Key Points
PF4 binds to nucleic acids and thereby exposes the epitope to which anti-PF4/heparin antibodies bind.
PF4/aptamer complexes can induce an immune response resembling heparin-induced thrombocytopenia.
Abstract
The tight electrostatic binding of the chemokine platelet factor 4 (PF4) to polyanions induces heparin-induced thrombocytopenia, a prothrombotic adverse drug reaction caused by immunoglobulin G directed against PF4/polyanion complexes. This study demonstrates that nucleic acids, including aptamers, also bind to PF4 and enhance PF4 binding to platelets. Systematic assessment of RNA and DNA constructs, as well as 4 aptamers of different lengths and secondary structures, revealed that increasing length and double-stranded segments of nucleic acids augment complex formation with PF4, while single nucleotides or single-stranded polyA or polyC constructs do not. Aptamers were shown by circular dichroism spectroscopy to induce structural changes in PF4 that resemble those induced by heparin. Moreover, heparin-induced anti-human–PF4/heparin antibodies cross-reacted with human PF4/nucleic acid and PF4/aptamer complexes, as shown by an enzyme immunoassay and a functional platelet activation assay. Finally, administration of PF4/44mer–DNA protein C aptamer complexes in mice induced anti–PF4/aptamer antibodies, which cross-reacted with murine PF4/heparin complexes. These data indicate that the formation of anti-PF4/heparin antibodies in postoperative patients may be augmented by PF4/nucleic acid complexes. Moreover, administration of therapeutic aptamers has the potential to induce anti-PF4/polyanion antibodies and a prothrombotic diathesis.
Introduction
The chemokine platelet factor 4 (PF4) is released from platelet α-granules during platelet activation1 and binds, due to its high positive charge, to many negatively charged polyanions, including heparin. PF4 forms large multimolecular complexes with heparin that are highly immunogenic.2,3 The resulting immunoglobulin G (IgG) antibodies are the cause of heparin-induced thrombocytopenia (HIT), a prothrombotic adverse drug effect.4 In HIT, multimolecular complexes composed of PF4, heparin, and anti-PF4/heparin IgG cross-link platelet FcγIIa receptors,5 triggering platelet activation, microparticle formation, and thrombin generation, with ∼50% of affected patients developing thrombosis.6
Recently, we showed that PF4 binds to polyanions on the surface of bacteria, thereby forming multimolecular complexes that are recognized by human anti-PF4/heparin antibodies. More specifically, we identified the PF4 binding site on gram-negative bacteria as the phosphate groups of lipid A.7 Based on this observation, we hypothesized that nucleic acids might also form multimolecular complexes with PF4 because they also expose multiple negatively charged phosphate groups. This idea was fostered by previous findings that salmon sperm DNA could substitute heparin in HIT-IgG–induced platelet activation.8
Plasma levels of extracellular nucleic acids are regulated by prompt nuclease degradation and renal clearance, resulting in their short half-life (4-30 minutes),9 with a slightly longer half-life for DNA than for RNA. In healthy individuals, extracellular nucleic acid concentrations in plasma range from 0 to >1000 ng/mL.10 However, under pathological conditions, cell-free nucleic acids can be generated from the breakdown of bacteria and viruses, tissue damage, cell apoptosis, and the release from blood cells such as the formation of neutrophil extracellular traps. Plasma levels of extracellular nucleic acids can rise up to 2000 µg/mL.11,12
With the recent introduction of aptamers, small DNA/RNA constructs designed as therapeutic oligonucleotides,13 the interaction of nucleic acids and PF4 becomes more relevant from a drug safety perspective. In this study, we characterized the structural features of RNA and DNA molecules, as well as aptamers, that are relevant for complex formation with PF4. We also demonstrated by circular dichroism (CD) spectroscopy that nucleic acids cause conformational changes in PF4 similar to those induced by heparin, and we showed that PF4/aptamer complexes can be highly immunogenic.
