Abstract
We characterized the full-length genomes of five distinct hepatitis C virus (HCV)-3 isolates. These represent the first complete genomes for subtypes 3g and 3h, the second such genomes for 3k and 3i, and of one novel variant presently not assigned to a subtype. Each genome was determined from 18–25 overlapping fragments. They had lengths of 9579–9660 nt and each contained a single ORF encoding 3020–3025 aa. They were isolated from five patients residing in Canada; four were of Asian origin and one was of Somali origin. Phylogenetic analysis using 64 partial NS5B sequences differentiated 10 assigned subtypes, 3a–3i and 3k, and two additional lineages within genotype 3. From the data of this study, HCV-3 full-length sequences are now available for six of the assigned subtypes and one unassigned. Our findings should add insights to HCV evolutionary studies and clinical applications.
The reconstruction of the epidemic history of the Hepatitis C virus (HCV) has been difficult due to an absence of historically archived samples that would allow earlier viral sequences to be determined. Thus, it relies on the determination of the worldwide geographical distribution of the contemporary circulating viral strains and the analysis of their genetic relationships by using traditional and newly developed approaches. For this purpose, the determined viral sequences are the sole source of information. Although our recent studies have described the genetic variation patterns for isolates representing various subtypes within genotypes 2, 4 and 6 (Li et al., 2006, 2009a, b, 2012; Lu et al., 2006, 2007a, b, 2008; Wang et al., 2009; Xia et al., 2008), such information is still in short supply for genotypes 1 and 3. For genotype 3, some strains (i.e. 3a and 3b) have spread worldwide mainly due to transmission via injection drug use and immigration, which are often seen in developed countries in Europe and North America (Bourlière et al., 2002; McCaw et al., 1997; Pawlotsky et al., 1995; Silini et al., 1995). In contrast, specific HCV-3 subtypes have been exclusively found in certain regions of Asia which reflects a long-term endemic circulation (Greene et al., 1995; Lole et al., 2003; Tokita et al., 1994a, b). At present, a total of 10 genotype 3 subtypes have been assigned. However, full-length genomes are available only for four: 3a, 3b, 3i and 3k. In this study, five genotype 3 isolates representing subtypes 3g, 3h, 3i and 3k, and a distinct lineage not yet assigned to a subtype were entirely sequenced.
Full-length genomes were characterized for isolates QC29, QC105, QC115, QC260 and QC268, each from 18–25 overlapping fragments. These genomes are 9579–9660 nt in length from the 5′ UTR termini to the highly conserved 3′ X tails. They each contain a single ORF of 9060–9075 nt. The 5′ UTRs are of 337–340 nt, while the 3′ UTRs vary from 180 to 269 nt. The sizes of the 10 protein-coding regions are as follows: core (573 nt/191 aa), E1 (576 nt/192 aa), E2 (1101–1110 nt/367-370 aa), P7 (189nt/63 aa), NS2 (651 nt/217 aa), NS3 (1893 nt/631 aa), NS4A (162 nt/54 aa), NS4B (783 nt/261 aa), NS5A (1353–1362 nt/451–454 aa) and NS5B (1776 nt/591 aa). Among these 10 regions, E2 and NS5A are variable in length (Table 1).
Table 1. Patient information for the five genotype 3 isolates and the number of nucleotides/amino acids in each genomic region.
The genotype 1 sequence H77 (GenBank accession no. AF009606) is included for comparison. Bold entries indicate that the genomic regions are variable in length. –, Data not known; F, female; M, male.
