the exon 1a transcriptional activity. The cells were transfected with 500 ng of the mutants described in (C). (E) Schematic representation of the reporter constructs, with wild type and mutated E boxes depicted as white and shaded rectangles, respectively. In the mutated E boxes, the central CACGTG sequence was changed to AGATCT. Reporter gene activity was determined and is shown in (F). Relative luciferase activity is shown as the data means and the error bars on each column show range of values of duplicate measurements from 1 of 3 similar experiments, based on the activity of +7/+266.
USF specifically recognizes both upstream and downstream E boxes within the exon 1a promoter and differentially regulates the exon 1a promoter activity
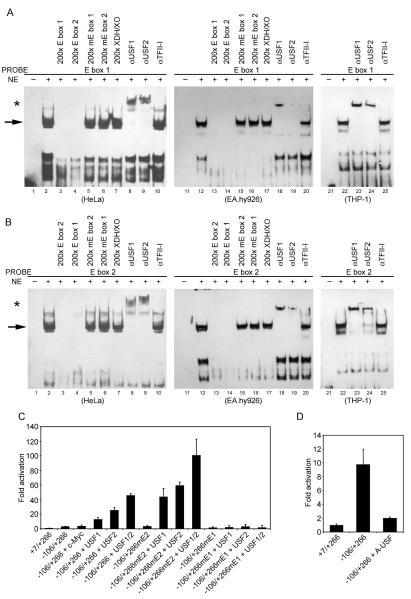
To show that the putative E boxes in the exon 1a promoter actually bind USF, EMSA experiments were performed. The results shown in Fig. 2A demonstrate that a shifted complex was observed when the DNA fragment containing upstream E box was incubated with HeLa, EA.hy926 or THP-1 nuclear extracts (lanes 2, 12 and 22). The band appears to be a doublet and this finding is consistent with published observations of alternative splicing of USF2 [23]. Unlabeled E box 1 (upstream) or E box 2 (downstream) oligonucleotide abolished the binding (lanes 3, 4, 13 and 14), whereas mutated E box 1 (mE box 1), mutated E box 2 (mE box 2) or a 33-bp nonspecific competitor oligonucleotide containing the sequence of the xanthine dehydrogenase/xanthine oxidase promoter (XDH/XO) did not affect binding (lanes 5, 6, 7, 15, 16 and 17). Antibodies against USF1 and USF2 were both capable of super-shifting the putative binding complex (lanes 8, 9, 18, 19, 23 and 24). As a control, TFII-I antibody did not cause any super-shift band or affect the binding intensity (lanes 10, 20 and 25). Similar results were also observed in HeLa, EA.hy926 and THP-1 nuclear extracts with the DNA fragment containing downstream E box (Fig. 2B). These results suggest that USF1 and USF2 were the major transcription factors binding to the upstream and downstream E boxes of the exon 1a promoter.
Figure 2.
USF1/2 binds to the exon 1a E boxes. (A) and (B) Nuclear extracts (NE) from HeLa, EA.hy926 or THP-1 cells were incubated with a labeled probe containing the exon 1a E box and surrounding nucleotides and the reaction mixture was then electrophoresed on a 4.5% non-denaturing gel to detect the specific retarded migrating band. The indicated excess unlabeled probes or antibodies were added in reactions shown for competition or super-shift analysis before the addition of a labeled probe. USF1 and USF2 super-shifts in (A, B) are indicated by an *. (C) and (D) HeLa cells were cotransfected with 100 ng of the exon 1a reporter plasmid and 400 ng of expression vectors encoding either c-Myc, USF1, USF2, USF1/2 (equimolar mixtures) or dominant-negative mutant A-USF, as indicated. The data presented are means of duplicate measurements from 1 of 3 similar experiments, and the error bars show range of values of duplicate measurements, based on the activity of +7/+266.
Involvement of specific transcription factors in the E box-dependent expression of the exon 1a promoter was also examined using co-transfection. First, responsiveness of the promoter to exogenous USF1, USF2, or c-Myc was assessed (Fig. 2C). This analysis revealed selectivity in the response of the promoter to different E box-binding factors. Reporter gene activity was increased by about 3-fold in USF1-overexpressing cells, by about 9-fold in USF2-overexpressing cells and by about 16-fold in USF1/2-overexpressing cells. In striking contrast, c-Myc had essentially no effect on the exon 1a promoter activity, consistent with the above results. Additionally, the mutation of the upstream E box abrogated USF1/2-mediated transcriptional activation, suggesting this E box and its bound USF play a critical role in the USF-induced response. Intriguingly, the construct with mutation of the downstream E box, which exhibited full promoter activity under basal conditions (Fig. 1F), had 2–3-fold more robust promoter activity in response to increased cellular USF than when both E boxes were present (Fig. 2C). Finding that USF-dependent promoter activity increases when the downstream E box is absent suggests that it suppresses the exon 1a gene transcription.