Methods
Platelets and sera
Platelet-rich plasma was prepared from 10 mL hirudinized whole blood (10 µg/mL lepirudin; Pharmion, Hamburg, Germany) obtained from healthy blood donors (120 g, 20 minutes, 30°C). Platelets were isolated by gel filtration as described in Krauel et al14 and adjusted to 50 × 109/L in Tyrode buffer (137 mM sodium chloride, 2.7 mM potassium chloride, 2 mM magnesium chloride × 6 water, 2 mM calcium chloride × 2 water, 12 mM sodium bicarbonate, 0.4 mM sodium phosphate monobasic, 0.4% bovine serum albumin [BSA], and 0.1% glucose, pH 7.2). Sera containing anti-PF4/heparin antibodies were leftovers from clinical specimens used for laboratory diagnosis of HIT.
Nucleic acids and aptamers
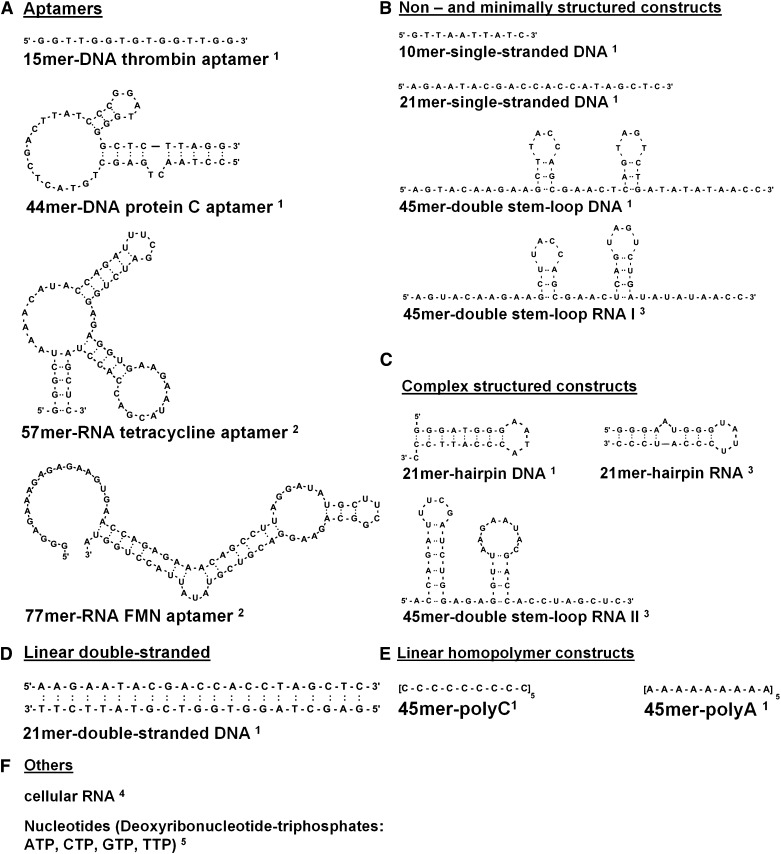
Sequences and sources of aptamers and nucleic acid compounds are given in Figure 1. Secondary structures were calculated by the mfold RNA and mfold DNA database of the University of Albany (NY, http://mfold.rna.albany.edu/?q=mfold). The single-stranded 15mer–DNA thrombin aptamer targets thrombin15; the 44mer–DNA protein C aptamer inhibits activated protein C16; the 57mer–RNA tetracycline aptamer binds tetracycline17; and the 77mer–RNA flavin mononucleotide (FMN) aptamer is an artificial FMN-responsive catalytic RNA.18 Cellular RNA and DNA were isolated from mouse vascular smooth muscle cells using an extraction kit (Sigma, Munich, Germany) or the DNAzol reagent (Invitrogen, Groningen, The Netherlands).
Figure 1.
The structures of aptamers, DNA and RNA constructs, and nucleotides are shown. Indicated compounds used in this study are1 (A-E) chemically synthesized by a commercial supplier (PURIMEX, Grebenstein, Germany),2 (A) enzymatically transcribed in vitro from oligonucleotide templates using T7 RNA polymerase,3 (B and C) chemically synthesized in house using the phosphoramidite method (Gene Assembler Special, Pharmacia Biotech, Freiburg, Germany),4 (F) isolated from mouse vascular smooth muscle cells using an extraction kit (Sigma, Munich, Germany) or the DNAzol reagent (Invitrogen, Groningen, The Netherlands),5 or (F) purchased from Fermentas (St. Leon-Rot, Germany).
