Abstract
Lampreys are representatives of an ancient vertebrate lineage that diverged from our own ~500 million years ago. By virtue of this deeply shared ancestry, the sea lamprey (P. marinus) genome is uniquely poised to provide insight into the ancestry of vertebrate genomes and the underlying principles of vertebrate biology. Here, we present the first lamprey whole-genome sequence and assembly. We note challenges faced owing to its high content of repetitive elements and GC bases, as well as the absence of broad-scale sequence information from closely related species. Analyses of the assembly indicate that two whole-genome duplications likely occurred before the divergence of ancestral lamprey and gnathostome lineages. Moreover, the results help define key evolutionary events within vertebrate lineages, including the origin of myelin-associated proteins and the development of appendages. The lamprey genome provides an important resource for reconstructing vertebrate origins and the evolutionary events that have shaped the genomes of extant organisms.
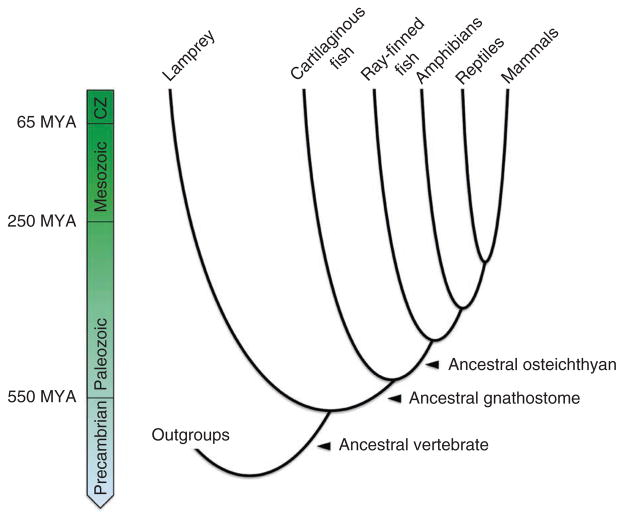
The fossil record shows that, during the Cambrian period, there was a great elaboration in the diversity of animal body plans. This included the emergence of a species with several characteristics shared with modern vertebrates, such as a cartilaginous skeleton that encases the central nervous system (cranium and vertebral column) and provides a support structure for the branchial arches and median fins. The cartilaginous cranium of this species housed a tripartite brain, with a forebrain for regulating neuroendocrine signaling via the pituitary gland, a midbrain (including an optic tectum) for processing sensory information from paired sensory organs and a segmented hindbrain for controlling unconscious functions, such as respiration and heart rate. These features in adults suggest that the corresponding embryos must have already possessed uniquely vertebrate cell types such as the skeletogenic neural crest and ectodermal placodes, both defining characters of modern-day vertebrates. Subsequent diversification of this lineage gave rise to the jawed vertebrates (gnathostomes), hagfish (for which genome-scale sequence data are currently limited), lamprey and several extinct lineages (Fig. 1 and Supplementary Note).
Figure 1.
An abridged phylogeny of the vertebrates. Shown is the timing of major radiation events within the vertebrate lineage. Extinct lineages and some extant lineages (for example, coelacanths, lungfish and hagfish) have been omitted for simplicity. Here, reptile is synonymous with sauropsid, ray-finned fish is synonymous with actinopterygian, and osteichthyan is synonymous with euteleostome. CZ, Cenozoic; MYA, million years ago.
Recent advances in developmental genetics methods for the lamprey and hagfish have advanced the reconstruction of several aspects of vertebrate evolution, although the interpretation of many of these findings is contingent on an understanding of genome structure, gene content and the history of gene and genome duplication events, areas that remain largely unresolved1. Given the critical phylogenetic position of the lamprey as an outgroup to the gnathostomes (Fig. 1), comparing the lamprey genome to gnathostome genomes holds the promise of providing insights into the structure and gene content of the ancestral vertebrate genome. Questions remain about the timing and subsequent elaboration of ancient genome duplication events and the elucidation of genetic innovations that may have contributed to the evolution and development of modern vertebrate features, including jaws, myelinated nerve sheaths, an adaptive immune system and paired appendages or limbs.
RESULTS
Sequencing, assembly and annotation
Approximately 19 million sequence reads were generated from genomic DNA derived from the liver of a single wild-captured adult female sea lamprey (P. marinus) (Supplementary Note). The lamprey genome project was initiated well before the discovery that the lamprey undergoes programmed genome rearrangements during early embryogenesis, which result in the deletion of ~20% of germline DNA from somatic tissues2,3, with the effects of rearrangement on the genic component of the genome not fully understood. We used raw sequence reads to examine large-scale sequence content and the repetitive structure of the lamprey genome. These analyses indicated that the lamprey genome is highly repetitive, rich in GC bases and highly heterozygous (Supplementary Figs. 1–3 and Supplementary Note). Although these features tend to encumber the assembly of long contiguous sequences, analyses of broad-scale structure enabled the optimization of the parameters used in assembly algorithms (Supplementary Note).
The current assembly was generated using Arachne4 and consisted of 0.816 Gb of sequence distributed across 25,073 contigs. Half of the assembly was in 1,219 contigs of 174 kb or longer, and the longest contig was 2.4 Mb. This assembly resolved multikilo-base- to megabase-scale structure over a majority of single-copy genomic regions (Supplementary Tables 1,2 and Supplementary Note), permitting the annotation of repetitive elements, genes and conserved intergenic features (Supplementary Note). Detection of extensive conserved synteny with gnathostome genomes indicates that the lamprey scaffolds accurately reflect the chromosomal organization of the lamprey genome. This assembly therefore provides unparalleled resolution of the gene content and structure of this evolutionarily informative genome.
Ab initio searches for repetitive DNA sequences showed that the lamprey genome contained abundant repetitive elements with high sequence identity. We identified 7,752 distinct families of repetitive elements, accounting for 34.7% of the assembly (Supplementary Fig. 4, Supplementary Tables 3,4 and Supplementary Note). Notably, this proportion is expected to be a significant underestimate, owing to the collapsing of repetitive elements during genome assembly. The large diversity of lamprey repetitive elements and the abundance of high-identity (presumably young) repeats represent a potentially rich resource for studies of the evolution and transposition of repetitive sequences.
