Abstract
Since the 1990s West Nile virus (WNV) has become an increasingly important public health problem and the cause of outbreaks of neurological disease. Genetic analyses have identified multiple lineages with many studies focusing on lineage 1 due to its emergence in New York in 1999 and its neuroinvasive phenotype. Until recently, viruses in lineage 2 were not thought to be of public health importance due to few outbreaks of disease being associated with viruses in this lineage. However, recent epidemics of lineage 2 in Europe (Greece and Italy) and Russia have shown the increasing importance of this lineage. There are very few genetic studies examining isolates belonging to lineage 2. We have sequenced the full-length genomes of four older lineage 2 WNV isolates, compared them to 12 previously published genomic sequences and examined the evolution of this lineage. Our studies show that this lineage has evolved over the past 300–400 years and appears to correlate with a change from mouse attenuated to virulent phenotype based on previous studies by our group. This evolution mirrors that which is seen in lineage 1 isolates, which have also evolved to a virulent phenotype over the same period of time.
Introduction
West Nile virus (WNV; Flaviviridae: Flavivirus) is a widely distributed mosquito-borne virus that can cause encephalitic disease in humans and other vertebrates. It is maintained in nature in a mosquito–bird cycle with Culex species mosquitoes being the primary vector. First isolated from a febrile woman in Uganda in 1937 (Smithburn et al., 1940), WNV caused sporadic outbreaks in humans and horses in parts of Africa, Australia and the Middle East with larger outbreaks in Israel (1951–52, 1957 and 1962), France (1962–65) and South Africa (1974 and 1983–84). In the 1990s, there was an increase in outbreaks of WNV with epidemics occurring in Algeria (1994 and 1997), Morocco (1996), Romania (1996), Tunisia (1997), Russia (1999), Israel (1998–2000) and France (2000) (Mackenzie et al., 2004; Murgue et al., 2001). These epidemics were associated with severe human and equine encephalitic disease. In 1999, WNV was first isolated in the Western hemisphere during an outbreak of encephalitis in New York. Since 2000, WNV rapidly spread across the United States and into Canada, the Caribbean, and Central and South America. Concurrently, there have been several severe epidemics of WNV in Israel, Russia, Tunisia, Hungary and most recently in Greece and Italy (Bagnarelli et al., 2011; Bakonyi et al., 2006; Papa, 2012; Platonov et al., 2011). Like the epidemics in North America, these involved large numbers of human, equine and avian cases with high incidence of severe encephalitic disease.
WNV is classified into at least five distinct lineages (Mackenzie & Williams, 2009). Lineage 1, the largest and most widespread, is found in Africa, Asia, the Middle East, Europe, Australia (Kunjin subtype) and the Americas (May et al., 2011; Scherret et al., 2001). Lineage 2 isolates are found primarily in sub-Saharan Africa and Madagascar with recent introductions into Europe (Greece, Hungary and Italy) and Russia (Bagnarelli et al., 2011; Bakonyi et al., 2006; Berthet et al., 1997; Papa et al., 2011a; Scherret et al., 2002). Lineage 3 is composed of a single isolate from the Rabensburg region of the Czech Republic (Bakonyi et al., 2005), while lineage 4 isolates are found in the Caucasus region of Russia (Lvov et al., 2004). Lineage 5 isolates have been found in India only (Bondre et al., 2007). Other putative lineages have been described, including a lineage based on a Koutango virus isolate from Africa, another lineage with isolates from Spain, and an additional lineage with the Kunjin virus strain from Sarawak, Malaysia (Mackenzie & Williams, 2009; Scherret et al., 2001, 2002; Vazquez et al., 2010).
While much recent work with WNV has focused primarily on lineage 1 isolates, particularly in the Americas, recent epidemics in Europe, primarily in Greece and Russia, due to lineage 2 isolates have shown that this lineage is of increasing public health importance (Papa et al., 2011b; Platonov et al., 2011). To investigate the evolution of lineage 2 we have determined the genomic sequences of four early lineage 2 isolates; three from Africa (Congo, South Africa and Madagascar) and one from the Mediterranean basin (Cyprus) to complement the published genomic sequences of recent isolates from Europe and phenotypic studies previously undertaken by our group (Beasley et al., 2002, 2004). These new sequence data together with our phenotypic studies have provided significant new insights into the evolution of WNV lineage 2.
Results
Sequence analyses
The full genome sequence of four lineage 2 isolates was determined in this study. Two isolates are from 1958 [Eyoku from Congo (CON58) and SA AN2842 from South Africa (SA58B)], one from 1968 [Q3574-5 from Cyprus (CYP68)] and one from 1988 [ArMg979 from Madagascar (MAD88)] (GenBank accession numbers: GQ903680, HM147822–HM147824). Details of these isolates are shown in Table 1. As of February 2012, there were a total of 18 published genomic sequences of lineage 2 WNV isolates, including the four isolates described here. The Sarafend and WNFC3 lineage 2 strains were omitted from these studies due to the uncertainty of passage history, isolation location and/or isolation date, leaving 16 lineage 2 strains analysed in these studies.
Table 1. Lineage 2 WNV isolates used in this study.