| ID | Age (years) | Gender | Origin | Full sequence | ORF | 5′ UTR | Core | E1 | E2 | P7 | NS2 | NS3 | NS4A | NS4B | NS5A | NS5B | 3′ UTR |
| H77 | – | – | – | 9646 | 9036/3011 | 341 | 573/191 | 576/192 | 1089/363 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1344/448 | 1776/591 | 269 |
| QC260_3 g | 31 | F | India | 9634 | 9075/3025 | 339 | 573/191 | 576/192 | 1110/370 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1362/454 | 1776/591 | 220 |
| QC29_3h | 44 | F | Somalia | 9579 | 9060/3020 | 339 | 573/191 | 576/192 | 1104/368 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1353/451 | 1776/591 | 180 |
| QC268_3i | 42 | M | Pakistan | 9646 | 9066/3022 | 340 | 573/191 | 576/192 | 1107/369 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1356/452 | 1776/591 | 240 |
| QC105_3k | 47 | F | Pakistan | 9660 | 9060/3020 | 339 | 573/191 | 576/192 | 1104/368 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1353/451 | 1776/591 | 261 |
| QC115_3 | 55 | M | Middle East | 9581 | 9066/3022 | 337 | 573/191 | 576/192 | 1101/367 | 189/63 | 651/217 | 1893/631 | 162/54 | 783/261 | 1362/454 | 1776/591 | 184 |
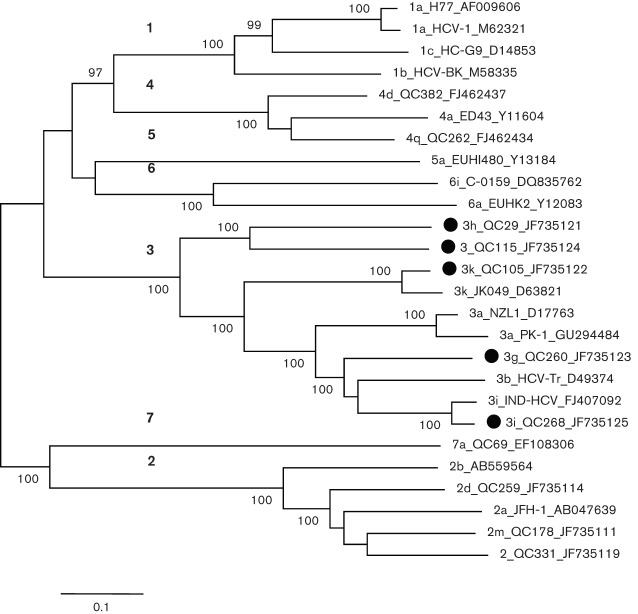
A maximum-likelihood (ML) tree was reconstructed using 26 full-length HCV genomes (Fig. 1). The tree showed seven major branches or clusters each representing one genotype. Major clusters comprising two or more sequences showed significant bootstrap support values of 100 %. Prior to the completion of this study, full-length HCV-3 sequences had been characterized for subtypes 3a, 3b, 3i and 3k. In the reconstructed tree these four subtypes were clearly differentiated. QC105 classified into 3k along with JK049, and QC268 into 3i along with IND-HCV. On the other hand, QC29, QC115 and QC260 grouped independently, representing three distinct subtypes. QC260 and QC29 have been provisionally assigned as subtypes 3g and 3h, respectively, based on partial C/E1 and NS5B region sequences (Murphy et al., 2007). In the present study, these two subtypes were confirmed as 3g and 3h based on the first determination of their full-length genomes. Similarly, QC115 also grouped independently from other type 3 sequences based on partial C/E1 and NS5B sequences and its complete genome sequence corroborates this finding.
Fig. 1.
ML phylogeny estimated from full-length nucleotide sequences. The five isolates described in this study (marked with black circles) and reference sequences from subtypes 1a, 1b, 1c, 2a, 2b, 2d, 2m, 2, 3a, 3b, 3k, 3i, 4a, 4d, 4q, 5a, 6a, 6i and 7a were included. Isolates are named with the following format: subtype_isolate_GenBank accession number. Regarding the format, ‘2’ and ‘3’ indicate that a subtype has not been assigned. Numbers in bold (on the left) indicate the seven major branches or clusters that each represent one genotype. Bootstrap support values (expressed as percentages) are shown at branch nodes. Bar, 0.1 nt substitutions per site.