The location of genes was determined by combining RNA sequencing (RNA-seq) mapping and exon linkage data with gene homologies and the prediction of coding sequences, splicing signals and repetitive elements using the MAKER pipeline5 (Supplementary Table 5 and Supplementary Note). The final set of annotated protein-coding genes contained a total of 26,046 genes. This number is similar to the numbers of predicted protein-coding genes in the other vertebrate genomes reported so far. Conserved noncoding elements (CNEs) were identified by homology to published sequences6,7. Searches identified a limited number of homologous CNEs in lamprey, 337 (5.0% of 6,670; ref. 6) and 287 (6.0% of 4,782; ref. 5), in close agreement with previous analyses8. For those lamprey CNEs that were linked to conserved homologous regions in the lamprey and gnathostome genomes, sequence identity typically extended over approximately half the length (53%) of the homologous gnathostome CNE (Supplementary Table 6 and Supplementary Note). Thus, either the lamprey lineage diverged from jawed vertebrates before most gnathostome CNE sequences became highly constrained or these CNEs have evolved much more rapidly in the lamprey genome than in jawed vertebrate genomes. Future work on additional lamprey and hagfish genomes should ultimately distinguish between these possibilities.
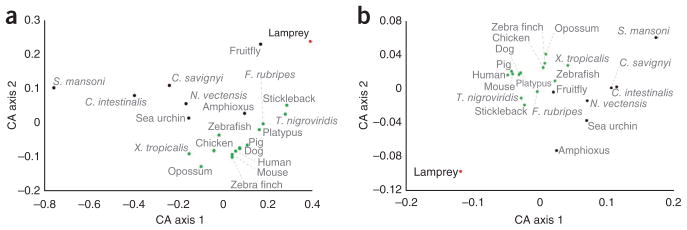
Variation in nucleotide content and substitution can strongly influence intragenomic functionality and intergenomic comparative analyses. Analysis of the lamprey genome showed that the GC content of the lamprey genome assembly was higher than that of most other vertebrate genome sequences that have been reported. Overall, 46% of the assembly was composed of GC bases, similar to the GC content of raw whole-genome sequencing reads (Supplementary Fig. 5 and Supplementary Note). Genome-wide analyses also showed patterns of intragenomic heterogeneity in GC content, similar to those of amniote species that possess isochore structures, but less variable. Moreover, the GC content of protein-coding regions (61%) was markedly higher than that of noncoding and repetitive regions. As expected, this content was highest in the third position of codons (75%) (Supplementary Fig. 6). Patterns of GC bias strongly affect codon usage and the amino-acid composition of lamprey proteins, imparting an underlying structure to lamprey coding sequences that differs substantially from those of all other sequenced vertebrate and invertebrate genomes (Fig. 2). Notably, we did not detect a significant correlation between the GC content of the third position of codons and the GC content of adjacent noncoding regions (Supplementary Fig. 7). Thus, it seems that the processes that lead to the patterns of intragenomic heterogeneity in lamprey GC content differ fundamentally from those in species that possess isochore structures. This raises a question regarding the adaptive value or other biological role of the observed variation of GC content within and among genomes.
Figure 2.
Genome-wide deviation of lamprey coding sequence properties from patterns observed in other vertebrate and invertebrate genomes. (a) Codon usage bias. Correspondence analysis (CA) on relative synonymous codon usage (RSCU) values was performed using the nucleotide sequences of all predicted genes concatenated for individual species. (b) Amino-acid composition. Red, lamprey; gray, invertebrates; green, jawed vertebrates.
To further explore the biological basis of high GC content and its intragenomic heterogeneity, we examined the relationship between the GC content of protein-coding regions and codon usage bias, amino-acid composition and the levels of gene expression. The results showed that genomic GC content strongly correlated with codon usage bias and amino-acid composition but not with the levels of gene expression (Supplementary Figs. 8–11, Supplementary Table 7 and Supplementary Note). These observations are consistent with a scenario in which high GC content results from broad-scale substitution bias rather than selection for specific GC-rich codons. As the lamprey is clearly an outlier among vertebrates, further dissection of coding GC content in the sea lamprey and other lamprey and hagfish species will help to identify the causes and consequences of the intragenomic heterogeneity of GC content in vertebrate genomes.
Duplication structure of the genome
It is generally accepted that two rounds of whole-genome duplication occurred early in the history of vertebrate evolution9. However, the timing of these defining duplication events has not been well supported by genome-wide sequence data thus far10. As the proximate outgroup to jawed vertebrates, the lamprey genome is uniquely suited for addressing several questions regarding the occurrence, timing and outcome of whole-genome duplication events. To identify gene and genome duplication events in the ancestral vertebrate lineage, we analyzed patterns of duplication within conserved syntenic regions of the lamprey and gnathostome genomes and compared these patterns to the entire lamprey genome assembly.
We estimated duplication frequencies by aligning all predicted lamprey protein-coding genes from the MAKER5 data set to the human (GRCh37, GCA_000001405.1) and chicken (Gallus_gallus-2.1, GCA_000002315.1) whole-genome assemblies. To account for the possibility that paralogs have been retained on one or both genomes, in a way that bypasses many confounding aspects of phylogenetic reconstruction (Supplementary Figs. 12–17, Supplementary Table 8 and Supplementary Note), regions were considered putative orthologs if they yielded the highest-scoring alignment between the two genomes or an alignment score (bit score) within 90% of the top-scoring alignment (Supplementary Note). Strong patterns of conserved synteny were observed between the lamprey and both the human and chicken genomes (Supplementary Figs. 18–21, Supplementary Tables 9–13 and Supplementary Note). For simplicity, we present comparisons to the chicken genome, as this genome is known to have undergone substantially fewer interchromosomal rearrangements than have mammalian genomes11,12.