Binding of biotinylated RNA to PF4
Microtiter plates were coated with 50-µL solutions of PF4 or BSA (each at 10 µg/mL) in 100 mM sodium carbonate (pH 9.5) at 4°C for 20 hours, then washed and blocked (Tris-buffered saline, 3% BSA; 2 hours). Different concentrations of biotinylated RNA (0.78-25 µg/mL), prepared from isolated cellular RNA using the Psoralen-PEO-Biotin reagent (Thermo Fisher Scientific [Pierce], Rockford, NY), were allowed to bind at 22°C for 2 hours, followed by 3 washes with Tris-buffered saline. Bound biotinylated RNA was detected using peroxidase-conjugated streptavidin (Dako, Glostrup, Denmark) and the immunopure 3,3′,5,5′-tetramethylbenzidine substrate kit (Pierce). In competition experiments, increasing concentrations of biotinylated RNA were mixed with unlabeled RNA, DNA, or heparin (each at 300 µg/mL) before binding assays were performed. To confirm that PF4 was not removed from the plate upon heparin addition, surface-coated PF4 was quantified by dot blot analysis and densitometry.
Influence of nucleic acids on PF4 binding to platelets
The impact of nucleic acids on the binding of human platelet–derived PF4 (25 µg/mL) (ChromaTec, Greifswald, Germany) to gel-filtered platelets (50 000/µL) was assessed by flow cytometry (Cytomics FC 500; Beckman Coulter, Krefeld, Germany) using a polyclonal fluorescein isothiocyanate–labeled rabbit anti-human PF4 antibody (Dianova, Marl, Germany),14 in the absence or presence of unfractionated heparin (UFH; activity ∼150 IU/mg; Braun, Melsungen, Germany) or different nucleic acid compounds (0.08-640 µg/mL; Figure 1). In parallel, RNA samples were treated with ribonuclease A (RNase A) (100 µg/mL, 90 minutes, 37°C) (Fermentas, St. Leon-Rot, Germany) before addition to the respective test solution to document RNA dependency. Antibody binding was quantified by geometric mean fluorescent intensity.
PF4/nucleic acid EIA
Binding of human anti-PF4/heparin IgG to PF4/nucleic acid complexes was assessed by enzyme immunoassay (EIA)19 using microtiter plates (Nunc, Langenselbold, Germany) coated (16 hours; 4°C) with preformed PF4/polyanion complexes: PF4 (20 µg/mL) preincubated (1 hour, room temperature) with either UFH as the positive control (3.33 µg/mL), 15mer–DNA thrombin aptamer, 44mer–DNA protein C aptamer, 57mer–RNA tetracycline aptamer, or 77mer–RNA FMN aptamer in concentrations ranging from 1.25 to 80 µg/mL. To find the optimal stoichiometric ratios for complex formation, we first titrated selected aptamers in the presence of a constant PF4 concentration in the EIA. As the control for specific antibody binding, UFH (660 µg/mL) was added to disrupt PF4/polyanion complexes and to inhibit antibody binding.