To further confirm the involvement of USF family members in the exon 1a promoter regulation, a specific dominant-negative mutant, A-USF [12], was employed to interfere with binding of the endogenous USF to the exon 1a reporter gene. A nearly fivefold decrease in the exon 1a promoter activity was observed in the presence of A-USF, indicating an essential role of endogenous USF in activating transcription from the exon 1a E box (Fig. 2D). Taken together, these results indicated a prominent role of USF1/2 in the regulation of the exon 1a promoter activity.
The −74/−70-bp Y box within the exon 1a promoter confers activity and binds the NF-Y protein
Sequence analysis of the proximal promoter (−106/+266) also revealed a canonical CCAAT box located in the −74/−70 region, 12 base pairs downstream to the upstream USF binding site (−92/−87) (Fig. 1A). To investigate the potential role of this cis-element in the regulation of the exon 1a promoter, the CCAAT sequence in the −106/+266-luc reporter construct was destroyed to generate the “−106/+266YD” construct (Fig. 3A). This construct eliminated most of basal promoter activity (Fig. 3A), suggesting an important role for the CCAAT box in maintaining the exon 1a promoter activity. Although a number of transcription factors are reported to recognize the CCAAT box [24–27], subsequent analyses reveal that there is divergence in the recognition sequence of CCAAT and that only NF-Y exclusively requires the bona fide CCAAT sequence [26;28;29]. To test whether NF-Y was capable of binding to this CCAAT motif, we performed EMSA using HeLa, EA.hy926 and THP-1 cell nuclear extracts with a probe bearing the CCAAT motif. As shown in Fig. 3B, DNA-protein complexes were formed (lanes 2, 8, 11 and 18), and were abolished by the addition of excessive unlabeled probe (lanes 3 and 12). An excess of unlabeled probe bearing a mutation that impairs the binding of NF-Y failed to compete (lanes 4 and 13). An antibody specific to NF-YA super-shifted a substantial amount of the formed DNA-protein complex (lane 5) and this was increased with the use of additional amounts of antibody (lanes 9, 14 and 19). On the other hand, normal rabbit serum and anti-YY1 antibody failed to super-shift the complex (lanes 6, 7, 15, 16, 20 and 21). These data demonstrated that the NF-Y trimer was present in the specific DNA-protein complex.
Figure 3.
NF-Y binding to the Y box within the exon 1a promoter is essential for the exon 1a promoter activity. (A) Reporter gene activity was evaluated by transient transfection assays in HeLa and EA.hy926 cells. In the mutated Y box, the central CCAAT sequence was changed to CCT. (B) HeLa, EA.hy926 or THP-1 nuclear extract (NE) was incubated with a labeled probe containing the Y box, and the indicated unlabeled probes or antibodies were added for competition or super-shift analysis. The super-shifted complex is indicated by an *. (C) and (D) HeLa cells were transfected with 100 ng of the exon 1a promoter reporter plasmid along with 400 ng of NF-YA/B/C plasmids (equimolar mixtures) or 300 ng of the exon 1a promoter construct with 200 ng of dominant-negative NF-YA13m29. Luciferase activities were measured 24 hours after transfection and normalized by Renilla reporter activities. The data are shown as means and range error bars of duplicate measurements, based on the activity of +7/+266. The experiment was repeated three times with similar results.
To investigate the function of NF-Y in regulating the exon 1a promoter activity, NFY constructs were co-transfected with the −106/+266 reporter construct into HeLa cells. As shown in Figure 3C and D, a doubling of transcription was observed. In contrast, the mutation of the Y box (−106/+266YD) abrogated any response to NF-Y. On the other hand, a dominant-negative mutant form of NF-YA, termed NF-YA13m29 [10], halved promoter activity stimulated by the −106/+266 construct. These observations demonstrated that the exon 1a promoter activity is activated by cellular NF-Y.
NF-Y and USF form an activating complex on the exon 1a promoter
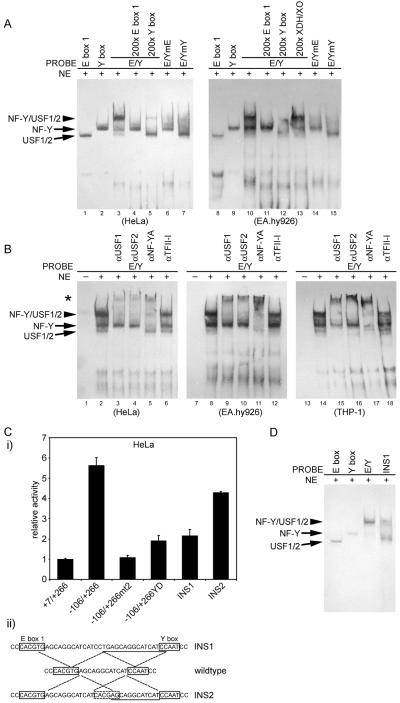
We next used EMSA to address the question of whether binding of NF-Y protein to the Y box complexes with USF bound to the adjacent E box. Using a 59-bp exon 1a probe containing both boxes (E/Y), we detected a band that migrated slower than when the probes of either box alone were used (Figure 4A, lanes 1–3 and 8–10). Formation of this band was blocked by inclusion of excessive amounts of either unlabeled E box 1, Y box, or XDH/XO probe (lanes 4, 5, 11, 12 and 13). The formation of the slowly migrating band was dependent on the integrity of both the Y box and the adjacent E box. An E/Y probe containing either a Y box mutation (E/YmY) or an E box mutation (E/YmE) failed to produce this specific band (lanes 6, 7, 14 and 15). Likewise, inclusion of antibodies against USF1, USF2, or NF-YA into the promoter/NE reaction caused a super-shift band, whereas an antibody to TFII-I did not (Figure 4B). Therefore, the DNA-binding protein complex responsible for this retarded mobility band contains not only NF-Y but also USF1/2, and these proteins bind specifically to E box 1 and Y box, respectively.