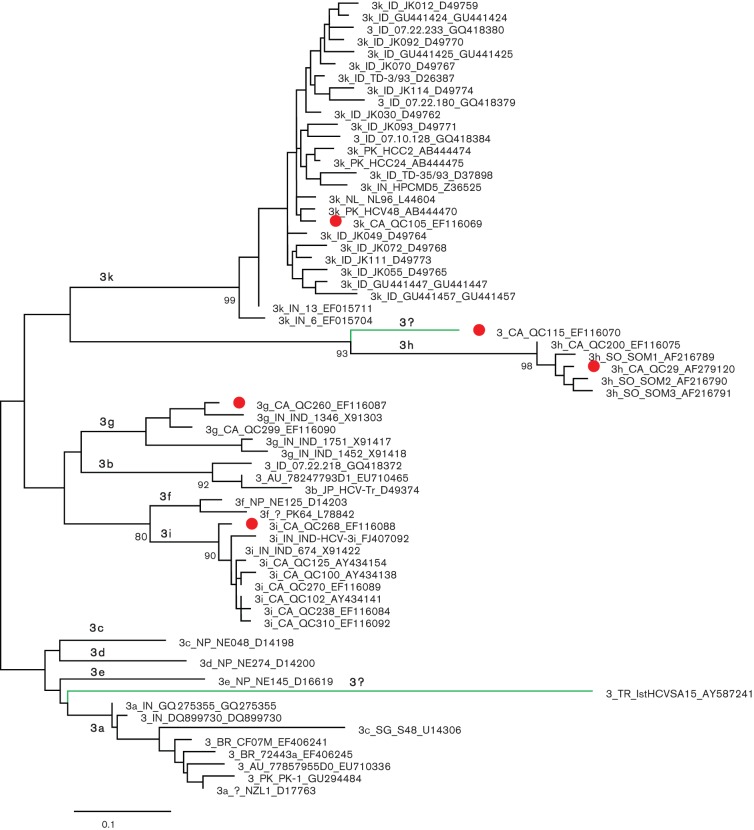
A segment of the NS5B region, corresponding to nucleotide numbering of 8276–8615 in the H77 genome, has been found to reliably differentiate HCV genotypes and subtypes and is therefore routinely used for proposing the provisional assignment of HCV isolates. To better understand the complexity of the HCV-3 lineage, a phylogenetic tree was reconstructed using 59 NS5B sequences retrieved from the HCV database and the five determined in this study. The tree displays 12 major branches or clusters corresponding to 12 subtypes or lineages within genotype 3 (Fig. 2). Among them, 10 have been assigned subtypes 3a-3i and 3k, while two (blue branches in the tree) have not. The five isolates from this study represent four of the 10 assigned subtypes and one of the two unassigned. Specifically, QC260/3g groups with four isolates, three from India (Panigrahi et al., 1996) and one from Canada (Murphy et al., 2007). QC29/3h also groups with four isolates, three from Somalia (Abid et al., 2000) and one from Canada (Murphy et al., 2007). QC268/3i groups with eight isolates, two from India (Panigrahi et al., 1996) and six from Canada (Murphy et al., 2007). QC105/3k groups with 26 isolates: 19 from Indonesia, three from India, three from Pakistan and one from a patient living in the Netherlands who contracted the infection in Indonesia (Hotta et al., 1994; Tokita et al., 1996; Valliammai et al., 1995; van Doorn et al., 1994, 1995). In contrast, QC115 represents an unclassified lineage equivalent to a new subtype. Although all living in Canada, the patients infected with the five isolates identified in this study originated from the Indian subcontinent and Somali. Not presented in Fig. 2, subtypes 3a and 3b each had more than 100 NS5B sequences available in the HCV database. Except for these two subtypes, other subtypes each had only a few sequences determined. For example, 3c, 3d and 3e each have only one sequence, all from Nepal, while 3f has two sequences, one from Pakistan and one from Nepal (Shrestha et al., 1994; Stuyver et al., 1996). Isolate 1stHCVSA15 from Turkey has not been formally published and constitutes the 12th lineage. In the HCV database, many genotype 3 sequences have been regarded as unclassified variants. However, our reanalysis showed that the majority of them can be grouped into subtypes 3a, 3b and 3k (data not shown). As a result, only isolates QC115, 1stHCVSA15, HCV-8 [3g] and AV69 (only the core region sequence available) represent three unclassified subtype equivalents, of which QC115 was entirely sequenced in the present study.
Fig. 2.
ML tree for 64 sequences of the NS5B region corresponding to nt 8276–8615 in the numbering of H77 genome. The five isolates completely sequenced in this study are marked with red circles. Green branches mark the two unassigned lineages; otherwise branches indicate the 10 assigned subtypes. Isolates are named with the following format: subtype_original country_isolate ID_GenBank accession number. Bootstrap support values (expressed as percentages) are shown at branch nodes. Bar, 0.1 nt substitutions per site. AU, Australia; BR, Brazil; CA, Canada; ID, Indonesia; IN, India; NL, the Netherlands; NP, Nepal; PL, Poland; PK, Pakistan; SG, Singapore; SO, Somalia.
To exclude the possibility of viral recombination, pairwise nucleotide similarity curves were plotted along HCV genomes. Comparing the five HCV-3 isolates against each other and against the 21 reference sequences shown in Fig. 1, no such evidence was detectable (data not shown).