+, Present; –S, serine mutant (S→P); –Δ, deletion mutant; nd, not determined.
| Designation | Strain | Origin | Year | Source | CHO* | Neurovirulence/LD50 intraperitoneal† | GenBank accession no. |
| UGA37 | B956 | Uganda | 1937 | Human | –Δ | Mild‡§/7.9|| | AY532665 |
| CON58 | Eyoku | Congo | 1958 | Human | –S | 0.8|| | HM147824 |
| CYP68 | Q3574-5 | Cyprus | 1968 | Sylvia nisoria | +/−S | ≥10 000|| | GQ903680 |
| SA58A | SAH-442 | South Africa | 1958 | Human | + | High/3.2 ‡|| | EF429200 |
| SA58B | SAAn2842 | South Africa | 1958 | Sylvietta rufescens | + | 125|| | HM147822 |
| MAD78 | DakAnMg798 | Madagascar | 1978 | Coracopsis vasa | –S | ≥10 000‡|| | DQ176636 |
| CAR82 | DakArB3573/82 | Central African Republic | 1982 | Amblyomma variegatum | + | 0.8|| | DQ318200 |
| MAD88 | DakArMg-979 | Madagascar | 1988 | Culex quinquefasciatus | + | 500|| | HM147823 |
| SA89 | SPU116-89 | South Africa | 1989 | Human | + | 5‡|| | EF429197 |
| SEN90 | DakArD-76104 | Senegal | 1990 | Mimomyia lascustris | –Δ | 50|| | DQ318019 |
| SA00 | SA381/00 | South Africa | 2000 | Human | + | Mild‡ | EF429199 |
| SA01 | SA93/01 | South Africa | 2001 | Human | + | High‡ | EF429198 |
| HUN04 | Goshawk-Hungary/04 | Hungary | 2004 | Goshawk | + | nd | DQ116961 |
| RUSV07 | Reb_VLG_07_H | Russia: Volgograd | 2007 | Human | + | nd | FJ425721 |
| GRE10 | Nea Santa-Greece-2010 | Greece | 2010 | Culex pipiens | + | nd | HQ537483 |
| ITA11B | Italy/2011/AN-2 | Italy | 2011 | Human | + | nd | JN858070 |
Indicates presence or absence of potential N-linked glycosylation site at E154–156 (NYS).
Neuroinvasiveness in mice: high, virus strain is virulent (low LD50); mild, virus strain has a relatively low virulence.
Yamshchikov et al. (2004).
Beasley et al. (2004).
As with most lineage 2 isolates, the ORFs of the isolates in this study are 10 302 nt long (3434 aa) with the exception of CYP68, which has only 10 296 nt (3432 aa). SEN90 and UGA37 are the only two lineage 2 isolates that have fewer nucleotides (10 290 nt; 3430 aa). CYP68 has amino acid deletions at envelope (E)-200 and NS3-265, while SEN90 and UGA37 lack the amino acids at E-153–156, which results in loss of the E protein glycosylation site. Pairwise comparison shows that nucleotide divergence ranges from 0.20 to 16.90 %, while amino acids differ by 0.30–4.30 % (Table 2). CYP68 and MAD78 are the most divergent with nucleotide and amino acid sequence differences of 16.90 and 4.30 %, respectively, while SEN90 and UGA37 are the most similar with nucleotide and amino acid divergence of 0.20 and 0.3 %, respectively.
Table 2. Pairwise comparisons of sequence identities of lineage 2 WNV isolates.
| MAD78 | MAD88 | SA00 | RUSV07 | ITA11b | GRE10 | HUN04 | CON58 | SA01 | SA89 | UGA37 | SEN90 | CAR82 | SA58A | CYP68 | SA58B | |
| Amino acid | ||||||||||||||||
| MAD78 | 0.966 | 0.965 | 0.966 | 0.965 | 0.967 | 0.966 | 0.968 | 0.965 | 0.964 | 0.965 | 0.966 | 0.967 | 0.966 | 0.957 | 0.962 | |
| MAD88 | 0.835 | 0.988 | 0.989 | 0.987 | 0.988 | 0.988 | 0.989 | 0.987 | 0.986 | 0.987 | 0.988 | 0.988 | 0.988 | 0.973 | 0.981 | |
| SA00 | 0.839 | 0.931 | 0.993 | 0.991 | 0.992 | 0.992 | 0.994 | 0.991 | 0.991 | 0.991 | 0.992 | 0.994 | 0.993 | 0.973 | 0.981 | |
| RUSV07 | 0.840 | 0.934 | 0.963 | 0.993 | 0.993 | 0.993 | 0.994 | 0.993 | 0.992 | 0.992 | 0.993 | 0.995 | 0.994 | 0.973 | 0.982 | |
| ITA11b | 0.836 | 0.933 | 0.958 | 0.960 | 0.996 | 0.996 | 0.993 | 0.992 | 0.991 | 0.991 | 0.993 | 0.993 | 0.993 | 0.971 | 0.980 | |