Figure 4.
Correlation of NF-Y/USF in binding to the exon 1a promoter with the exon 1a promoter activity. (A) Note a slowly migrating band (arrow head) detected behind USF1/2 and NF-Y bands when the intact E/Y probe (59 bp) containing both E box 1 and Y box was used as the probe with HeLa or EA.hy926 cell nuclear extracts (lanes 3 and 10). In lanes 1, 2, 8 and 9, E box 1 or Y box probe was used in the absence of competing probe. In lanes 4, 5, 11, 12 and 13, a 200× molar excess of unlabeled wild-type probe of E box 1, Y box or XDH/XO was pre-incubated with HeLa or EA.hy926 cell NEs. In lanes 6, 7, 14 and 15, an E/Y probe containing either a mutated E box 1 (E/YmE) or a mutated Y box (E/YmY) was used as the probe, in the absence of competing probe. (B) EMSA, in which HeLa, EA.hy926 or THP-1 NEs were pretreated with specific antibodies prior to being incubated with the exon 1a promoter probe. (C) Proper spacing between E box 1 and Y box is required for the full exon 1a promoter activity. HeLa cells were transfected with 500 ng of indicated reporter plasmids, and luciferase activities were measured 24 hours later (i). (ii) INS1 and INS2 were constructed by inserting a 16-bp sequence (underlined) within the interval between E box 1 and Y box, without altering the flanking sequences. Data represent means of duplicate measurements from 1 of 3 similar experiments and the range error bars are shown, based on the activity of +7/+266. (D) The insertion mutation between E box 1 and the Y box disrupts the cooperative in vitro binding of USF1/2 and NF-Y to the exon 1a promoter, as detected by EMSA.
To further examine the relationship between NF-Y/USF complex formation and the exon 1a activation, we performed both EMSA and transient transfections by using a promoter construct in which an additional 16-bp sequence was inserted between the E box and Y box in the exon 1a promoter (Fig. 4C. ii). Increasing the distance between the NF-Y- and USF binding sites without disturbing their surrounding sequences resulted in 62% reduction in reporter activity (Fig. 4C. i). In contrast, the introduction of a non-canonical USF binding site (CACGAG) at the normal distance from the Y box restored most of the exon 1a promoter activity (Fig. 4C. i). Addition of this same 16-bp sequence to the exon 1a promoter oligonucleotide probe also greatly inhibited the formation of the retarded mobility band representing most of promoter-bound NF-Y and USF signals (Fig. 4D). Taken together, these results suggest that full functioning of the exon 1a promoter requires the formation of the protein complex including both NF-Y and USF1/2 on the promoter.
The findings that the formation of NF-Y/USF complex is prerequisite for the exon 1a promoter activity suggest that their association could be important in some aspect of their binding to the exon 1a promoter. To test if the binding of USF1/2 to the E box were stabilized by the binding of NF-Y to the Y box, or vice versa, we incubated serial dilutions of unlabeled E box 1 or Y box oligonucleotide together with the labeled exon 1a probe containing both boxes (E/Y), and measured the strength of USF/NF-Y binding in EMSA. As shown in Figure 5A, USF1/2 or NF-Y in NF-Y/USF complex clearly had higher binding affinity for their respective target sequence because USF1/2 or NF-Y binding in the absence of adjacent NF-Y or USF1/2 binding was fully competed at a lower concentration than was required for USF1/2 or NF-Y binding in NF-Y/USF complex. Similarly, the unlabeled E/Y oligonucleotide competed more efficiently for the formation of the NF-Y/USF complex on the promoter than the oligonucleotides mutated at either the Y box (E/YmY) or the E box (E/YmE) (Figure 5B). These data clearly demonstrate NF-Y and USF1/2 amplify each other's ability to bind to the exon 1a promoter in a cooperativity-dependent manner.
Figure 5.