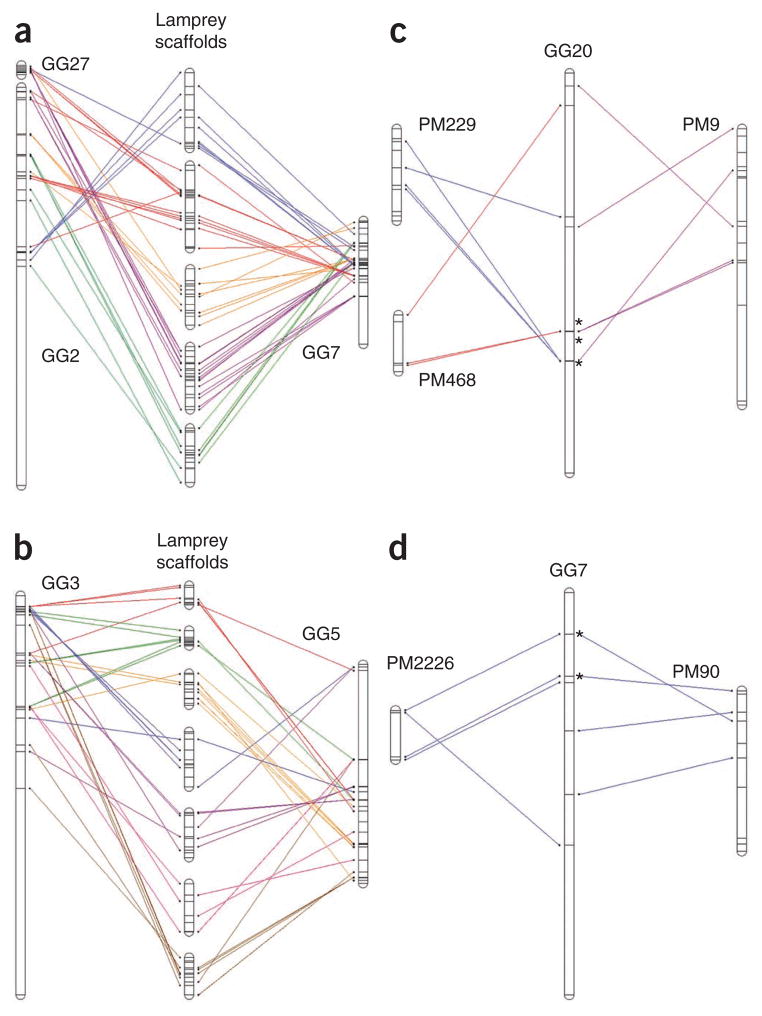
Our analyses indicate that most lamprey and gnathostome genes currently do not possess two copies in their respective genomes resulting from the two rounds of whole-genome duplication (Supplementary Note), presumably owing to the frequent loss of one paralog after duplication. Accordingly, we used the lamprey genome to search for a signature of large-scale duplication that does not rely on the retention of duplicated genes but can be informed by their presence. Specifically, we searched for cases in which a single lamprey scaffold contained interdigitated homologies from two distinct regions of a gnathostome genome (Fig. 3). Such patterns are consistent with large-scale duplication followed by random loss of either paralogous copy. Nearly all lamprey scaffolds showed patterns of interdigitated conserved synteny of gnathostome orthologs (Supplementary Tables 9 and 10). Moreover, homologs from individual pairs of gnathostome chromosomes were recurrently observed in interdigitated syntenic blocks on several lamprey scaffolds. Notably, some of the individual homologous markers that contributed to these conserved syntenic blocks were mapped to duplicate positions within gnathostome genomes, being present on the two homologous gnathostome chromosomes. Although these duplicates constituted a relatively modest fraction of the conserved syntenic homologs (14.5%, Fig. 3a; 18.2%, Fig. 3b; not counting redundant copies), we interpret these as strong evidence that large-scale (whole-genome) duplication has had a major role in shaping gnathostome genome architecture.
Figure 3.
Conserved synteny and duplication in the lamprey and gnathostome (chicken) genomes. (a–d) The locations of presumptive lamprey-chicken orthologs (including duplicates) are plotted relative to their physical positions on chromosomes and scaffolds and are connected by colored lines. (a,b) Pairs of chicken chromosomes that correspond to a series of lamprey scaffolds. (a) Ten lamprey loci are present as duplicate copies in the chicken genome, and 59 are present as single copies. (b) Twelve lamprey loci are present as duplicate copies in the chicken genome, and 54 are present as single copies. (c,d) Pairs of lamprey scaffolds that correspond to individual chicken chromosomes. (c) Three chicken loci are present as duplicate copies on syntenic lamprey scaffolds. (d) Two chicken loci are present as duplicate copies on syntenic lamprey scaffolds. Asterisks indicate duplicates.
Similar duplication patterns on lamprey scaffolds also seem to support the notion that large-scale (whole-genome) duplication has had a major role in shaping lamprey genome architecture. Although lamprey scaffolds do not yet provide chromosome-scale resolution, several cases were identified in which two large lamprey scaffolds contained predicted paralogs and patterns of interdigitated conserved synteny (two defining signatures of large-scale duplication; Fig. 3c,d and Supplementary Note). To further assay for patterns indicative of ancient whole-genome duplication events (for example, two rounds) within the lamprey genome, we manually examined all lamprey scaffolds that possessed ten or more gnathostome homologs. These 83 scaffolds accounted for 10% of the comparative map (10% of homology-informative genes) and possessed a duplication frequency (0.463, including redundant copies of duplicates) that was similar to that of the genome at large (0.448). Among these scaffolds, we identified 29 gene pairs that were present as duplicates on two large scaffolds and one trio that was present on three large scaffolds. For a majority of duplicates, scaffolds contained at least one additional ortholog on the chicken chromosome that harbored an ortholog of the duplicate (specifically, both scaffolds (59.3%), one scaffold (29.6%) and no scaffold (11.1%) contained an additional syntenic ortholog). On average, these scaffolds contained 2.98 additional conserved syntenic genes for each individual lamprey duplicate (including the 11.1% with no syntenic markers). These patterns are consistent with the existence of patterns of interdigitated synteny in the lamprey genome that are highly similar to those in gnathostome genomes, indicating that the most recent (two-round) whole-genome duplication event likely occurred in the common ancestral lineage of lampreys and gnathostomes.