| GRE10 | 0.838 | 0.933 | 0.958 | 0.960 | 0.995 | 0.997 | 0.994 | 0.993 | 0.992 | 0.992 | 0.994 | 0.994 | 0.993 | 0.972 | 0.981 | |
| HUN04 | 0.838 | 0.935 | 0.959 | 0.961 | 0.995 | 0.995 | 0.995 | 0.993 | 0.992 | 0.993 | 0.994 | 0.994 | 0.994 | 0.972 | 0.981 | |
| CON58 | 0.840 | 0.939 | 0.965 | 0.969 | 0.966 | 0.965 | 0.967 | 0.994 | 0.993 | 0.994 | 0.995 | 0.995 | 0.995 | 0.974 | 0.983 | |
| SA01 | 0.840 | 0.933 | 0.962 | 0.960 | 0.973 | 0.973 | 0.974 | 0.968 | 0.994 | 0.992 | 0.993 | 0.993 | 0.993 | 0.971 | 0.980 | |
| SA89 | 0.838 | 0.934 | 0.963 | 0.963 | 0.975 | 0.976 | 0.977 | 0.970 | 0.986 | 0.991 | 0.992 | 0.993 | 0.992 | 0.971 | 0.980 | |
| UGA37 | 0.839 | 0.940 | 0.968 | 0.972 | 0.975 | 0.975 | 0.976 | 0.979 | 0.976 | 0.979 | 0.997 | 0.993 | 0.993 | 0.971 | 0.980 | |
| SEN90 | 0.840 | 0.941 | 0.969 | 0.973 | 0.975 | 0.976 | 0.977 | 0.980 | 0.977 | 0.980 | 0.998 | 0.995 | 0.994 | 0.972 | 0.981 | |
| CAR82 | 0.842 | 0.935 | 0.969 | 0.971 | 0.962 | 0.962 | 0.963 | 0.972 | 0.964 | 0.966 | 0.974 | 0.975 | 0.995 | 0.973 | 0.983 | |
| SA58A | 0.841 | 0.939 | 0.975 | 0.976 | 0.967 | 0.967 | 0.968 | 0.979 | 0.969 | 0.972 | 0.982 | 0.983 | 0.986 | 0.972 | 0.982 | |
| CYP68 | 0.831 | 0.883 | 0.885 | 0.887 | 0.885 | 0.885 | 0.886 | 0.887 | 0.886 | 0.885 | 0.888 | 0.888 | 0.888 | 0.890 | 0.975 | |
| SA58B | 0.837 | 0.894 | 0.899 | 0.900 | 0.897 | 0.897 | 0.898 | 0.903 | 0.897 | 0.898 | 0.901 | 0.902 | 0.900 | 0.904 | 0.910 | |
| Nucleotide | ||||||||||||||||
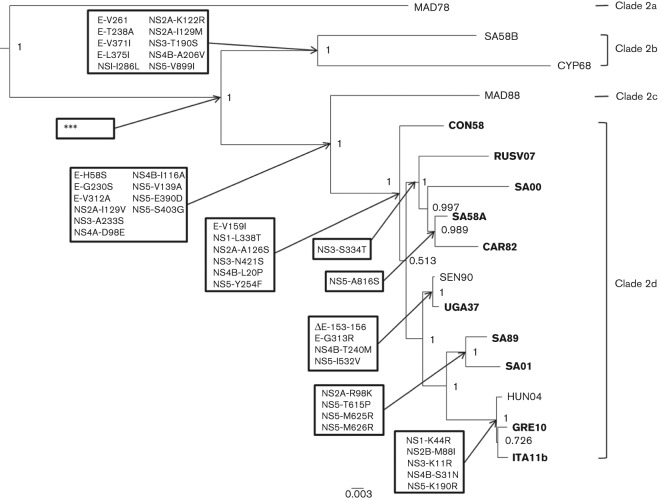
Five of the 16 lineage 2 isolates (MAD78, CYP68, CON58, UGA37 and SEN90) have a modified E protein glycosylation site (Table 1). The conserved sequence for the glycosylation site amongst all WNV strains is NYS at residues 154–156 in the E protein. CYP68, MAD78 and CON58 have the motif NYP, which ablates glycosylation, and UGA37 and SEN90 each have a deletion of four amino acids (E-153–156). All other lineage 2 viruses examined have the NYS motif at the E protein glycosylation site. Fig. 1 shows a Bayesian phylogenetic tree composed of lineage 2 isolates with the pertinent amino acid substitutions that define each node. The lineage 2 nodes are defined by one to ten amino acid substitutions distributed across the E and non-structural proteins. Several of these residues are exposed on the surface of the E protein including E-26, 153–156, 230, 312 and 313. The node between MAD78 and the other lineage 2 isolates is defined by 90 aa substitutions, illustrating the significant divergence of this strain.
Fig. 1.
Bayesian phylogenetic tree of lineage 2 WNV isolates. Posterior values and pertinent amino acid substitutions that define each node are shown. ***Amino acids defining the changes from MAD78 to remaining isolates were omitted from the figure due to the fact that there were 90 changes. Isolates in bold have been shown to be virulent in mice or were isolated from humans.
The 5′- and 3′-UTR’s of the newly sequenced isolates were also aligned and compared. There were no differences seen within the 5′-UTR of any lineage 2 strain. Within the 3′-UTR, there was a 14 nt insertion for both CYP68 and SA58B at position 10 441–10 454. This is a variable region in the 3′-UTR and contains a variety of deletions in different lineage 1 strains (data not shown). SA58A, SA58B, CYP68 and MAD88 also encoded different single nucleotide insertions within the 3′-UTR. Not all published sequences contained the full-length 5′- and 3′-UTR sequences so these regions were not included in the phylogenetic analyses.