Platelet activation assay
Platelet activation by human anti-PF4/polyanion antibodies was assessed by the heparin-induced platelet activation assay, as described in Warkentin and Greinacher,20 using 0.2 µg/mL heparin (positive control: reviparin; Abbott GmbH & Co KG, Wiesbaden, Germany), deoxynucleotidetriphosphates (0.05-1000 µg/mL), 21mer–double-stranded DNA (0.05-1000 µg/mL), 21mer–hairpin DNA (0.05-1000 µg/mL), cellular RNA (0.05-5 µg/mL), 15mer–DNA thrombin aptamer (0.5-40 µg/mL), 44mer–DNA protein C aptamer (0.05-1000 µg/mL), 57mer–RNA tetracycline aptamer (0.5-5 µg/mL), and 77mer–RNA FMN-ribozyme (0.5-5 µg/mL). To find the optimal concentrations for platelet activation, titration with the respective nucleic acid constructs in the heparin-induced platelet activation test with 1 or 2 sera was performed, and then additional sera were assessed using this concentration. Specificity of platelet activation was confirmed by its inhibition using high concentrations of heparin (660 µg/mL), which disrupts PF4/polyanion complexes,21 as well as the monoclonal antibody IV.3 (2.5 µg/mL; LGC Promochem, Wesel, Germany), which blocks FcγIIa receptor–mediated platelet activation.22
CD spectroscopy of PF4/polyanion complexes
Conformational changes of PF4 (40 µg/mL) were measured by far-UV CD spectroscopy (200-260 nm; Chirascan CD spectrometer; Applied Photophysics, Leatherhead, United Kingdom) at increasing concentrations of heparin (6.66-73.33 µg/mL), or 44mer–DNA protein C aptamer (0.31-40 µg/mL) in a 101-QS quartz precision cell (path length: 10 mm) (Hellma, Muellheim, Germany). Baseline spectra of phosphate-buffered saline, heparin, and 44mer–DNA protein C aptamer were subtracted to estimate secondary structures of the respective PF4/heparin or PF4/aptamer complexes (CDNN CD Spectra Deconvolution Software).23
Immunization of mice by PF4/aptamer complexes
C57BL/6 mice (8-10 weeks of age; Charles River Laboratories Europe, Kisslegg, Germany) were anesthetized (2% xylazine/10% ketamine) and immunized as described in Suvarna et al24 with recombinant murine PF4 (mPF4) (ChromaTec, Greifswald, Germany), 44mer–DNA protein C aptamer, mPF4/44mer–DNA protein C aptamer complexes (each at 200 µg/mL), or complexes of mPF4 (200 µg/mL) and heparin (33 µg/mL) in a final volume of 100 µL via retroorbital injection daily for 5 days. After another 10 days, blood was withdrawn and serum was analyzed for anti-mPF4/aptamer or anti-mPF4/heparin antibodies, respectively, by EIA using mPF4/heparin and mPF4/44mer–DNA protein C aptamer complexes as antigens.
Ethics
All blood donors gave informed consent. The use of antibodies from leftovers of clinical diagnostic material was approved by the ethical board of Greifswald University. The animal studies were performed and approved according to the regulations of Greifswald University.
Statistical analysis
Binding of biotinylated RNA to PF4 in the presence of competing polyanions was compared using paired samples Student t test. The significance of differences in the impact on PF4 binding to platelets between nucleic acids was tested by analysis of variance and paired samples Student t test (RNase experiment). Reduction of antibody binding in the PF4/polyanion immunoassays after the heparin inhibition step was assessed by paired samples Student t test. Murine immune responses to mPF4/44mer–DNA protein C aptamer complexes and mPF4 were compared by Wilcoxon 2-sample test. The significance level was .05.
Results
RNA binds to PF4
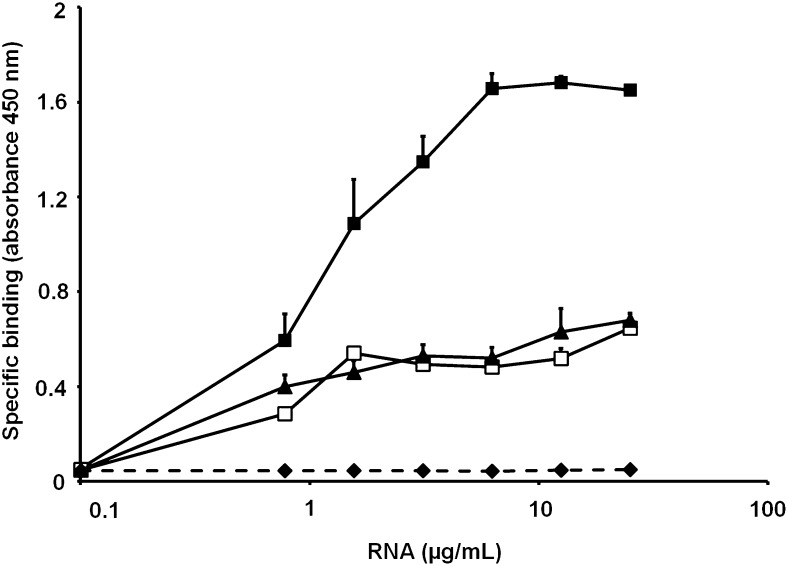
The binding of RNA to PF4 was assessed using biotinylated RNA, which bound to PF4 in a saturable manner, reaching a maximum at 6.25 µg/mL (Figure 2). High concentrations of unlabeled RNA, DNA, or heparin decreased the binding of biotinylated RNA to PF4, with heparin showing the strongest inhibitory effect, followed by RNA and DNA. This indicates charge-dependent binding of RNA to PF4.