Additional genome-wide analyses showed that (i) the number of ancestral loci with retained duplicates in gnathostome genomes was not significantly different from the number with retained duplicates in lamprey (lamprey = 0.271, chicken = 0.262; χ2 = 2.94, P = 0.08; Supplementary Note); (ii) the frequency of shared duplications was higher than would be expected by chance (observed = 0.150, expected = 0.022; χ2 = 6179, P(χ2) < 1 × 10−100, P(Fisher’s exact test) < 1 × 10−100; Supplementary Note); (iii) a model invoking recurrent selection against small-scale duplicates across a majority of the genome was not sufficient to explain genome-wide patterns of shared duplication (Supplementary Figs. 18–21 and Supplementary Note); and (iv) inclusion of the lamprey in phylogenetic analyses resolved gene families consistent with two rounds of whole-genome duplication (Supplementary Figs. 12–17 and Supplementary Note). Moreover, targeted analyses of Hox clusters and gonadotropin-releasing hormone (GnRH) syntenic regions showed that the loss of paralogs after duplication occurred largely independently in the lamprey and gnathostome genomes, consistent with the divergence of the two lineages shortly after the last whole-genome duplication event (Fig. 4, Supplementary Figs. 22–24, Supplementary Table 14 and Supplementary Note). Although the less parsimonious scenario involving one or two independent and ancient whole-genome duplication events in gnathostome and lamprey lineages cannot be completely ruled out, neither a gnathostome-specific genome duplication nor persistent selection to retain a subset of independent duplicates is likely to explain the subtle differences in the duplication structures of the lamprey and gnathostome genomes. It seems exceedingly unlikely that such genomic arrangements and distributions of synteny blocks would arise by chance or mechanisms other than an ancient shared whole-genome duplication event. We therefore propose that genome-wide patterns of duplication are indicative of a shared history of two rounds of genome-wide duplication before lamprey-gnathostome divergence.
Figure 4.
The effect of genome duplication and independent paralog loss on the evolution of lamprey-gnathostome conserved syntenic regions. (a) Conserved synteny among the GnRH2, GnRH3 and (previously proposed) GnRH4 genes in lamprey, chicken and humans, including the medaka region for GnRH3, which is absent in tetrapods. The orientation of each chromosome (chr.) and scaffold (scf.) is indicated with line arrows. A pointed box represents the orientation of each gene. Open rectangles with red X’s indicate lost GnRH loci. The presumptive ancestral state of the gene region is shown at the bottom. (b) Assembled lamprey Hox scaffolds and patterns of conserved synteny relative to human Hox clusters (human Hox clusters rather than chicken are used because all four human Hox syntenic regions are integrated into the human genome assembly). Three additional conserved syntenic genes, located adjacent to the PM2Hox cluster, are omitted owing to space limitations (retinoic acid receptor (RAR), heterogeneous nuclear ribonucleoprotein (HNRNP) and thyroid hormone receptor (THR)). Symbols indicate representative family members of lamprey-gnathostome homology groups.
Ancestral vertebrate biology
It has been suggested that many of the morphological and physiological features that characterize vertebrates evolved through the modification of preexisting regulatory regions and gene networks13. However, we reasoned that the lamprey genome might enable us to identify genes that evolved within the ancestral vertebrate lineage and infer how these new genes might have contributed to specific innovations in ancestral vertebrates that contributed to their arguably successful evolutionary trajectory. Toward this end, we searched for lamprey genes that (i) had homologs in at least one sequenced gnathostome genome and (ii) had no identifiable invertebrate homolog in annotated sequence databases and genome project–based resources (including but not limited to invertebrate deuterostomes: sea urchin, sea limpet, acorn worm, lancelet and sea squirt). In total, this search identified 224 gene families that presumably trace their evolutionary origin to the ancestral vertebrate lineage (Supplementary Table 15 and Supplementary Note). Notably, these included many gene families whose taxonomic distribution was previously thought to be more restricted (for example, APOBEC4 was previously reported to be a tetrapod-specific gene)14. Thus, roughly 1.2–1.5% of the protein-coding landscape in the human genome (263 genes from 224 families out of ~20,000 genes) originated from new genes that emerged at the base of vertebrate evolution. Phylogenetic analyses also showed expansions and reductions of gene families within vertebrate lineages (Supplementary Table 8 and Supplementary Note). These included the specific loss of clotting-related genes in the lamprey lineage and the differential contraction and expansion of gene families related to neural function and inflammation in the lamprey versus gnathostome lineages, which reflect broad parallels in the evolution of lamprey and gnathostome immunity (Supplementary Figs. 25–30, Supplementary Tables 16–22 and Supplementary Note).
To better understand how new genes might have contributed to the evolution of the vertebrate ancestor, we collected gene ontology (functional) information for the 224 vertebrate-specific gene families (Supplementary Fig. 31 and Supplementary Note). Comparing these gene ontologies to the genome-wide distribution of lamprey ontologies showed that these vertebrate-specific gene families were significantly enriched in functions related to myelination and neuropeptide and neurohormone signaling (Fig. 5). These findings suggest that the elaboration of signaling in the vertebrate central nervous system might have been facilitated by the advent of new vertebrate genes. Ontology analyses were also consistent with the broadly held view that most genes involved in the regulation of morphogenesis are of ancient origin and are common throughout animals.
Figure 5.
Enrichment of gene ontologies among vertebrate-specific gene families. Horizontal bars show the frequencies of ontology classes among vertebrate-specific gene families and in the entire set of lamprey gene models. Data are shown for all ontologies that are over-represented with P < 0.005 (Fisher’s exact test). Most over-represented ontologies are related to neural development and neurohormone signaling.