Cooperation of NF-Y and USF1/2 in binding to the exon 1a promoter in vitro. (A) EMSAs were performed using a constant amount of HeLa nuclear extract, a constant amount of biotin end-labeled E box 1, Y box or E/Y probe, and increasing quantities of unlabeled E box 1 (left panel) or Y box (right panel) oligonucleotide. (B) HeLa nuclear extract was incubated with labeled E/Y probe, and the indicated increasing quantities of unlabeled probes (E/Y, E/YmY or E/YmE) were added for competition. For each reaction, 10 fmol of fixed labeled probe was used and concentrations of the variable unlabeled oligonucleotide included 20 fmol (2×), 50 fmol (5×), 100 fmol (10×), and 200 fmol (20×), increasing from left to right.
Binding of USF and NF-Y to the proximal exon 1a promoter in cells
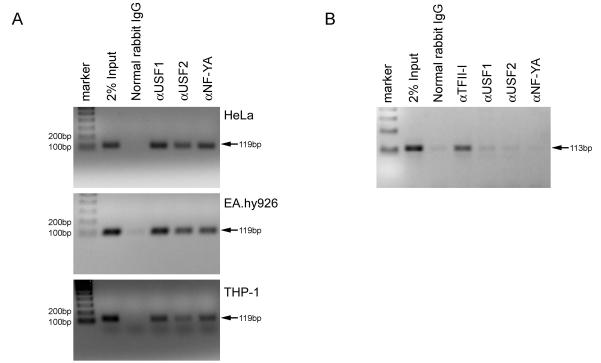
Chromatin immunoprecipitation (ChIP) of cell extracts was used to directly assess the presence or absence of USF and NF-Y bound to the proximal promoter of the endogenous exon 1a gene. As shown in Fig. 6A, a 119-bp fragment spanning the exon 1a proximal promoter was detected by PCR in 2% Input, but no specific band was detected when ChIP was performed with normal rabbit IgG. When DNA from HeLa, EA.hy926 and THP-1 cells was immunoprecipitated with antibodies to USF1/2 and NF-YA, the 119-bp region was detected. As a control for non-specific protein-DNA interactions, we also amplified a genomic fragment containing a TFII-I site where no sites for USF and NF-Y are detectable by sequence analysis. The resulting 113 bp fragment from HeLa cells was immunoprecipitated by an antibody to TFII-I, but not significantly by antibodies to USF or NF-Y (Fig. 6B). These results show that all of these transcription factors are bound to their cognate sites on the endogenous SVCT2 exon 1a promoter in intact cells.
Figure 6.
ChIP analysis of transcription factor bindings to the exon 1a promoter. (A) ChIP assay was performed using HeLa, EA.hy926 and THP-1 cells. Antibodies for USF1/2 and NF-Y or normal rabbit IgG were used. Immunoprecipitated DNA fragments and 2% of total sample DNA were amplified by PCR using primers specific for the human exon 1a promoter (−106 to +13). PCR products were separated on a 2% agarose gel and stained by ethidium bromide. (B) Amplification of an unrelated region does not show a detectable signal from HeLa cell-derived genomic DNA.
CpG methylation at the upstream USF binding site contributes to cell-specific expression of the exon 1a
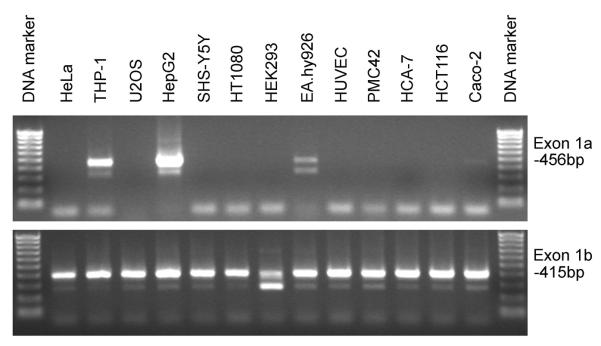
To assess whether the SVCT2 exon 1a is widely expressed, we measured its mRNA expression in 13 cell lines, with results shown in Fig. 7. In contrast to the ubiquitously expressed SVCT2 exon 1b (bottom panel), the exon 1a mRNA transcript was specifically expressed in only four of thirteen cell lines (top panel; THP-1 monocytes, HepG2 liver cells, EA.hy926 endothelial cells, and faintly in Caco-2 colonic intestinal cells). This is the first evidence that the expression of the SVCT2 exon 1a exhibits cell-specificity. Since USF and NF-Y are ubiquitously expressed transcription factors, they are not likely to account for the cell-specific expression of the exon 1a.
Figure 7.
Expression pattern of SVCT2 mRNA in 13 cell lines. Gel electrophoresis of RT-PCR products shows the exon 1a transcripts in HepG2, THP-1, EA.hy926 and Caco-2 cells, while exon 1b transcript is ubiquitously expressed in all cell lines.