Figure 2.
The graph shows the binding of biotinylated RNA to PF4 and the influence of competitors. The binding of increasing doses of biotinylated RNA to surface-coated PF4 was performed in the absence (▪) or presence of excess DNA (▲), nonbiotinylated RNA (□), or heparin (♦). Values represent mean ± standard deviation (SD) of 3 independent experiments.
RNA and DNA enhance PF4 binding to platelets
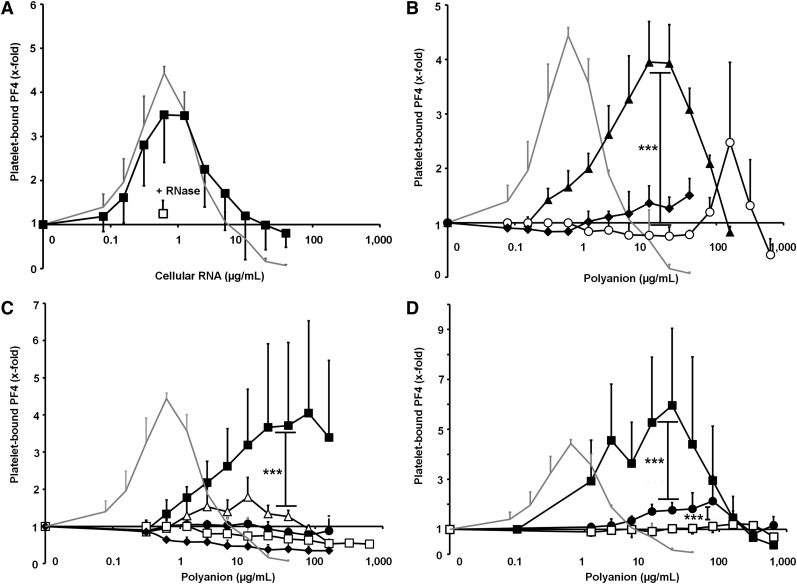
Next we assessed the interaction of PF4 and different nucleic acid constructs with gel-filtered platelets by flow cytometry, as previously described in Krauel et al14 for PF4-polysaccharide-platelet interactions. To analyze the structure-function relationship, RNA and DNA constructs of different length and structural complexity were tested. Figure 3 shows the influence of these constructs on PF4 binding to the platelet surface, whereby baseline binding was defined as PF4 binding to platelets in the absence of nucleic acids. Heparin, the prototype compound for PF4-polyanion interaction, increased PF4 binding to platelets in a dose-dependent manner, represented by a bell-shaped curve with a maximal 4.44-fold increase (±0.15, shown as the gray curve in Figure 3A-D).
Figure 3.
The influence of differently structured nucleic acid compounds on PF4 binding to platelets is shown. (For details of the nucleic acid constructs, please compare with Figure 1B-F.) Enhancement of PF4 binding is expressed as the x-fold increase compared with PF4 binding to platelets in buffer only. Heparin (gray line) was used as the positive control. (A) PF4 binding to platelets was performed in the presence of increasing doses of cellular RNA (▪). Pretreatment with RNase A (□) significantly reduced PF4 binding to platelets (P = .0037). (B) PF4 binding to platelets was evaluated in the presence of increasing doses of nucleic acids with different lengths: 45mer–double-stem-loop RNA II (▲), 21mer–hairpin RNA (○), and 10mer–single-stranded DNA (♦). (C) PF4 binding to platelets was performed with increasing doses of 45mer–nucleic acid compounds, comprising different structures: 45mer–double-stem-loop RNA I (▪), 45mer–double stem-loop DNA (△), 45mer–polyA (●), 45mer–polyC (♦), and single nucleotides (□). (D) PF4 binding to platelets was measured in the presence of increasing doses of double-stranded vs single-stranded 21mer–DNA compounds: 21mer–double-stranded DNA (▪), 21mer–hairpin DNA (●), and 21mer–single-stranded DNA (□). Enhancement of PF4 binding is expressed as the x-fold increase of PF4 binding compared with platelets in buffer only. Negative values indicate detachment of PF4 from the platelet surface below the baseline value. All data represent mean ± SD of 3 independent experiments. ***P < .001.