Phylogenetic analyses
As of February 2012, there were over 350 complete WNV genome sequences published within GenBank with the majority of these sequences being highly related lineage 1 isolates from North America. The majority of these sequences were not included in these analyses since they would have little effect on the dating of the phylogenetic nodes of interest for this study. Therefore, 84 sequences were chosen to represent all lineages (lineages 1–5) and geographical regions where WNV has been isolated (Table S1, available in JGV Online). All methods of phylogenetic analyses [neighbour-joining (NJ), maximum-likelihood (ML) and Bayesian] used in these studies confirmed the placement of the isolates sequenced within lineage 2 as had been previously determined by partial sequencing studies (Beasley et al., 2002, 2004). With the addition of the new sequences, lineage 2 is composed of four clades (2a–2d) as determined by the genetic diversity between the strains (Fig. 1, Table 2). Clade 2a is composed of the most divergent isolate, MAD78. Clade 2b is composed of SA58b and CYP68, while clade 2c is composed of MAD88. Clade 2d is the largest and is composed of CON58, isolates from Russia (RUSV07), Africa (SA00, SA58A, CAR82, UGA37, SEN90, SA89 and SA01) and Europe (HUN04, ITA11b and GRE10).
Recombination detection and selection analyses
Recombination analyses performed using programs within the Datamonkey server (sbp and gard) and rdp3 identified no evidence of recombination within any of the lineage 2 isolates. Selection analyses using programs within the Datamonkey server (fel and rel) identified one site of potential positive selection within the lineage 2 isolates. Residue 312 in the E protein encodes a valine in MAD78, SA00, SA58B and CYP68 and an alanine in all other isolates. E-312 is a surface exposed amino acid in domain III of the E protein. E-L312A has been linked to subtle effects on neutralization by mAbs (Li et al., 2005). This is one of the significant changes identifying the node in Fig. 1, which includes clades 2c and d.
Evolutionary rates
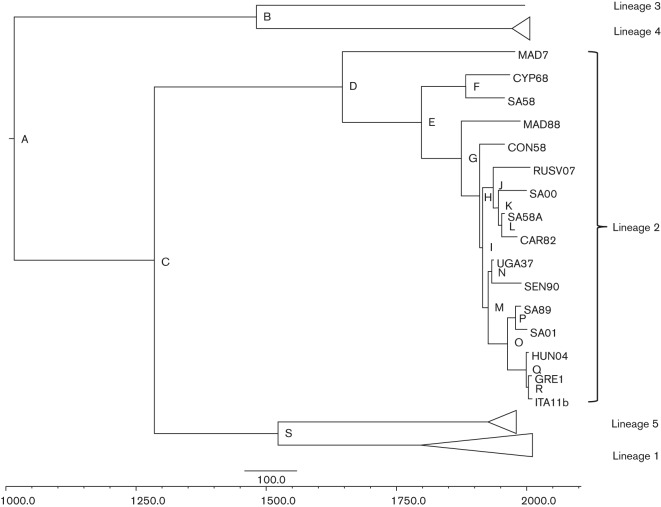
beast analyses were performed to determine the evolutionary rates and dates of the major nodes of the WNV lineages in addition to the nodes within lineage 2. The Bayesian maximum-clade credibility (MCC) tree produced confirms the placement of the WNV isolates within each lineage, clade and cluster as determined previously (Fig. 2) (Beasley et al., 2002; Botha et al., 2008; May et al., 2011). Posterior values for each node are shown in Table 3. Lower posterior values for the nodes for the most recent common ancestor (MRCA) for lineages 2, 3 and 4 are most likely low due to the relatively few numbers of available isolates for each of these lineages.
Fig. 2.
Bayesian MCC phylogenetic tree of all WNV lineages. Posterior values and divergence dates for each node are provided in Table 3.
Table 3. Posterior probabilities and divergence dates from the WNV MCC phylogenetic tree. Nodes correspond to those listed on Fig. 2.
| Node | Lineage | Divergence | Posterior probability | |
| Year | 95 % HPD | |||
| A | All | 1016 | 90–1524 | 1.0000 |
| B | 3.4 | 1481 | 966–1770 | 0.8261 |
| C | 1.2.5 | 1285 | 789–1600 | 0.4951 |
| D | 2 | 1646 | 1375–1815 | 0.9963 |
| E | 2 | 1798 | 1668–1881 | 0.9910 |
| F | 2 | 1883 | 1805–1932 | 0.9975 |
| G | 2 | 1875 | 1807–1915 | 0.9929 |
| H | 2 | 1910 | 1875–1930 | 0.9854 |
| I | 2 | 1915 | 1890–1931 | 0.6501 |
| J | 2 | 1936 | 1914–1951 | 0.9911 |
| K | 2 | 1946 | 1929–1955 | 0.9911 |
| L | 2 | 1952 | 1941–1957 | 0.9983 |
| M | 2 | 1926 | 1908–1935 | 0.9931 |
| N | 2 | 1933 | 1924–1936 | 0.9975 |
| O | 2 | 1963 | 1936–1981 | 0.9943 |
| P | 2 | 1979 | 1962–1987 | 0.9947 |
| Q | 2 | 1999 | 1986–2004 | 0.9979 |
| R | 2 | 2003 | 1995–2009 | 0.8942 |
| S | 1.5 | 1553 | 1163–1768 | 0.9625 |
The mean nucleotide substitution rate for all lineages of WNV is 3.74×10−4 substitutions per site per year [95 % high posterior density (HPD): 2.21×10−4 to 5.34×10−4] and the substitution rate of lineage 2 is 2.73×10−4 substitutions per site per year (95 % HPD: 5.23×10−5 to 5.44×10−4). These rates are comparable to previously published nucleotide substitution rates for WNV and other flaviviruses (Baillie et al., 2008; Bryant et al., 2007; Hanada et al., 2004; Jenkins et al., 2002; May et al., 2011).