Since some mammalian genes exhibit an inverse correlation between the extent of DNA methylation and gene activity [30–32], we examined CpG methylation and its correlation with cell-specific expression of the exon 1a. To accomplish this, two of the non-expressing cell lines were treated with the demethylating agent decitabine (5-aza-2'-deoxycytidine; DAC) for 96 h. Expression of the exon 1a was significantly induced in both HeLa and HT1080 cells (Fig. 8A), suggesting that methylation of CpG sites in the exon 1a promoter region silences the exon 1a gene expression. The upstream USF binding site in the exon 1a promoter has a CpG site, a potential target for DNA methylation in mammals. Thus, we prepared chromosomal DNA and analyzed the methylation status of the CpG site by bisulfite genomic sequencing. As shown in Fig. 8B and Fig. S1, bisulfite genomic sequencing revealed that the CpG at the upstream USF binding site was methylated in HeLa, U2OS, SHS-Y5Y, HCA-7 and HCT116 cells (no exon 1a expression), but not in HepG2, THP-1, EA.hy926 and CaCo-2 cells (exon 1a expression with various levels). This indicates that the methylation status of the CpG dinucleotide at the upstream USF binding site reflected the cell-specificity of the exon 1a expression.
Figure 8.
CpG methylation analysis at the upstream USF binding site. (A) Effects of the demethylating reagent DAC on the exon 1a transcription. Cells were treated with 5 μM DAC for 4 days, and the RNA was isolated and amplified using exon 1a-specific primers. (B) Correlation between the upstream E box methylation and the exon 1a transcript expression in various cells.
To further investigate whether CpG methylation at the upstream USF binding site affected USF binding, EMSA was carried out. CpG methylation at the upstream USF binding site abolished USF binding (Fig. 9A), whereas NF-Y binding to the Y box was not affected in the absence of the CpG site. Furthermore, CpG methylation at the upstream USF binding site apparently impaired the formation of functional NF-Y/USF complex, although USF was still able to form NF-Y/USF complex on the exon 1a promoter in the presence NF-Y binding (lanes 5, 6, 11, 12, 17 and 18). This observation also further confirmed that NF-Y can stabilize USF1/2 binding to the exon 1a promoter.
We also evaluated the effect of CpG methylation on the exon 1a promoter activity. Since traditional pGL3 reporter vector contains a large number of backbone CpG residues and significantly represses a CpG-free promoter when methylated, we employed a novel luciferase reporter vector, pCpGL, which completely lacks CpG dinucleotides and can be used to study the effect of promoter DNA methylation in transfection assays [14]. As shown in Fig. 9B, CpG methylation of the proximal promoter (−106/+266) significantly inhibited the exon 1a promoter activity, and CpG methylation at the USF binding site (a second CpG site in −106/+100G is mutated) also impaired the promoter activity. NF-Y over-expression failed to restore the methylated promoter activity (Fig. 9C).
Together, these results demonstrate that: (i) NF-Y can indeed stabilize the ability of USF1/2 to bind to the exon 1a promoter; (ii) CpG methylation at the upstream USF binding site is associated with cell-specific expression of the exon 1a by disrupting the cooperation of NF-Y with USF1/2.
Discussion
In this report, we mapped the discrete cis-acting elements responsible for the transcriptional activity of the SVCT2 exon 1a promoter. Specific protein binding sites were identified by the introduction of mutations within these elements. The transcription factors USF1/2 and NF-Y were shown to bind to active elements of the promoter. This represents the first characterization of the upstream elements important to SVCT2 gene expression related to this promoter, while site-specific DNA methylation appears to be associated with cell-specific expression of the gene.
The trimeric transcription factor NF-Y was identified as an activator of the expression of the SVCT2 exon 1a through its binding to the CCAAT Y box motif on the exon 1a promoter in cooperation with USF1/2 binding to the upstream E box. The NF-Y complex consists of 3 subunits encoded by 3 different genes. Current models suggest that NF-YB and NF-YC first form heterodimers, which then serve as a platform to recruit NF-YA proteins. Once recruited to this trimeric complex, the DNA-binding domain of NF-YA can specifically bind to the consensus sequence. Consistent with this, our in vitro EMSA studies showed the presence of the key NF-YA subunit on the exon 1a promoter. Furthermore, NF-YA-linked chromatin immunoprecipitation confirmed NF-Y binding to the endogenous exon 1a promoter, along with USF1/2. EMSA experiments demonstrated that NF-Y and USF1/2 can significantly increase each other's DNA binding ability to the promoter in a cooperativity-dependent manner when NF-Y and USF1/2 bind to the Y box and E box, respectively. Although this cooperation may occur due to either altered DNA structure following NF-Y/USF protein binding or a direct protein interaction between the NF-Y/USF proteins at the promoter, the detailed mechanism still remains to be determined. Our co-transfection studies also showed that the NF-Y trimer and USF heterodimer activated the exon 1a promoter constructs in HeLa cells. Most importantly, the exon 1a promoter activity depends on the presence of the USF and NF-Y binding sites in the exon 1a promoter as well as on the formation of a functional transcriptional regulatory complex including NF-Y and USF1/2. Correct spacing between the two binding sites is also a crucial aspect in the exon 1a promoter proficiency. Disruption of either the USF or NF-Y site severely impaired the activity of the exon 1a promoter. These results support and extend previous transcriptional assays of NF-Y, which suggested that the regulatory roles of NF-Y may depend on the transcriptional regulatory partners to which it binds in a given genomic and cellular context [15;33–35]. Note that in these reporter assays, three expression constructs, NF-YA, NF-YB, and NF-YC were simultaneously co-transfected along with a reporter construct. The fact that only cells that have taken up all three plasmids can produce a complete form of NF-Y that can activate the promoter suggests that the increase of the exon 1a promoter activity obtained in our co-transfection assay system may represent only a fraction of naturally occurring NF-Y regulated the exon 1a expression.