Cellular RNA exhibited a similar effect on PF4 binding to platelets as heparin, with a maximal 3.69-fold increase (±1.09; Figure 3A) at 0.63 µg/mL. This was almost completely abrogated by pretreatment with RNase A (1.25-fold increase ±0.29; P = .0037, n = 3; Figure 3A).
Enhancement of PF4 binding to the platelet surface by DNA or RNA was dependent on the size of the nucleic acid construct: shorter constructs (10mer–single-stranded DNA; 21mer–hairpin RNA) showed a weaker effect and required higher concentrations than longer constructs (45mer–double-stem-loop RNA II; Figure 3B). RNA compounds appeared to have a stronger effect on PF4 binding than DNA homologs, as exemplified by the 45mer–double-stem-loop RNA I, which induced binding of more PF4 to platelets, compared with the 45mer–double-stem-loop DNA (P < .0001; Figure 3C), although both 45mers had the same nucleotide sequence (RNA, however, contained uridine and ribose instead of thymidine and deoxyribose).
In contrast to the structured 45mers, the unpaired homopolymers 45mer–polyA and 45mer–polyC, or single nucleotides, did not promote PF4 binding to platelets (Figure 3C). Furthermore, the influence of 3 DNA 21mers with distinct secondary structures (Figure 1B-D) on PF4 binding was analyzed: the 21mer–double-stranded DNA showed the strongest enhancement on PF4 binding to platelets (P < .0001; Figure 3D), while the 2 single-stranded constructs were far less effective. PF4 binding increased with the structural complexity of the nucleic acid construct (compare 21mer–hairpin vs 21mer–single-stranded DNA, P < .0001; Figure 3D), indicating that interactions of nucleic acids with PF4 were not only charge dependent but determined by their conformation.
Aptamers enhance PF4 binding to platelets
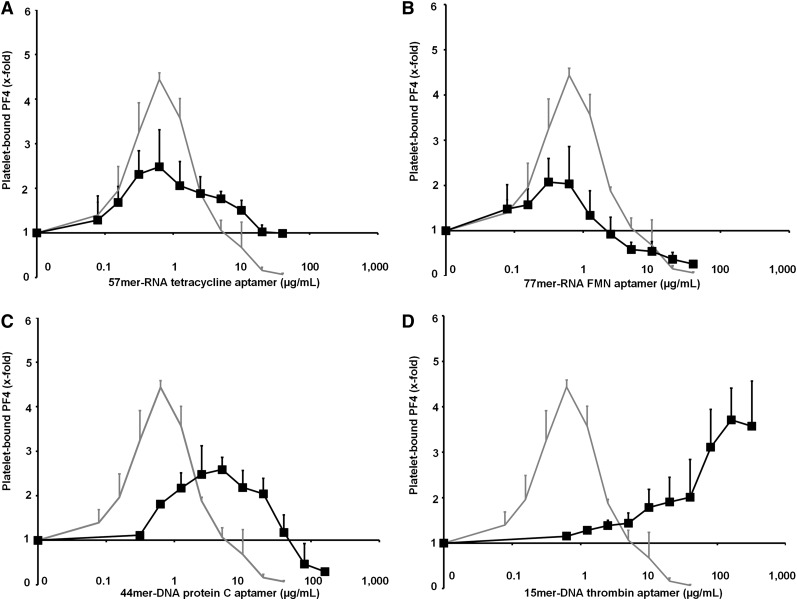
We then investigated the influence of 4 different aptamers (Figure 1A) on PF4 binding to the platelet surface. All 4 aptamers enhanced PF4 binding to gel-filtered platelets; however, maximal PF4 binding in each case occurred at different concentrations of the respective aptamer: the 57mer–RNA tetracycline aptamer (2.48-fold increase ±0.63; Figure 4A) and the 77mer–RNA FMN aptamer (2.07-fold increase ±0.52; Figure 4B) induced maximal PF4 binding to platelets at similar concentrations (0.63 µg/mL). However, maximal enhancement of PF4 binding by these aptamers was lower than that induced by heparin. The 44mer–DNA protein C aptamer induced maximal enhancement of PF4 binding (2.59-fold increase ±0.35) at a greater than 10-fold higher concentration of 5 µg/mL (Figure 4C), whereas the short 15mer–DNA thrombin aptamer enhanced PF4 binding only at very high concentrations (at 320 µg/mL: 3.57-fold increase ±1.1; Figure 4D).