Conclusively, full-length genome sequences were characterized for five HCV-3 isolates: QC260, QC29, QC268, QC105 and QC115. The former four represent subtypes 3g, 3h, 3i and 3k, respectively, while the last one indicates a distinct variant. Subtype 3g was first proposed in 1996 from partial core (451 nt) and NS5B (249 nt) sequences obtained from three patients in India (Panigrahi et al., 1996). Following this report, several other studies have also detected 3g sequences in patients or immigrants from India (Lole et al., 2003; Murphy et al., 2007; Verma & Chakravarti, 2008). Currently, the Los Alamos HCV database contains partial sequences for a total of 20 isolates of 3g. Hence, the characterization of the full-length genome sequence for this subtype was one of our primary tasks.
An isolate of subtype 3i was identified in India as early as 1996, but a formal designation was not given (Panigrahi et al., 1996). In 2003, five additional isolates of 3i were detected in India. Based on a partial sequence in the core region, it was proposed that these isolates be classified as subtype 3i (Lole et al., 2003). In 2007, seven isolates of 3i were detected in Canada. They were obtained from five Caucasians and two immigrants, one (QC270) from Lebanon and one (QC268) from Pakistan (Table 1) (Murphy et al., 2007). In that study, partial C/E1 and NS5B region sequences were determined for all isolates. In addition, the complete core region sequence was determined for three of the seven isolates. Comparison of the core region sequences of these three isolates with those reported by Lole et al. (2003) showed that these belonged to the same subtype. In this study, QC268 was selected to have its entire genome sequenced. However, in 2009 a full-length 3i sequence (GenBank accession no. FJ407092) from India was released in the Los Alamos HCV database, but it has not been formally published.
Isolates of subtype 3k were first reported in 1994 in more than 10 patients from Indonesia (Hotta et al., 1994; Okamoto et al., 1994) and subsequently in a study in 1995 in two patients from India (Valliammai et al., 1995). In 1996, it was proposed that these sequences be classified as subtype 10a (Tokita et al., 1996), only to be later reclassified into genotype 3 (Simmonds et al., 1996). In 2005 when the HCV nomenclature was updated, a consensus was made to reassign this subtype as 3k (Simmonds et al., 2005). Currently, in the Los Alamos HCV database a total of 66 sequences are classified as subtype 3k, representing 45 independent isolates, with the majority from Indonesia, followed by India, Pakistan and a few from other countries. For this subtype, a full-length genome sequence (JK049) has been characterized (Tokita et al., 1996) and in this study a second sequence (QC105) was determined from a Pakistani immigrant living in Canada (Murphy et al., 2007).
Subsequent to subtypes 3g, 3i and 3k, the sequences of an isolate (QC29) corresponding to subtype 3h was first described in 1996 in a Somali patient who had emigrated to Canada (Bernier et la., 1996). However, a formal designation was only proposed in 2000 following the identification of 3h sequences in three Somali refugees (Abid et al., 2000). In 2007, another subtype 3h sequence was identified in a Congolese emigrant in Canada (Murphy et al., 2007). In the Los Alamos HCV database, 3h sequences of two additional isolates (GenBank accession nos AY766911 and AY768137) have also been recorded. Although the submission was from the UK, the countries where the samples were collected are unknown. In the HCV database, two unclassified HCV-3 isolates, 52735 and 8332488, also belong to 3h. The former was from a Somali refugee in Denmark (Corbet et al., 2003), while the latter was from an immigrant in France originating from Djibouti, which neighbours Somalia (Colson et al., 2011). Due to the lack of a full-length genome in confirming the provisional designation of this subtype, the earliest identified 3h isolate, QC29, was selected for complete sequencing.
In this study, phylogenetic analysis demonstrated that the full-length genomes of three isolates, QC29, QC115 and QC260, were genetically distinct from each other and from subtypes 3a, 3b, 3i and 3k (Figs 1 and 2). Further analysis of nucleotide similarity curves along these genomes excluded the possibility of viral recombination. The results are consistent with the notion that each of these isolates represents three distinct subtypes within genotype 3. Since QC29 and QC260 have been provisionally classified into subtypes 3h and 3g (Murphy et al., 2007), respectively, the present study now confirms their designations for the first time by providing their full-length genomes. Phylogenetic analysis of the full-length genome sequence of QC115 reveals that it corresponds to a distinct subtype. However, we are not able to make a formal subtype assignment for this isolate, because based on the proposed criteria this requires the identification of at least three epidemiologically unlinked isolates (Simmonds et al., 2005).