In contrast to the ubiquitously expressed SVCT2 exon 1b, our current study showed that the expression of the exon 1a mRNA transcript was cell-specific. The cell-specificity of gene expression is generally thought to be regulated by the interaction of basal and cell-specific transcription factors with cis-acting elements on promoters [36]. However, NF-Y/USF transcription factors are present in cells whether or not they express the exon 1a, and our results seem unlikely to account for their involvement in cell-specific expression. This prompted us to investigate whether DNA methylation was involved in the mechanisms underlying the silencing of the exon 1a in non-expressing cells. In the present study, we showed that (i) the exon 1a transcription was induced in cell culture treated with demethylating agent decitabine; (ii) the CpG at the upstream USF binding site is demethylated in exon 1a-expressing cells and the methylation status is inversely correlated with the exon 1a expression among cells; (iii) site-specific in vitro methylation at the USF binding site suffices to repress the exon 1a promoter activity in cells containing the transcription factors required for expression. These results clearly indicate that the SVCT2 exon 1a transcription is mediated by the ubiquitously expressed transcription factors USF and NF-Y in exon 1a-expressing cells, and that, in other non-expressing cells, CpG methylation at the USF binding site located in the exon 1a promoter shows a strong correlation between expression and demethylation.
In our current studies, the SVCT2 exon 1a promoter activity was assessed in both exon 1a-expressing cells (EA.hy926 and THP-1) and non-expressing cells (HeLa). The exon 1a promoter clearly exhibited transcriptional activity in the cell lines that do not express the exon 1a gene, indicating that these cells contain transcription factors capable of inducing the exon 1a promoter activity. In this regard, it should be mentioned that the functional complex formation in the exon 1a-expressing cells was also observed when the probes were incubated with nuclear extracts from cells not expressing the exon 1a. Previous studies indicated that tumor cell lines, whether or not they express MAGE-A1, contain transcription factors capable of interacting with the unmethylated MAGE-A1 promoter to induce significant transcriptional activity[37–39]. This implied that the total lack of MAGE-A1 transcription requires a local repression mechanism to prevent activation by these ubiquitous transcription factors. The studies indicated that in these cells there is a process that inhibits de novo methylation within the 5' region of MAGE-A1, since unmethylated MAGE-A1 transgenes undergo remethylation at all CpGs except those located within the 5' region. Conversely, the impaired MAGE-A1 promoter activity led to remethylation of the 5' region, suggesting that this local inhibition of methylation appears to depend on MAGE-A1 promoter activity[40]. The protection of the 5' region of MAGE-A1 against remethylation is probably related to the transcriptional activity of the promoter. The ability of transcription factors to inhibit methylation has also been described previously [41–43]. Transcription factors may inhibit methylation either directly by preventing access of the DNA methyltransfrase or indirectly by inducing histone modifications that exclude these methylation enzymes [44–46]. In keep with this, our recent observations confirmed that the transcriptional co-activator p300, but not its closely related family member CBP, is involved in the transcriptional regulation of the exon 1a. This protein is capable of interacting withNF-Y/USF functional complex and synergistically increased the exon 1a promoter activity via the interaction with NFY/USF (H Qiao and J May, submitted for publication). The p300 protein can function by relaxing the chromatin structure at the gene promoter through their intrinsic histone acetyltransferase (HAT) activity. Thus, the exon 1a 5' region could be protected against remethylation in transient transfection studies due to the presence of these transcription factors.
The methylation of genomic DNA is a major epigenetic modification of the mammalian genome and has been linked inevitably with transcriptional repression [47;48]. It is believed to ensure gene silencing in non-expressing cells [32]. So far, most investigations of the role of DNA methylation have focused on CpG islands. In these cases, CpG methylation is generally involved in transcriptional suppression, either by inhibiting association between DNA-binding factors and their cognate DNA recognition sequences, resulting in direct inhibition of transcriptional activation [32] or by recruiting proteins that recognize methyl-CpG such as methyl-CpG-binding proteins, which elicit repressive potential of methylated DNA by recruiting co-repressor molecules to silence gene transcription and to modify surrounding chromatin such as histone modification [49]. In the case of the SVCT2 exon 1a, the first mechanism most likely plays a critical role in silencing expression of the gene. To date, it is still not clear whether the mechanisms whereby DNA methylation operates on genes with CpG-rich promoters are similar to those preventing the transcription of genes with CpG-poor promoters. The current study provides an example in which CpG methylation at the USF binding sites is associated with its cell-specific transcription.