Lineage 2 emerged from lineages 1 and 5 in approximately 1285AD (95 % HPD 789–1600AD), and lineage 5 diverged from lineage 1 in approximately 1553AD (95 % HPD 1163–1768AD). Nodes within lineage 1 were not determined as they have been described elsewhere (May et al., 2011) and the primary focus of this study was lineage 2. The divergence dates for the MRCA at the node of each of the eight clades of lineage 2 are shown in Table 3. Overall, the progenitor for lineage 2 occurred around 1646AD (95 % HPD 1375–1815AD), while the progenitor for the most recent isolates, ITA11b and GRE10, occurred very recently around 2003AD (95 % HPD 1995–2009AD).
Discussion
With the increasing importance of lineage 2 WNV in recent outbreaks and epidemics in Europe and Russia, our study provides a better understanding of the phylogenetic and evolutionary relationships of WNV lineage 2. We have sequenced the entire genome of four older WNV isolates (CON58, SA58b, CYP68 and MAD88) from Africa and Cyprus isolated between 1958 and 1988, and these are among the most divergent lineage 2 isolates sequenced thus far. As determined in previous studies, these isolates are naturally attenuated for mouse neuroinvasiveness with LD50 values ranging from 125 p.f.u. for SA58B to >10 000 p.f.u. for CYP68 (Beasley et al., 2002, 2004) (Table 1). These previous studies carried out in our laboratory and others (Botha et al., 2008) show that beginning with CON58, there is a change from an attenuated to virulent phenotype in terms of mouse neuroinvasiveness that is associated with genetic clade d and not with their chronological year of isolation (see Fig. 1). This modification in mouse neuroinvasive phenotype appears to coincide with the increase of outbreaks and epidemics associated with more recent isolates within lineage 2. The mouse neuroinvasiveness phenotypes of the most recent isolates (HUN04, GRE10 and ITA11b) have not been reported, but can be assumed to be virulent since the latter two have been isolated from humans during large outbreaks and epidemics within those regions (Bagnarelli et al., 2011; Papa et al., 2011a).
As compared to lineage 1, lineage 2 WNV isolates have a smaller geographical range, with isolates being found in Africa with recent introductions into Europe and the Volgograd region of Russia. Thus, the MRCA of WNV probably occurred in Africa and spread to Europe, Russia and the Middle East, most probably through migratory birds. Our phylogenetic analysis suggests that lineage 2 WNV emerged out of Africa on at least three separate occasions (Fig. 2 and Table 3). The first occurred during the 19th century/early 20th century (node F: 95 % HPD1805–1932) when the virus spread to an area, most likely in the Mediterranean region, that eventually led to virus isolation in Cyprus in 1968. The second and third occurrences were during the middle of the 20th century, when the virus emerged in south-western Russia (node J: 95 % HPD 1914–1951), and into Europe and was later isolated in recent outbreaks in Hungary, Italy and Greece (node O: 95 % HPD 1936–1981). While our studies suggest that WNV has emerged out of Africa on at least three separate occasions, it is possible that there is continual movement of WNV between the two regions each year, but has only been identified as the virus has become more virulent and/or entered into new regions. Clearly, a better understanding of how lineage 2 WNV has moved between Africa and Eurasia would be gained with more genomic sequences of isolates.
As the virus has spread and continued to evolve, it appears to have become more virulent leading to increased numbers of cases of avian, equine and human disease. This evolutionary pattern is similar to that being seen with lineage 1 isolates, which have also evolved from a naturally attenuated to virulent phenotype over the same 300–400 years. For lineage 2 isolates, the change from the attenuated to virulent phenotype, based on the mouse neuroinvasive phenotype, is characterized by six amino acid substitutions: E-V159I, NS1-L338T, NS2A-A126S, NS3-N421S, NS4B-L20P and NS5-Y254F (Fig. 1). At most of these positions, these amino acids vary greatly between the different lineages (Table 4). At E-159, an isoleucine is found in almost all lineage 1b isolates except for those in cluster 4, which includes the virulent North America isolates and their ancestors (TUN97, ISR98 and HUN03). These isolates have a valine at position E-159. Starting in 2002, an alanine is found at this position corresponding to the change from the NY99 to NA/WN02 genotype in North America (Beasley et al., 2003; Davis et al., 2005; Ebel et al., 2004). Lineage 2 clades a and b also contain a valine at this residue, while clades c and d contain an isoleucine. At position 338 in NS1, the attenuated clades of lineage 2 contain a leucine, which is also found in the attenuated lineage 3 and 4 isolates. Virulent lineage 2 strains (clade d) have a threonine at this position. NS3-S421 is a conserved residue in all lineages, except for attenuated lineage 2 clades a–c, which contain an asparagine. The NS5-Y254 substitution to phenylalanine is the only mutation found in virulent lineage 2 isolates. This position is located in the methyltransferase (MTase) domain of NS5 proximal to the RNA-binding site (Zhou et al., 2007). Mutating the residue to an alanine has been shown to suppress 2′-O methylation activity (Dong et al., 2008a, b). The substitution of a tyrosine to a phenylalanine results in the loss of a hydroxyl group. This change could potentially result in an increased activity of the MTase, which could aid in viral propagation, leading to the increase in virulence phenotype exhibited by the clades containing the mutation. Finally, one additional amino acid substitution to note is the substitution to a proline at NS3-249, which is found concurrently in the lineage 2 strain GRE10 (H249P) and in lineage 1a, cluster 4 isolates from the Middle East and the Americas (T249P). This substitution has been shown to lead to an increased virulence and pathogenesis in American crows and other birds (Brault et al., 2004, 2007; Papa, 2011a). Interestingly, the GRE10 strain was isolated during the largest lineage 2 epidemic recorded to date and phylogenetic analysis suggests that this isolate originated during the period 1995–2009 (Fig. 2 and Table 3, node R), which is approximately the same time period as when lineage 1a, cluster 4 originated (1990–1996) (May et al., 2011) with the substitution at NS3-249.