Figure 4.
The influence of aptamers on PF4 binding to platelets is shown. (For details of aptamer structure, please compare with Figure 1A.) The following 4 aptamers (▪) were tested in increasing doses for promoting PF4 binding to platelets in comparison with heparin (gray line) as the positive control: (A) 57mer–RNA tetracycline aptamer, (B) 77mer–RNA FMN aptamer, (C) 44mer–DNA protein C aptamer, and (D) 15mer–DNA thrombin aptamer. All data represent mean ± SD of 3 independent experiments. The figures indicate that PF4 binding to platelets correlates with the length and the size of double-stranded domains of the aptamers.
Aptamer-induced changes in PF4 resemble those induced by heparin
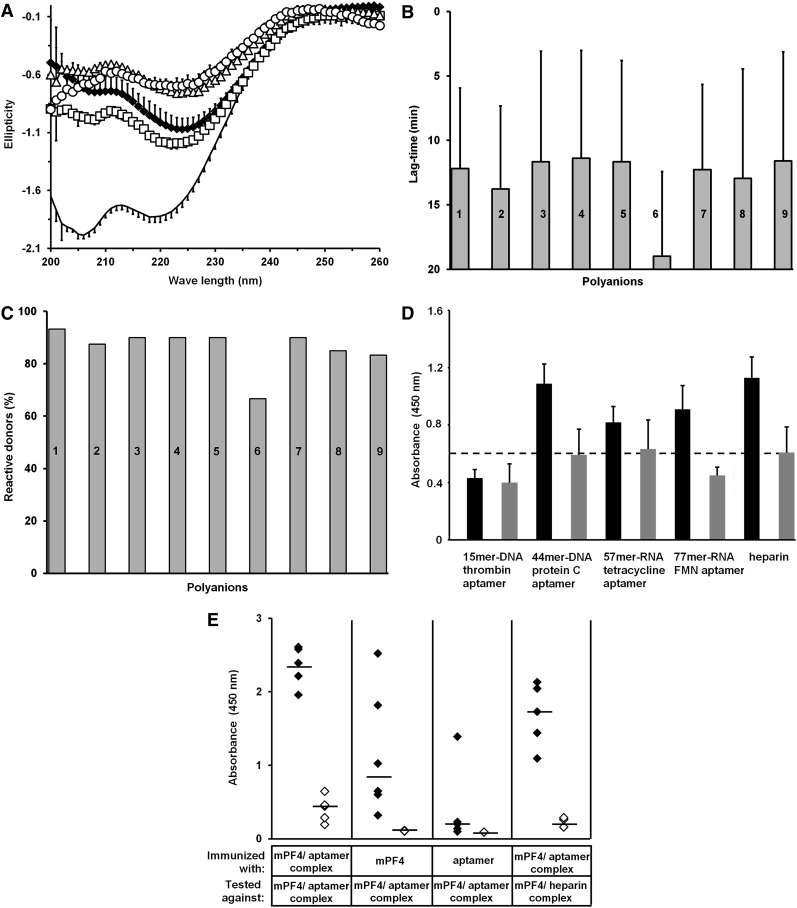
The impact of nucleic acids on the secondary structure of PF4 was analyzed using CD spectroscopy. The 44-mer–DNA protein C aptamer was used due to its nuclease resistance. Although changes in the PF4 structure induced by complex formation with the 44-mer–DNA protein C aptamer were similar to those induced by heparin, the most pronounced alterations of PF4 were observed at broader ranges of 44mer–DNA protein C aptamer (5-20 µg/mL) compared with that seen with heparin (6.9–7.2 µg/mL; Figure 5A).
Figure 5.