In fact, cell-specific gene expression may be accomplished by the activation of transcription by ubiquitous transcription factors such as c-Myb [50], c-Myc/Myn [51], E2F [52], CREB [53], AP2 [54] and NF-κB [55], which are incapable of binding to methylated forms of their recognition sequences. For some CpG island promoters, CpG methylation contributes to cell-specific gene expression by blocking the binding of transcription factors [56;57]. Such a mechanism also works to regulate the cell-specificity of the exon 1a expression. This is established by CpG methylation that prevents the ubiquitous transcriptional factor USF from binding to the exon 1a promoter and inhibits the formation of crucial NF-Y/USF complex in non-expressing cells. However, we could not exclude the possibility that other CpG methylation also contributes to transcriptional suppression.
Since the over-expression of MeCP2 is able to suppress CpG-poor promoter activity [58;59], it is of interest to investigate whether the regulation of higher-order chromatin structures by DNA methylation contribute to the exon 1a silencing in non-expressing tissues. Such information would also give further insight into the role of CpG methylation in the cell-specific expression of genes with a CpG-poor promoter.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (NS 057674) and by the Vanderbilt Diabetes Research and Training Center (DK 020593).
Abbreviations
- USF
Upstream Stimulating Factor
- NF-Y
Nuclear Factor-Y
- ChIP
Chromatin immunoprecipitation
- EMSA
electrophoretic mobility-shift assay
References
- 1.Tsukaguchi H, Tokui T, Mackenzie B, Berger UV, Chen XZ, Wang Y, Brubaker RF, Hediger MA. A family of mammalian Na+-dependent L-ascorbic acid transporters. Nature. 1999;399:70–75. doi: 10.1038/19986. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou S, Gispert S, Cheng J, Wang Y, Chen A, Hoogstraten-Miller S, Miller GF, Kwon O, Levine M, Guttentag SH, Nussbaum RL. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat.Med. 2002;8:514–517. doi: 10.1038/0502-514. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Itoh N, Taniguchi T, Nakanishi T, Tatsu Y, Yumoto N, Tanaka K. Zinc-induced sodium-dependent vitamin C transporter 2 expression: potent roles in osteoblast differentiation. Arch.Biochem.Biophys. 2003;420:114–120. doi: 10.1016/j.abb.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Wu X, Itoh N, Taniguchi T, Nakanishi T, Tanaka K. Requirement of calcium and phosphate ions in expression of sodium-dependent vitamin C transporter 2 and osteopontin in MC3T3-E1 osteoblastic cells. Biochim.Biophys.Acta. 2003;1641:65–70. doi: 10.1016/s0167-4889(03)00065-x. [DOI] [PubMed] [Google Scholar]
- 5.Qiao H, May JM. Macrophage differentiation increases expression of the ascorbate transporter (SVCT2) Free Radic.Biol.Med. 2009;46:1221–1232. doi: 10.1016/j.freeradbiomed.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita I, Hirano J, Itoh N, Nakanishi T, Tanaka K. Dexamethasone induces sodium-dependant vitamin C transporter in a mouse osteoblastic cell line MC3T3-E1. Br.J.Nutr. 2001;86:145–149. doi: 10.1079/bjn2001406. [DOI] [PubMed] [Google Scholar]
- 7.Biondi C, Pavan B, Dalpiaz A, Medici S, Lunghi L, Vesce F. Expression and characterization of vitamin C transporter in the human trophoblast cell line HTR-8/SVneo: effect of steroids, flavonoids and NSAIDs. Mol.Hum.Reprod. 2007;13:77–83. doi: 10.1093/molehr/gal092. [DOI] [PubMed] [Google Scholar]
- 8.Savini I, Rossi A, Catani MV, Ceci R, Avigliano L. Redox regulation of vitamin C transporter SVCT2 in C2C12 myotubes. Biochem.Biophys.Res.Commun. 2007;361:385–390. doi: 10.1016/j.bbrc.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Rubin SA, Dey S, Reidling JC. Functional analysis of two regulatory regions of the human Na+ -dependent vitamin C transporter 2, SLC23A2, in human vascular smooth muscle cells. Biochim.Biophys.Acta. 2005;1732:76–81. doi: 10.1016/j.bbaexp.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Mantovani R, Li XY, Pessara U, Hooft van HR, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J.Biol.Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 11.Okada Y, Matsuura E, Tozuka Z, Nagai R, Watanabe A, Matsumoto K, Yasui K, Jackman RW, Nakano T, Doi T. Upstream stimulatory factors stimulate transcription through E-box motifs in the PF4 gene in megakaryocytes. Blood. 2004;104:2027–2034. doi: 10.1182/blood-2003-09-3107. [DOI] [PubMed] [Google Scholar]
- 12.Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol.Cell Biol. 1999;19:1508–1517. doi: 10.1128/mcb.19.2.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara T, Matsumura-Arioka Y, Ohtani K, Nakamura M. Role of human T-cell leukemia virus type I Tax in expression of the human telomerase reverse transcriptase (hTERT) gene in human T-cells. Cancer Sci. 2008;99:1155–1163. doi: 10.1111/j.1349-7006.2008.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 15.Giannola DM, Shlomchik WD, Jegathesan M, Liebowitz D, Abrams CS, Kadesch T, Dancis A, Emerson SG. Hematopoietic expression of HOXB4 is regulated in normal and leukemic stem cells through transcriptional activation of the HOXB4 promoter by upstream stimulating factor (USF)-1 and USF-2. J.Exp.Med. 2000;192:1479–1490. doi: 10.1084/jem.192.10.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 17.Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol.Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendall AJ, Molloy PL. Base preferences for DNA binding by the bHLHZip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato GJ, Lee WM, Chen LL, Dang CV. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- 20.Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol.Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher F, Crouch DH, Jayaraman PS, Clark W, Gillespie DA, Goding CR. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12:5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szentirmay MN, Yang HX, Pawar SA, Vinson C, Sawadogo M. The IGF2 receptor is a USF2-specific target in nontumorigenic mammary epithelial cells but not in breast cancer cells. J.Biol.Chem. 2003;278:37231–37240. doi: 10.1074/jbc.M305791200. [DOI] [PubMed] [Google Scholar]
- 23.Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J.Biol.Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 24.Landschulz WH, Johnson PF, Adashi EY, Graves BJ, McKnight SL. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 25.Jones KA, Yamamoto KR, Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 26.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 27.Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 28.Kim CG, Sheffery M. Physical characterization of the purified CCAAT transcription factor, alpha-CP1. J.Biol.Chem. 1990;265:13362–13369. [PubMed] [Google Scholar]
- 29.Hooft van Huijsduijnen RA, Bollekens J, Dorn A, Benoist C, Mathis D. Properties of a CCAAT box-binding protein. Nucleic Acids Res. 1987;15:7265–7282. doi: 10.1093/nar/15.18.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr.Biol. 1997;7:R305–R307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 31.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 32.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr.Opin.Genet.Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Giannola DM, Zhang Y, Rivera AJ, Emerson SG. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- 34.Radomska HS, Satterthwaite AB, Taranenko N, Narravula S, Krause DS, Tenen DG. A nuclear factor Y (NFY) site positively regulates the human CD34 stem cell gene. Blood. 1999;94:3772–3780. [PubMed] [Google Scholar]
- 35.Jabrane-Ferrat N, Nekrep N, Tosi G, Esserman LJ, Peterlin BM. Major histocompatibility complex class II transcriptional platform: assembly of nuclear factor Y and regulatory factor X (RFX) on DNA requires RFX5 dimers. Mol.Cell Biol. 2002;22:5616–5625. doi: 10.1128/MCB.22.15.5616-5625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem.Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 37.De SC, Courtois SJ, Faraoni I, Lurquin C, Szikora JP, De BO, Boon T. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics. 1995;42:282–290. doi: 10.1007/BF00176446. [DOI] [PubMed] [Google Scholar]
- 38.De SC, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol.Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De SC, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5' region of gene MAGE-A1 in tumor cells. Mol.Cell Biol. 2004;24:4781–4790. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De SC, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5' region of gene MAGE-A1 in tumor cells. Mol.Cell Biol. 2004;24:4781–4790. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 42.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat.Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 43.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 44.Lin IG, Tomzynski TJ, Ou Q, Hsieh CL. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol.Cell Biol. 2000;20:2343–2349. doi: 10.1128/mcb.20.7.2343-2349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 47.Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 48.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat.Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 49.Ordway JM, Curran T. Methylation matters: modeling a manageable genome. Cell Growth Differ. 2002;13:149–162. [PubMed] [Google Scholar]
- 50.Klempnauer KH. Methylation-sensitive DNA binding by v-myb and c-myb proteins. Oncogene. 1993;8:111–115. [PubMed] [Google Scholar]
- 51.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 52.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc.Natl.Acad.Sci.U.S.A. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weih F, Nitsch D, Reik A, Schutz G, Becker PB. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 1991;10:2559–2567. doi: 10.1002/j.1460-2075.1991.tb07796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat.Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 56.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet.Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- 57.Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J.Cell Biochem. 2003;88:899–910. doi: 10.1002/jcb.10464. [DOI] [PubMed] [Google Scholar]
- 58.Cho JH, Kimura H, Minami T, Ohgane J, Hattori N, Tanaka S, Shiota K. DNA methylation regulates placental lactogen I gene expression. Endocrinology. 2001;142:3389–3396. doi: 10.1210/endo.142.8.8347. [DOI] [PubMed] [Google Scholar]
- 59.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic.Biol.Med. 2010;48:895–904. doi: 10.1016/j.freeradbiomed.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.