Table 4. Virulent lineage 2 WNV amino acid substitutions.
| Position | Lineage | ||||||
| 1a | 1b | 2, clades a–c (attenuated)* | 2, clade d (virulent)† | 3 | 4 | 5 | |
| E-159 | T | V/A/I | V | I | M | N | Q |
| NS1-338 | Q | Q | L | T | L | L | Q |
| NS2A-126 | L | L | A/L | S | G | N | Q |
| NS3-421 | S | S | N | S | S | S | S |
| NS4B-20 | I | I | L/S/V | P | P | T | L |
| NS5-254 | Y | Y | Y | F | Y | Y | Y |
Attenuated lineage 2, clades a–c WNV isolates: MAD78, CYP68, SA5B and MAD88.
Virulent lineage 2, clade d WNV isolates: CON58, RUSV07, SA00, SA58A, CAR82, UGA37, SEN90, SA89, SA01, HUN04, GRE10 and ITA11B.
WNV lineage 2 isolates pose a continuing threat to Europe and potentially other regions around the world. A better understanding of how these isolates have evolved can provide insights into the future evolution of this lineage, and allow us to better predict how these isolates will lead to future outbreaks and epidemics.
Methods
Virus strains.
Four WNV strains (Eyoku: Congo 1958, SA AN2842: South Africa 1958, Q3574-5: Cyprus 1968 and ArMg979: Madagascar 1988) were obtained from the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. Viruses were passaged once in Vero cells and stored at −80 °C. Properties for these viruses are shown in Table 1.
Genome sequencing.
Viral RNA was extracted directly from WNV-infected Vero cell culture supernatants by using the QIAamp viral RNA extraction kit (Qiagen Sciences), as per manufacturer’s instructions. PCR products encompassing the entire ORF of each isolate were obtained using primers designed on the published sequence of lineage 2 isolate B956 (GenBank accession no. AY532665). RT-PCR was performed with the Titan One-tube RT-PCR system (Roche) as per manufacturer’s instructions (primer sequences and PCR conditions available from authors on request). Briefly, RT-PCR was performed in a 50 µl volume containing 5 µl viral RNA, 1 µl of each primer, 10 µl of 5× RT buffer, 1 µl 10 mM dNTPs, 1 µl enzyme mix, 2.5 µl 100 mM DTT, 0.25 µl of inhibitor (10 U µl−1), and 28.25 µl HPLC water. PCR products were electrophoresed and analysed on 1 % agarose gels. DNA bands were purified by using the QIAquick gel extraction kit (Qiagen Sciences). PCR products were directly sequenced in both directions to generate consensus sequences at the University of Texas Medical Branch’s Protein Chemistry Core Laboratory. In some cases where RT-PCR amplification was insufficient, products were cloned into the pGEM-T (Easy) vector (Promega), and the cDNA clones for each were sequenced in both directions. Sequences were assembled and analysed using the Vector NTI suite of programs (Invitrogen) and SeaView v4.2 (Gouy et al., 2010).
Phylogenetic analyses.
Genomic sequences of 12 lineage 2 WNV isolates were downloaded from GenBank. The Sarafend and WNFC3 strains were not used for this study due to unavailability of published isolation dates. Additionally, genomic sequences of 83 isolates representing the remaining lineages were also downloaded. Properties for the strains used for the phylogenetic analyses are listed in Table 3. Nucleotide sequences were aligned and edited using the muscle algorithm in Seaview v4.2 (Gouy et al., 2010) keeping gaps consistent within the reading frame. NJ and ML trees were constructed using phylip (Felsenstein, 1989) and PhyML (Guindon & Gascuel, 2003), respectively, while a Bayesian tree was constructed using beast v1.6.1 package (Drummond & Rambaut, 2007). To determine the robustness of the trees, 100 bootstrap replicates were performed. Evolutionary rates and times to MRCA for all of WNV and for lineage 2 individually were estimated using the Bayesian Markov Chain Monte Carlo (MCMC) method in beast. The GTR+I+Γ6 model was used with a relaxed uncorrelated exponential molecular clock and the exponential model. The Bayes factors for all population models available in the beast package were compared to choose the best-fit model. The MCMC analysis was run for 330 million chains (11 runs of 30 million each) to attain convergence with 10 % of the runs discarded as ‘burn-in’. Using TreeAnnotator, within the beast package, an MCC tree was created. beast runs were performed using the cipres Science Gateway (Miller et al., 2010).