In the Los Alamos HCV database, a total of 36 full-length genomes have been classified into HCV-3. The sampling geographical region is available for 33 of these sequences. These include 29 sequences of subtype 3a from Europe, Asia and America, and one sequence for each of subtypes 3b, 3i and 3k from Asia (accessed up until the end of December 2011). While 10 genotype 3 subtypes have been designated, these 36 full-length genomes only represent four subtypes. Although this study provides the full-length genomes of two additional subtypes (3g and 3h) and of one unclassified variant, this information is still lacking for subtypes 3c, 3d, 3e, 3f and 3l (which lacks a sequence to analyse in Fig. 2). Since this information constitutes a basic condition in achieving complete HCV nomenclature and is critical for HCV prevention and treatment strategies, one goal of our future studies will be to completely sequence isolates representing these subtypes.
We first reviewed the patient data that had genotype 3 nucleotide sequences previously reported from 1993 to 2005 (Murphy et al., 2007). Age, gender, sampling time and country of birth or ethnic origin were collected for 29 patients (Table S1, available in JGV Online). Five were selected for this study, for which full-length HCV genomes were determined using the approaches we have recently described (Li et al., 2012) and the primers listed in Table S2.
To reconstruct the ancestral relationship, a total of 21 full-length HCV sequences were retrieved from a HCV database and used as references. They not only represented genotype 3, but also genotypes 1, 2, 4, 5, 6 and 7 sequences (Kuiken et al., 2005). To further comprehend the genetic complexity within genotype 3, a set of NS5B sequences representing assigned subtypes and unclassified variants were also selected for analysis. The resulting dataset contained 64 partial NS5B sequences, each having 340 nt corresponding to nt 8276–8615 in the H77 genome. For both the sequence datasets, ML phylogenies were reconstructed and the recent virus recombination events were excluded as recently described (Li et al., 2012).
Acknowledgements
The study described was supported by a grant from NIAID/NIH (5 R01 AI080734-03A).
Footnotes
Two supplementary tables are available with the online version of this paper.
References
- Abid K., Quadri R., Veuthey A. L., Hadengue A., Negro F. (2000). A novel hepatitis C virus (HCV) subtype from Somalia and its classification into HCV clade 3. J Gen Virol 81, 1485–1493 [DOI] [PubMed] [Google Scholar]
- Bernier L., Willems B., Delage G., Murphy D. G. (1996). Identification of numerous hepatitis C virus genotypes in Montreal, Canada. J Clin Microbiol 34, 2815–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlière M., Halfon P., Portal I. (2002). [Treatment of chronic hepatitis C in special groups]. Gastroenterol Clin Biol 26 (Spec No 2), B238–B247 (in French). [PubMed] [Google Scholar]
- Colson P., Gayet S., Gerolami R. (2011). NS3 protease of genotype 3 subtype h HCV identified in southeastern France. Antivir Ther 16, 615–619 10.3851/IMP1765 [DOI] [PubMed] [Google Scholar]
- Corbet S., Bukh J., Heinsen A., Fomsgaard A. (2003). Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol 41, 1091–1100 10.1128/JCM.41.3.1091-1100.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene W. K., Cheong M. K., Ng V., Yap K. W. (1995). Prevalence of hepatitis C virus sequence variants in South-East Asia. J Gen Virol 76, 211–215 10.1099/0022-1317-76-1-211 [DOI] [PubMed] [Google Scholar]
- Hotta H., Handajani R., Lusida M. I., Soemarto W., Doi H., Miyajima H., Homma M. (1994). Subtype analysis of hepatitis C virus in Indonesia on the basis of NS5b region sequences. J Clin Microbiol 32, 3049–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C., Yusim K., Boykin L., Richardson R. (2005). The Los Alamos hepatitis C sequence database. Bioinformatics 21, 379–384 10.1093/bioinformatics/bth485 [DOI] [PubMed] [Google Scholar]