In all extant gnathostomes, myelinating oligodendrocytes wrap axons in a layer of proteins and lipids, increasing the efficiency and speed of neuronal conduction. In humans, disorders of myelination have many manifestations that range from cognitive to movement disorders. Notably, analysis of the lamprey genome identified the specific enrichment of genes associated with myelin formation in the central and peripheral nervous systems of jawed vertebrates (Fig. 5, Supplementary Fig. 32, Supplementary Tables 15,23,24 and Supplementary Note), despite the fact that extant jawless vertebrates are thought to completely lack myelinating oligodendrocytes15. These genes include Pmp22 (encoding peripheral myelin protein 22) and Mpz (encoding myelin protein zero), as well as Plp (encoding myelin proteolipid protein), Mal (encoding myelin and lymphocyte protein) and Myt1l (encoding myelin transcription factor 1-like). Homologs of Mal and Pmp22 were reported to be present in Ciona intestinalis, an invertebrate chordate16, and putative Ciona homologs of Myt1l and Plp1 are identifiable in Ensembl17. Unexpectedly, analysis of the lamprey genome identified three myelination-related genes that might have evolved specifically within the ancestral vertebrate lineage (Mbp (encoding myelin basic protein), Mpz and CNP (encoding 2′,3′-cyclic nucleotide 3-phosphodiesterase); Supplementary Tables 15,23 and Supplementary Note). This suggests that the molecular components of myelin already existed in the vertebrate ancestor and were later recruited in the evolution of myelinating oligodendrocytes in the gnathostome lineage, perhaps through the evolution of regulatory systems18. Alternatively, oligodendrocyte-like cells might have been present in the vertebrate ancestor but were secondarily lost in the lamprey lineage, although it retained genes encoding myelin proteins. Dissecting the function of myelination-related genes in lamprey and hagfish should continue to shed light on the origin of gnathostome myelin.
By virtue of its basal phylogenetic position, the lamprey also serves as a key comparative model for understanding the evolution of the vertebrate immune system. Lamprey possess two major immune cell types that are similar to the T and B lymphocytes of gnathostomes but possess adaptive immune receptors that are unrelated to gnathostome immunoglobulins, perhaps instead reflecting the receptor of the ancestral vertebrate19,20. The lamprey genome harbors several genes that impart unique functionality to gnathostome T and B lymphocytes. Annotation of other components of the immune system showed that the reduced complexity in vertebrate innate immune receptors might have coincided with the evolution of adaptive immune receptors (Supplementary Figs. 25–30, Supplementary Tables 16–22 and Supplementary Note). Analysis of the lamprey genome assembly and end-mapped BAC clones showed that each rearranging lamprey immune receptor locus (encoding variable lymphocyte receptors, VLRs) extends for several hundred contiguous kilobases. For example, the VLRB locus extends for at least 717 kb, with components of the receptor face being drawn from regions distributed across practically the entire length of the current scaffold (Supplementary Fig. 25).
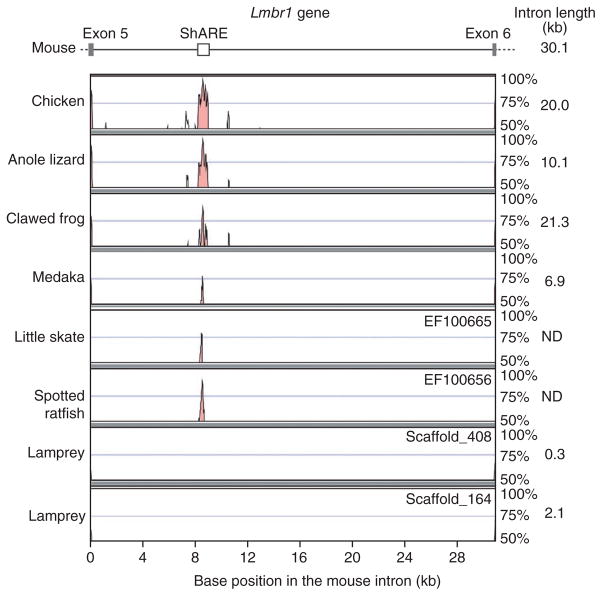
The lamprey genome also sheds light on the evolutionary events that occurred early in the evolution of the gnathostome lineage, after the lamprey-gnathostome split. Paired appendages (pelvic and pectoral fins in fish, hind- and forelimbs in tetrapods) are a major evolutionary innovation of gnathostome vertebrates, as they permitted additional forms of locomotion and behavior. The lamprey has well-developed dorsal and caudal fins but lacks paired fins. Despite different embryonic origins, the signaling pathways involved in the development and positioning of median fins were reused for paired fin development21, raising the question of whether these pathways were already present in the limbless ancestral vertebrate (Supplementary Note). During fin and limb development, Shh is required to pattern the anteroposterior axis of appendages. It has been shown that the limb-specific expression of Shh is coordinated by a long-range cis-acting enhancer. This Shh appendage-specific regulatory element (ShARE) is found in homologous positions in tetrapods, teleosts and chondrichthyans22–24. In all vertebrate species analyzed so far, this element is found in intron 5 of the Lmbr1 gene (encoding limb region 1) that lies up to 1 Mb away from the transcription start site of Shh. Notably, the presence of ShARE is correlated with the presence of paired appendages, at least within the tetrapod lineage, as snakes and caecilians seem to have lost this element secondarily25. Because of the conserved genomic position of the element in other vertebrates, we focused our analysis on the lamprey orthologs of the Lmbr1 gene. Directed analysis of intron 5 in the Lmbr1 orthologs showed that these introns were much shorter and had no similarity to ShAREs (Fig. 6 and Supplementary Fig. 33). Searches of the entire genome assembly and raw sequence reads also did not detect any regions similar to ShARE, suggesting that this regulatory region evolved within the gnathostome lineage.
Figure 6.
Absence of sequence conservation for a limb Shh enhancer in lamprey. Comparison of an intronic region in the Lmbr1 gene, focusing on the intron containing the Shh cis-regulatory element (ShARE, also known as MFCS1)22,24. Note that two genomic regions were identified in the lamprey harboring potential Lmbr1 orthologs. The lengths of this intron for individual species are listed on the right. ND, not determined.
DISCUSSION
The lamprey genome provides unique insight into the origin and evolution of the vertebrate lineage. Here, we present a few examples of its use in dissecting the evolution of vertebrate genomes and aspects of ancestral vertebrate biology. As examples, we (i) provide genome-wide evidence for two whole-genome duplication events in the common ancestral lineage of lampreys and gnathostomes, (ii) identify new genes that evolved within this ancestral lineage, (iii) link vertebrate neural signaling features to the advent of new genes, (iv) uncover parallels in immune receptor evolution and (v) provide evidence that a key regulatory element in limb development evolved within the gnathostome lineage. This genomic resource holds the promise of providing insights into many other aspects of vertebrate biology, especially with continued refinements in the assembly and the capacity for direct functional analysis in lamprey26,27.