Recombination and selection analyses.
Recombination analyses were performed to determine the presence of recombination within the nucleotide sequence alignment. Using the RDP3 vα44 program (Martin et al., 2010), analyses were performed using the rdp, geneconv, Chimaera, MaxChi and Bootscan methods. Additional recombination analyses were performed using the sbp and gard methods in the Datamonkey server (Kosakovsky Pond et al., 2006; Pond & Frost, 2005). The Datamonkey server was also used for selection analyses. Positive selection was determined using the alignment containing 16 lineage 2 isolates and the fel and rel methods (Kosakovsky Pond & Frost, 2005). Sites were considered positive when dN/dS >1 with a P-value <0.1.
Acknowledgements
This work was supported in part by NIH grant AI 067847 (to A. D. T. B.) and contract HHSN272201000040I/HHSN27200004/D04. A. R. M. is supported by NIH T32 training grant AI 07526. A special thanks to Amy Schuh and Andrew Beck for their help with the beast phylogenetic analyses.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Bagnarelli P., Marinelli K., Trotta D., Monachetti A., Tavio M., Del Gobbo R., Capobianchi M., Menzo S., Nicoletti L. & other authors (2011). Human case of autochthonous West Nile virus lineage 2 infection in Italy, September 2011. Euro Surveill 16, 20002. [PubMed] [Google Scholar]
- Baillie G. J., Kolokotronis S. O., Waltari E., Maffei J. G., Kramer L. D., Perkins S. L. (2008). Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Mol Phylogenet Evol 47, 717–728 10.1016/j.ympev.2008.02.015 [DOI] [PubMed] [Google Scholar]
- Bakonyi T., Hubálek Z., Rudolf I., Nowotny N. (2005). Novel flavivirus or new lineage of West Nile virus, central Europe. Emerg Infect Dis 11, 225–231 10.3201/eid1102.041028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakonyi T., Ivanics E., Erdélyi K., Ursu K., Ferenczi E., Weissenböck H., Nowotny N. (2006). Lineage 1 and 2 strains of encephalitic West Nile virus, central Europe. Emerg Infect Dis 12, 618–623 10.3201/eid1204.051379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley D. W., Li L., Suderman M. T., Barrett A. D. (2002). Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296, 17–23 10.1006/viro.2002.1372 [DOI] [PubMed] [Google Scholar]
- Beasley D. W., Davis C. T., Guzman H., Vanlandingham D. L., Travassos da Rosa A. P., Parsons R. E., Higgs S., Tesh R. B., Barrett A. D. (2003). Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology 309, 190–195 10.1016/S0042-6822(03)00150-8 [DOI] [PubMed] [Google Scholar]
- Beasley D. W., Davis C. T., Whiteman M., Granwehr B., Kinney R. M., Barrett A. D. (2004). Molecular determinants of virulence of West Nile virus in North America. Arch Virol Suppl 18, 35–41 [DOI] [PubMed] [Google Scholar]
- Berthet F. X., Zeller H. G., Drouet M. T., Rauzier J., Digoutte J. P., Deubel V. (1997). Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J Gen Virol 78, 2293–2297 [DOI] [PubMed] [Google Scholar]
- Bondre V. P., Jadi R. S., Mishra A. C., Yergolkar P. N., Arankalle V. A. (2007). West Nile virus isolates from India: evidence for a distinct genetic lineage. J Gen Virol 88, 875–884 10.1099/vir.0.82403-0 [DOI] [PubMed] [Google Scholar]
- Botha E. M., Markotter W., Wolfaardt M., Paweska J. T., Swanepoel R., Palacios G., Nel L. H., Venter M. (2008). Genetic determinants of virulence in pathogenic lineage 2 West Nile virus strains. Emerg Infect Dis 14, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Langevin S. A., Bowen R. A., Panella N. A., Biggerstaff B. J., Miller B. R., Komar N. (2004). Differential virulence of West Nile strains for American crows. Emerg Infect Dis 10, 2161–2168 10.3201/eid1012.040486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Huang C. Y., Langevin S. A., Kinney R. M., Bowen R. A., Ramey W. N., Panella N. A., Holmes E. C., Powers A. M., Miller B. R. (2007). A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet 39, 1162–1166 10.1038/ng2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J. E., Holmes E. C., Barrett A. D. (2007). Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog 3, e75. 10.1371/journal.ppat.0030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. T., Ebel G. D., Lanciotti R. S., Brault A. C., Guzman H., Siirin M., Lambert A., Parsons R. E., Beasley D. W. & other authors (2005). Phylogenetic analysis of North American West Nile virus isolates, 2001-2004: evidence for the emergence of a dominant genotype. Virology 342, 252–265 10.1016/j.virol.2005.07.022 [DOI] [PubMed] [Google Scholar]
- Dong H., Ren S., Zhang B., Zhou Y., Puig-Basagoiti F., Li H., Shi P.-Y. (2008a). West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol 82, 4295–4307 10.1128/JVI.02202-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Zhang B., Shi P.-Y. (2008b). Flavivirus methyltransferase: a novel antiviral target. Antiviral Res 80, 1–10 10.1016/j.antiviral.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel G. D., Carricaburu J., Young D., Bernard K. A., Kramer L. D. (2004). Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am J Trop Med Hyg 71, 493–500 [PubMed] [Google Scholar]