- Li C., Fu Y., Lu L., Ji W., Yu J., Hagedorn C. H., Zhang L. (2006). Complete genomic sequences for hepatitis C virus subtypes 6e and 6g isolated from Chinese patients with injection drug use and HIV-1 co-infection. J Med Virol 78, 1061–1069 10.1002/jmv.20663 [DOI] [PubMed] [Google Scholar]
- Li C., Lu L., Wu X., Wang C., Bennett P., Lu T., Murphy D. (2009a). Complete genomic sequences for hepatitis C virus subtypes 4b, 4c, 4d, 4g, 4k, 4l, 4m, 4n, 4o, 4p, 4q, 4r and 4t. J Gen Virol 90, 1820–1826 10.1099/vir.0.010330-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lu L., Zhang X., Murphy D. (2009b). Entire genome sequences of two new HCV subtypes, 6r and 6s, and characterization of unique HVR1 variation patterns within genotype 6. J Viral Hepat 16, 406–417 10.1111/j.1365-2893.2009.01086.x [DOI] [PubMed] [Google Scholar]
- Li C., Cao H., Lu L., Murphy D. (2012). Full-length sequences of 11 hepatitis C virus genotype 2 isolates representing five subtypes and six unclassified lineages with unique geographical distributions and genetic variation patterns. J Gen Virol 93, 1173–1184 10.1099/vir.0.038315-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S., Jha J. A., Shrotri S. P., Tandon B. N., Prasad V. G., Arankalle V. A. (2003). Comparison of hepatitis C virus genotyping by 5′ noncoding region- and core-based reverse transcriptase PCR assay with sequencing and use of the assay for determining subtype distribution in India. J Clin Microbiol 41, 5240–5244 10.1128/JCM.41.11.5240-5244.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Nakano T., Li C., Fu Y., Miller S., Kuiken C., Robertson B. H., Hagedorn C. H. (2006). Hepatitis C virus complete genome sequences identified from China representing subtypes 6k and 6n and a novel, as yet unassigned subtype within genotype 6. J Gen Virol 87, 629–634 10.1099/vir.0.81400-0 [DOI] [PubMed] [Google Scholar]
- Lu L., Li C., Fu Y., Gao F., Pybus O. G., Abe K., Okamoto H., Hagedorn C. H., Murphy D. (2007a). Complete genomes of hepatitis C virus (HCV) subtypes 6c, 6l, 6o, 6p and 6q: completion of a full panel of genomes for HCV genotype 6. J Gen Virol 88, 1519–1525 10.1099/vir.0.82820-0 [DOI] [PubMed] [Google Scholar]
- Lu L., Li C., Fu Y., Thaikruea L., Thongswat S., Maneekarn N., Apichartpiyakul C., Hotta H., Okamoto H. & other authors (2007b). Complete genomes for hepatitis C virus subtypes 6f, 6i, 6j and 6m: viral genetic diversity among Thai blood donors and infected spouses. J Gen Virol 88, 1505–1518 10.1099/vir.0.82604-0 [DOI] [PubMed] [Google Scholar]
- Lu L., Murphy D., Li C., Liu S., Xia X., Pham P. H., Jin Y., Hagedorn C. H., Abe K. (2008). Complete genomes of three subtype 6t isolates and analysis of many novel hepatitis C virus variants within genotype 6. J Gen Virol 89, 444–452 10.1099/vir.0.83460-0 [DOI] [PubMed] [Google Scholar]
- McCaw R., Moaven L., Locarnini S. A., Bowden D. S. (1997). Hepatitis C virus genotypes in Australia. J Viral Hepat 4, 351–357 10.1046/j.1365-2893.1997.00060.x [DOI] [PubMed] [Google Scholar]
- Murphy D. G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. (2007). Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol 45, 1102–1112 10.1128/JCM.02366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Sakamoto M., Iizuka H., Hadiwandowo S., Suwignyo S., Miyakawa Y., Mayumi M. (1994). The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol 75, 629–635 10.1099/0022-1317-75-3-629 [DOI] [PubMed] [Google Scholar]
- Panigrahi A. K., Roca J., Acharya S. K., Jameel S., Panda S. K. (1996). Genotype determination of hepatitis C virus from northern India: identification of a new subtype. J Med Virol 48, 191–198 [DOI] [PubMed] [Google Scholar]