URLs
CodonW, http://codonw.sourceforge.net/; RECON, http://www.repeatmasker.org/; Repbase, http://www.girinst.org/repbase; Rebuilder, http://www.broadinstitute.org/crd/wiki/index.php/Improving_Assemblies.
METHODS
Methods and any associated references are available in the online version of the paper.
Accession codes
The lamprey genome assembly has been deposited under GenBank accession AEFG01. Improved assemblies for Hox clusters have been deposited under GenBank accessions JQ706314–JQ706327. Transcript sequencing data have been deposited under GenBank Short Read Archive accessions SRX109761.3, SRX109762.3, SRX109764.3, SRX109765.3, SRX109766.3, SRX109767.3, SRX109768.3, SRX109769.3, SRX109770.3, SRX110023.2, SRX110024.2, SRX110025.2, SRX110026.2, SRX110027.2, SRX110028.2, SRX110029.2, SRX110030.2, SRX110031.2, SRX110032.2, SRX110033.2, SRX110034.2 and SRX110035.2. Additional information is provided in Supplementary Table 5.
ONLINE METHODS
Genome sequencing
Sea lamprey DNA for whole-genome shotgun sequencing and fosmid and BAC libraries was derived from a liver dissected from a single female lamprey captured from the Great Lakes. Production of BAC library CHORI-303 was described previously28. Other libraries were cloned into bacterial vectors, arrayed individually into the wells of growth trays and sequenced as previously described11,29–31.
Preassembly analyses
Several analyses were performed before initiating the assembly. These provided insight as to the selection of the assembler. Initial characterization of the repetitive content of the genome was performed by selecting a subset of 10,000 high-quality shotgun sequence reads (>500 bp at Q20) and aligning these to the complete data set of 18.5 million whole-genome shotgun sequence reads (Q20 trimmed). A complementary analysis was also performed by aligning 10,000 trimmed whole-genome shotgun sequence reads from a single human genome32 to a complete data set of 12.1 million whole-genome shotgun sequence reads (Q20 trimmed). All reads were downloaded from the NCBI Trace Archives in .scf format and processed with phred33,34 to generate base calls and quality scores. Alignments to human and lamprey whole-genome shotgun sequence data sets were performed using Megablast35.
To gain insight into the potential influence of allelic polymorphism, we estimated the depth of coverage by processing Megablast35 alignments between a subset of reads and the entire whole-genome shotgun sequencing effort, as described above, but with varying thresholds for percent nucleotide identity between aligning sequences. Distributions of coverage depth were estimated using sequence identity thresholds of 90%, 95%, 97% and 99%.
Genome assembly
Assembly of the lamprey genome was performed using a total of ~19 million sequence reads with Arachne36 parameterized for the assembly of an outbred diploid genome (Supplementary Note). After assembly by the Assemblez module, contigs corresponding to divergent haplotypes were assembled together using the Rebuilder module, parameterized with liberal settings that permitted the merger of divergent haplotypes (see URLs), and haplotypes were then joined using linkage information from end-read mapping information. End-mapping information was incorporated via the ExtendHaploSupers module in a series of steps that prioritized the number of end reads supporting linkages between contigs and the source of end-mapping information (shotgun reads versus large-insert clones). Specifically, paired-end mapping information was incorporated in the following steps, where subsequent linkages might not supplant linkages that had been previously identified at a more stringent threshold: at least four paired-end linkages from large-insert clones, at least four paired-end linkages from large-insert clones or whole-genome shotgun sequence clones, three paired-end linkages from large-insert clones, three paired-end linkages from large-insert clones or whole-genome shotgun sequence clones, two paired-end linkages from large-insert clones, two paired-end linkages from large-insert clones or whole-genome shotgun sequence clones, a single paired-end linkage from a large-insert clone and, finally, a single paired-end linkage from a whole-genome shotgun sequence clone.
Characterization of repetitive sequences
Repetitive sequences were collected with RECON (v1.06; see URLs)37, with a cutoff of ten copies, and sequences were further curated to verify their identity, individuality and 5′ and 3′ boundaries. Each sequence was searched against the sea lamprey genomic sequences, and at least ten hits (BLASTN38 E < 1 × 10−10) plus 100 bp of 3′ and 5′ flanking sequence were recovered. If a particular lamprey sequence was found to be similar to a known transposon at the nucleotide or protein level (BLASTN or BLASTX, respectively; E < 1 × 10−5; RepBase 14.12), it was assigned to that repeat class. Recovered sequences were then aligned using dialign 2 (ref. 39), with the resulting output examined for the presence of possible boundaries between putative elements and the possible presence of target site duplications. Repeats were additionally searched for homology to known repeat classes in Repbase 14.12 (see URLs)40, using RepeatMasker and BLAST (BLASTX E < 1 × 10−5) to identify elements similar to other known transposable elements.
Gene annotation
Annotations for the lamprey genome assembly were generated using the automated genome annotation pipeline MAKER5, which aligns and filters EST and protein homology evidence, identifies repeats, produces ab initio gene predictions, infers 5′ and 3′ UTRs and integrates these data to produce final downstream gene models along with quality control statistics. Inputs for MAKER included the P. marinus genome assembly, P. marinus ESTs, a species-specific repeat library and protein databases containing all annotated proteins for 14 metazoans (Supplementary Note) combined with the Uniprot/Swiss-Prot41 protein database and all sequences for Chondrichthyes (cartilaginous fishes) and Myxinidae (hagfishes) in the NCBI protein database42,43. Ab initio gene predictions were produced inside of MAKER by the programs SNAP44 and Augustus45. MAKER was also passed P. marinus RNA-seq data processed by the programs tophat and cufflinks (Supplementary Note)46.
Identification of CNEs
The lamprey assembly was searched for sequences homologous to conserved noncoding sequences previously identified in comparisons between human and Fugu47 and human and Callorhinchus milii6 genomes. BLASTN (2.2.25+) was used with the word size set to 5 and with gap existence and extension penalties of 1.