- Felsenstein J. (1989). phylip – phylogeny inference package (version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27, 221–224 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hanada K., Suzuki Y., Gojobori T. (2004). A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol 21, 1074–1080 10.1093/molbev/msh109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. M., Rambaut A., Pybus O. G., Holmes E. C. (2002). Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol 54, 156–165 10.1007/s00239-001-0064-3 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Frost S. D. W. (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22, 1208–1222 10.1093/molbev/msi105 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. W. (2006). Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23, 1891–1901 10.1093/molbev/msl051 [DOI] [PubMed] [Google Scholar]
- Li L., Barrett A. D. T., Beasley D. W. C. (2005). Differential expression of domain III neutralizing epitopes on the envelope proteins of West Nile virus strains. Virology 335, 99–105 10.1016/j.virol.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Lvov D. K., Butenko A. M., Gromashevsky V. L., Kovtunov A. I., Prilipov A. G., Kinney R., Aristova V. A., Dzharkenov A. F., Samokhvalov E. I. & other authors (2004). West Nile virus and other zoonotic viruses in Russia: examples of emerging-reemerging situations. Arch Virol Suppl 18, 85–96 [DOI] [PubMed] [Google Scholar]
- Mackenzie J. S., Williams D. T. (2009). The zoonotic flaviviruses of southern, south-eastern and eastern Asia, and Australasia: the potential for emergent viruses. Zoonoses Public Health 56, 338–356 10.1111/j.1863-2378.2008.01208.x [DOI] [PubMed] [Google Scholar]
- Mackenzie J. S., Gubler D. J., Petersen L. R. (2004). Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med 10 (Suppl.), S98–S109 10.1038/nm1144 [DOI] [PubMed] [Google Scholar]
- Martin D. P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. (2010). rdp3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463 10.1093/bioinformatics/btq467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May F. J., Davis C. T., Tesh R. B., Barrett A. D. T. (2011). Phylogeography of West Nile virus: from the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J Virol 85, 2964–2974 10.1128/JVI.01963-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the cipres Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1–8. New Orleans, LA. [Google Scholar]
- Murgue B., Murri S., Triki H., Deubel V., Zeller H. G. (2001). West Nile in the Mediterranean basin: 1950-2000. Ann N Y Acad Sci 951, 117–126 10.1111/j.1749-6632.2001.tb02690.x [DOI] [PubMed] [Google Scholar]
- Papa A. (2012). West Nile infections in Greece: an update. Expert Reviews of Anti-Inpective Therapy 10, 743–750 [DOI] [PubMed] [Google Scholar]
- Papa A., Bakonyi T., Xanthopoulou K., Vázquez A., Tenorio A., Nowotny N. (2011a). Genetic characterization of West Nile virus lineage 2, Greece, 2010. Emerg Infect Dis 17, 920–922 10.3201/eid1705.101759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A., Xanthopoulou K., Gewehr S., Mourelatos S. (2011b). Detection of West Nile virus lineage 2 in mosquitoes during a human outbreak in Greece. Clin Microbiol Infect 17, 1176–1180 [DOI] [PubMed] [Google Scholar]
- Platonov A. E., Karan’ L. S., Shopenskaia T. A., Fedorova M. V., Koliasnikova N. M., Rusakova N. M., Shishkina L. V., Arshba T. E., Zhuravlev V. I. & other authors (2011). [Genotyping of West Nile fever virus strains circulating in southern Russia as an epidemiological investigation method: principles and results]. Zh Mikrobiol Epidemiol Immunobiol 2, 29–37 [PubMed] [Google Scholar]
- Pond S. L. K., Frost S. D. W. (2005). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 10.1093/bioinformatics/bti320 [DOI] [PubMed] [Google Scholar]
- Scherret J. H., Poidinger M., Mackenzie J. S., Broom A. K., Deubel V., Lipkin W. I., Briese T., Gould E. A., Hall R. A. (2001). The relationships between West Nile and Kunjin viruses. Emerg Infect Dis 7, 697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherret J. H., Mackenzie J. S., Hall R. A., Deubel V., Gould E. A. (2002). Phylogeny and molecular epidemiology of West Nile and Kunjin viruses. Curr Top Microbiol Immunol 267, 373–390 10.1007/978-3-642-59403-8_18 [DOI] [PubMed] [Google Scholar]
- Smithburn K. C., Hughes T. P., Burke A. W., Paul J. H. (1940). A neurotropic virus isolated from the blood of a native of Uganda. Am J Trop Med 20, 471–492 [Google Scholar]
- Vazquez A., Sanchez-Seco M. P., Ruiz S., Molero F., Hernandez L., Moreno J., Magallanes A., Tejedor C. G., Tenorio A. (2010). Putative new lineage of West Nile virus, Spain. Emerg Infect Dis 16, 549–552 10.3201/eid1603.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamshchikov G., Borisevich V., Seregin A., Chaporgina E., Mishina M., Mishin V., Kwok C. W., Yamshchikov V. (2004). An attenuated West Nile prototype virus is highly immunogenic and protects against the deadly NY99 strain: a candidate for live WN vaccine development. Virology 330, 304–312 10.3201/eid1603.091033 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K. A., Shi P. Y., Li H. (2007). Structure and function of flavivirus NS5 methyltransferase. J Virol 81, 3891–3903 10.1128/JVI.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]