- Pawlotsky J. M., Dhumeaux D., Bagot M. (1995). Hepatitis C virus in dermatology. A review. Arch Dermatol 131, 1185–1193 10.1001/archderm.1995.01690220091017 [DOI] [PubMed] [Google Scholar]
- Shrestha S. M., Tsuda F., Okamoto H., Tokita H., Horikita M., Tanaka T., Miyakawa Y., Mayumi M. (1994). Hepatitis B virus subtypes and hepatitis C virus genotypes in patients with chronic liver disease in Nepal. Hepatology 19, 805–809 10.1002/hep.1840190402 [DOI] [PubMed] [Google Scholar]
- Silini E., Bono F., Cividini A., Cerino A., Maccabruni A., Tinelli C., Bruno S., Bellobuono A., Mondelli M. (1995). Molecular epidemiology of hepatitis C virus infection among intravenous drug users. J Hepatol 22, 691–695 10.1016/0168-8278(95)80225-8 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Mellor J., Sakuldamrongpanich T., Nuchaprayoon C., Tanprasert S., Holmes E. C., Smith D. B. (1996). Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J Gen Virol 77, 3013–3024 10.1099/0022-1317-77-12-3013 [DOI] [PubMed] [Google Scholar]
- Simmonds P., Bukh J., Combet C., Deléage G., Enomoto N., Feinstone S., Halfon P., Inchauspé G., Kuiken C. & other authors (2005). Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973 10.1002/hep.20819 [DOI] [PubMed] [Google Scholar]
- Stuyver L., Wyseur A., van Arnhem W., Hernandez F., Maertens G. (1996). Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol 34, 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita H., Okamoto H., Tsuda F., Song P., Nakata S., Chosa T., Iizuka H., Mishiro S., Miyakawa Y., Mayumi M. (1994a). Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc Natl Acad Sci U S A 91, 11022–11026 10.1073/pnas.91.23.11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita H., Shrestha S. M., Okamoto H., Sakamoto M., Horikita M., Iizuka H., Shrestha S., Miyakawa Y., Mayumi M. (1994b). Hepatitis C virus variants from Nepal with novel genotypes and their classification into the third major group. J Gen Virol 75, 931–936 10.1099/0022-1317-75-4-931 [DOI] [PubMed] [Google Scholar]
- Tokita H., Okamoto H., Iizuka H., Kishimoto J., Tsuda F., Lesmana L. A., Miyakawa Y., Mayumi M. (1996). Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol 77, 293–301 10.1099/0022-1317-77-2-293 [DOI] [PubMed] [Google Scholar]
- Valliammai T., Thyagarajan S. P., Zuckerman A. J., Harrison T. J. (1995). Diversity of genotypes of hepatitis C virus in southern India. J Gen Virol 76, 711–716 10.1099/0022-1317-76-3-711 [DOI] [PubMed] [Google Scholar]
- van Doorn L. J., Kleter B., Stuyver L., Maertens G., Brouwer H., Schalm S., Heijtink R., Quint W. (1994). Analysis of hepatitis C virus genotypes by a line probe assay and correlation with antibody profiles. J Hepatol 21, 122–129 10.1016/S0168-8278(94)80148-7 [DOI] [PubMed] [Google Scholar]
- van Doorn L. J., Kleter G. E., Stuyver L., Maertens G., Brouwer J. T., Schalm S. W., Heijtink R. A., Quint W. G. (1995). Sequence analysis of hepatitis C virus genotypes 1 to 5 reveals multiple novel subtypes in the Benelux countries. J Gen Virol 76, 1871–1876 10.1099/0022-1317-76-7-1871 [DOI] [PubMed] [Google Scholar]
- Verma V., Chakravarti A. (2008). Comparison of 5′ noncoding-core with 5′ noncoding regions of HCV by RT-PCR: importance and clinical implications. Curr Microbiol 57, 206–211 10.1007/s00284-008-9175-z [DOI] [PubMed] [Google Scholar]
- Wang Y., Xia X., Li C., Maneekarn N., Xia W., Zhao W., Feng Y., Kung H. F., Fu Y., Lu L. (2009). A new HCV genotype 6 subtype designated 6v was confirmed with three complete genome sequences. J Clin Virol 44, 195–199 10.1016/j.jcv.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Xia X., Lu L., Tee K. K., Zhao W., Wu J., Yu J., Li X., Lin Y., Mukhtar M. M. & other authors (2008). The unique HCV genotype distribution and the discovery of a novel subtype 6u among IDUs co-infected with HIV-1 in Yunnan, China. J Med Virol 80, 1142–1152 10.1002/jmv.21204 [DOI] [PMC free article] [PubMed] [Google Scholar]