Codon usage
Genome-wide assessment of codon usage bias and aminoacid composition in lamprey genes was performed using predicted coding sequences after discarding all but the longest transcript variant for each gene. To avoid any bias imparted by small sequences, sequences shorter than 300 bp were excluded from analyses of GC content, leaving a total of 18,444 coding sequences. Overall GC content and GC content at third codon positions were calculated for each protein-coding gene, and the GC content was calculated for the 10-kb fragment harboring the gene(s). To investigate the possible influence of gene expression levels on codon usage bias and amino-acid composition, we compared the GC content of 50 highly expressed and 50 lowly expressed genes on the basis of RNA-seq reads. To analyze codon usage bias and aminoacid composition, we performed correspondence analysis (COA) on RSCU values48 and on amino-acid composition values using the software CodonW49 (see URLs).
To assess the possible deviation of the sequence properties of lamprey protein-coding regions relative to other species, we downloaded genome-wide protein-coding sequences for diverse vertebrates and invertebrates from Ensembl17 and the archives for individual genome projects. Using species-by-species concatenated protein-coding sequences, we calculated RSCU values and performed a correspondence analysis.
Phylogenetic analysis of lamprey genes
A genome-wide phylogenetic analysis including 50 vertebrate genomes, 2 additional chordates and 3 outgroups was performed using the Ensembl tree reconstruction pipeline and the Ensembl compara database, Build 64 (ref. 50). All genes were clustered with hcluster_sg51 according to their sequence similarity52. A multiple-sequence alignment was built for each cluster using MCoffee53, and TreeBeST51 was then used to reconstruct a consensus tree for each family using two maximum-likelihood and three neighbor-joining trees. The software package CAFE54 was used to study the evolution of gene families in the lamprey and the gnathostomes.
Comparative genomics
Regions were considered putative orthologs if they yielded the highest-scoring alignment between the two genomes or an alignment score (bit score) within 90% of the top-scoring alignment (TBLASTN38; comparison of lamprey gene models to the human or chicken genomes). This convention permits some variation in the divergence rate and can be applied uniformly to the genome but may not identify some duplicates that have undergone exceedingly rapid diversification after duplication. Second, analyses were limited to single-copy genes and duplicates that were broadly distributed throughout the genome and present at relatively low copy number by removing redundant copies of tandemly duplicated genes (lineage-specific gene amplifications) and homology groups that contained more than six homologs in either of the two species being compared in any pairwise analysis.
Hox genes
To supplement the assembly of Hox gene–containing regions, we selected a series of BACs via hybridization to a Hox2 probe designed from a known lamprey transcript (GenBank accession AY497314). Another series of BACs were selected by hybridization to Hox4 or Hox9 homeodomain probes and were pooled and sequenced by 454 sequencing.
Identification of vertebrate-specific genes
All P. marinus predicted peptides were aligned to peptides of all gnathostome species (Ensembl version 58; ref. 55) using BLASTP38. All gnathostome peptide sequences that showed a maximal bit score of no less than 50 were used as query in a BLASTP search against invertebrate peptide sequences. This invertebrate database included all sequences available in GenBank and Ensembl for invertebrates, as well as all peptides predicted in the genomes of Schistosoma japonicum56, Schistosoma mansoni57 and Lottia gigantea42. All gnathostome query sequences with identifiable homologs in lamprey but not in any invertebrate were considered candidate vertebrate-specific genes. Candidates with bit scores between 50 and 60 were regarded as valid if the best hit from a reciprocal BLASTP search was the starting query sequence itself or its homolog with a bit score of no less than 50.
Immunity-related gene families
To understand the relationships among members of individual gene families, neighbor-joining trees were constructed in MEGA5 (ref. 58) using complete gap deletion.
The Shh enhancer ShARE
The genomic sequences of jawed vertebrates and the lamprey were compared with mVISTA59 using the mouse as a reference.
Supplementary Material
Acknowledgments
We thank the Genome Institute, Washington University School of Medicine, Production Sequencing group for all sample procurement and genome sequencing work, the Michigan State University Genomic Core for transcriptome sequencing and the US Geological Survey, Lake Huron Biological Station for providing lamprey samples for sequencing. We thank F. Antonacci and E.E. Eichler (University of Washington) for performing FISH and providing access to computational facilities, respectively. We thank M. Robinson for bioinformatic analysis of immune system–related genes and conversion of GFF files for BAC end mapping. A portion of this research was conducted at the Marine Biological Laboratory (Woods Hole, Massachusetts). We acknowledge the support of the Stowers Institute for Medical Research (SIMR) and technical support from the SIMR Molecular Biology Core, particularly K. Staehling, A. Perera and K. Delventhal for BAC screening and sequencing. We acknowledge the Center for High-Performance Computing at the University of Utah for the allocation of computational resources toward gene annotation. We recognize all the important work that could not be cited owing to space limitations. The lamprey genome project was funded by the National Human Genome Research Institute (U54HG003079 (R.K.W.)). Additional support was provided by grants from the US National Institutes of Health (R24GM83982 (W.L.)) and the Great Lakes Fisheries commission (W.L.). Partial funding was provided by several additional sources, including grants from the US National Institutes of Health (F32GM087919 and T32HG00035 (J.J.S.); DE017911 (M.E.B.); R03NS078519 (O.E.B.); R01HG004694 (M.D.Y.); GM079492, GM090049 and RR014085 (C.T.A.); and R37HD032443 (C.J.T.)), the National Science Foundation (MCB-0719558 (C.T.A.); IOS-0849569 (S.A.S.); IBN-0208138 (L. Holland); and IOS-1126998 (M.D.Y.)), the New Hampshire Agricultural Experiment Station (Scientific Contribution Number 2471 (S.A.S.)), the Charles Evans Research Award (O.E.B., J.D.B. and J.R.M.), the Wellcome Trust (WT095908 (P.F.) and WT098051), the Canadian Institutes of Health Research (MOP74667 (J.P.R.)) and the Natural Sciences and Engineering Research Council of Canada (312221 (J.P.R.)).
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.J.S. developed the assembly, coordinated analyses, performed analyses of genome structure and conserved synteny, coordinated the manuscript, and wrote and edited the manuscript. S.K. contributed to analyses of GC content, assembly completeness, vertebrate-specific genes, myelin-related genes and limb development, and to preparation of the manuscript and supplements. C.H. compiled molecular data sets and developed the consortium gene annotations and annotation pipeline. T.S.-S. developed the protocol for the preparation of BACs, identified the sequenced individual, and prepared genomic DNA for sequencing and BAC library construction. N.J. performed computational identification and analysis of transposable elements. M.D.Y. and M.S.C. contributed to the development of the consortium gene annotations and the annotation pipeline. T.M. and A.M. performed analysis of vertebrate-specific gene families, codon usage bias and amino-acid composition, and contributed to the writing of the manuscript. S.D. and M.H. contributed to analysis of codon usage bias and amino-acid composition. O.E.B., J.R.M., J.D.B. and R.S. performed experiments generating neuronal transcriptomes and data, and sequence analysis related to the vertebrate central nervous system. C.S., L.M.W., A.S.G., M.C. and R.K. performed experiments and data analysis related to the identification and annotation of Hox genes, led and prepared by L.M.W. S.A.S., W.A.D. and J.A.H. performed analyses related to the evolution of neuroendocrine genes, led by S.A.S. and prepared by W.A.D. C.T.A., N.R.S., K.M.B., J.P.R., S.D. and M.H. performed analyses related to the evolution of immune system genes, led and prepared by C.T.A., K.M.B., J.P.R. and M.H. N.R. and C.J.T. performed analyses related to the evolution and development of appendages. P.P. performed BLAST analyses of the noncoding portion of the lamprey genome, and G.E. analyzed BLAST output and wrote the corresponding sections. M.R., B.L.A. and S.M.J.S. developed the Ensembl gene set, led and prepared by M.R. M.M., M.P. and J.H. performed GeneTree and CAFE analysis for the study of whole-genome duplications at the stem of the vertebrate lineage and prepared the corresponding sections. T.S.-S., M.J., J.A.L. and D.W.M. developed the protocol for the preparation of cDNA. N.M. and P.G. provided isolated leukocyte RNA. C.T.B. and K.G.N. performed transcriptome assemblies. W.L., Y.-W.C.-D., S.V.L., C.-Y.Y. and D.W.M. contributed to next-generation transcriptome sequencing. Z.P. provided lamprey leukocyte RNA and cDNA samples and libraries, and evaluated the first draft assembly of the genome. B.F. contributed to the development of neurodevelopment-related text. P.J.d.J. and B.Z. generated the BAC library used for genome sequencing and assembly. L.L.F., W.C.W. and S.W.C. contributed to sequencing project management. B.T. coordinated the cDNA sequencing projects. P.F. supervised the Ensembl annotation efforts. M.E.B. contributed to the conception of the sea lamprey genome project and the development of the manuscript. R.K.W. provided supervision of the genome sequencing project. W.L. provided coordination of the consortium and analysis of the assembly, and contributed to the development of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Shimeld SM, Donoghue PC. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish) Development. 2012;139:2091–2099. doi: 10.1242/dev.074716. [DOI] [PubMed] [Google Scholar]
- 2.Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel BL, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolfe A, et al. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatesh B, et al. Ancient noncoding elements conserved in the human genome. Science. 2006;314:1892. doi: 10.1126/science.1130708. [DOI] [PubMed] [Google Scholar]
- 8.McEwen GK, et al. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS Genet. 2009;5:e1000762. doi: 10.1371/journal.pgen.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin Cell Dev Biol. 1999;10:517–522. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- 10.Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 11.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 12.Smith JJ, Voss SR. Gene order data from a model amphibian (Ambystoma): new perspectives on vertebrate genome structure and evolution. BMC Genomics. 2006;7:219. doi: 10.1186/1471-2164-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 15.Bullock TH, Moore JK, Fields RD. Evolution of myelin sheaths: both lamprey and hagfish lack myelin. Neurosci Lett. 1984;48:145–148. doi: 10.1016/0304-3940(84)90010-7. [DOI] [PubMed] [Google Scholar]
- 16.Gould RM, Morrison HG, Gilland E, Campbell RK. Myelin tetraspan family proteins but no non-tetraspan family proteins are present in the ascidian (Ciona intestinalis) genome. Biol Bull. 2005;209:49–66. doi: 10.2307/3593141. [DOI] [PubMed] [Google Scholar]
- 17.Flicek P, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. doi: 10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha NR, Smith J, Amemiya CT. Evolution of adaptive immune recognition in jawless vertebrates. Semin Immunol. 2010;22:25–33. doi: 10.1016/j.smim.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freitas R, Zhang G, Cohn MJ. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature. 2006;442:1033–1037. doi: 10.1038/nature04984. [DOI] [PubMed] [Google Scholar]
- 22.Dahn RD, Davis MC, Pappano WN, Shubin NH. Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature. 2007;445:311–314. doi: 10.1038/nature05436. [DOI] [PubMed] [Google Scholar]
- 23.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 24.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 25.Sagai T, et al. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh) Mamm Genome. 2004;15:23–34. doi: 10.1007/s00335-033-2317-5. [DOI] [PubMed] [Google Scholar]
- 26.Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc Natl Acad Sci USA. 2008;105:20083–20088. doi: 10.1073/pnas.0806009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikitina N, Bronner-Fraser M, Sauka-Spengler T. The sea lamprey Petromyzon marinus: a model for evolutionary and developmental biology. In: Behringer RR, Johnson AD, Krumlauf EE, editors. Emerging Model Organisms: A Laboratory Manual. Vol. 1. CSHL Press; Cold Spring Harbor, New York: 2009. pp. 405–429. [DOI] [PubMed] [Google Scholar]
- 28.Osoegawa K, et al. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- 29.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 30.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 34.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 35.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 36.Jaffe DB, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 41.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenyon EJ, McEwen GK, Callaway H, Elgar G. Functional analysis of conserved non-coding regions around the short stature hox gene (shox) in whole zebrafish embryos. PLoS ONE. 2011;6:e21498. doi: 10.1371/journal.pone.0021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp PM, Li WH. The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:81–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peden JF. DNA Repair. University of Nottingham; 2000. Analysis of codon usage. [Google Scholar]
- 50.Vilella AJ, et al. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan J, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 53.Wallace IM, O′Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 55.Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 59.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.