Aptamer-induced changes in PF4 lead to expression of PF4/heparin-like epitopes. (A) Shown are structural changes in PF4 induced by the 44mer–DNA protein C aptamer. Changes in the CD pattern of PF4 (20 µg/mL) are shown in the absence (n = 4, black line) or presence of either heparin (n = 3, 6.9 µg/mL, ♦) or 44mer–DNA protein C aptamer at different doses (n = 2 each, 2.5 µg/mL, □; 5 µg/mL, Δ; 20 µg/mL, ○). Note that the changes in PF4 spectra are rather similar, regardless whether heparin or the aptamer was used. All data represent mean ± SD of n independent experiments. Each experiment consists of at least 5 measurements. (B-C) Human anti-PF4/heparin antibodies induce platelet activation in the presence of nucleic acids and aptamers. (B) Mean lag time until aggregation of donor platelets (at least n = 15 for each polyanion) and (C) reactivity of donor platelets expressed as the percentage of all tested donor platelets were analyzed in the presence of different anti-PF4/heparin antibody–containing sera (at least 4 for each polyanion) and either reviparin (0.2 µg/mL, bar 1), cellular RNA (0.5 µg/mL, bar 2), 21mer–double-stranded DNA (0.5 µg/mL, bar 3), 21mer–hairpin DNA (0.5 µg/mL, bar 4), deoxynucleotidetriphosphates (0.5 µg/mL, bar 5), 15mer–DNA thrombin aptamer (20 µg/mL, bar 6), 44mer–DNA protein C aptamer (8 µg/mL, bar 7), 57mer–RNA tetracycline aptamer (5 µg/mL, bar 8), or 77mer–RNA FMN–ribozyme (2.5 µg/mL, bar 9). Except for the short 15mer–DNA thrombin aptamer, which induces platelet aggregation after 19.0 ± 0.67 minutes and only in 66.6% of all donors, lag time and reactivity of platelets show only minor differences between the constructs. (D) The graph shows the binding of anti-PF4/heparin antibodies to PF4/aptamer complexes. Anti-PF4/heparin antibodies bind to complexes generated with 20 µg/mL PF4 and either 44mer–DNA protein C aptamer (20 µg/mL, n = 9), 57mer–RNA tetracycline aptamer (10 µg/mL, n = 4), 77mer–RNA FMN aptamer (10 µg/mL, n = 4), or heparin (3.3 µg/mL, n = 9, black bars), while no binding to complexes of PF4 and the 15mer–DNA thrombin aptamer (40 µg/mL, n = 4) occurred. Gray bars show the inhibition of binding by high heparin (660 µg/mL). All data represent mean ± SD of at least 4 independent experiments. (E) Shown is the immune response to PF4/aptamer complexes in mice. Mice were immunized with either mPF4/44mer–DNA protein C aptamer complexes (n = 5), mPF4 alone (n = 6), or 44mer–DNA protein C aptamer alone (n = 6), and the binding of antibodies from the respective sera to mPF4/44mer–DNA protein C aptamer complexes as well as mPF4/heparin complexes was assessed by EIA in the absence (♦) or presence (◊) of 660 µg/mL heparin. Median values are marked by black lines.
Deconvolution of the spectra of PF4 obtained after the addition of 5 µg/mL 44mer–DNA protein C aptamer or 6.9 µg/mL heparin showed an increase in anti-parallel β sheet content from 24.2% to 34.3% for heparin and to 35.8% for the 44mer–DNA protein C aptamer. This refolding was balanced by a decrease of the α helix fraction from 12.2% to 8.9% (heparin) and to 8.1% (44mer–DNA protein C aptamer) and a decrease in the β-turn fraction from 22.8% to 19.8% (heparin) and 20.5% (44mer–DNA protein C aptamer). Parallel β sheet and random coil content did not change significantly.
Human anti-PF4/heparin antibodies bind to PF4/nucleic acid complexes
To assess whether complex formation between aptamers and PF4 also results in exposure of the same epitopes as on PF4/heparin complexes, we tested human sera known to contain anti-PF4/heparin IgG by EIA. Antibody binding to PF4/heparin (positive control) resulted in a mean optical density (OD) of 1.130 ± 0.143. Anti-PF4/heparin antibodies bound differently to complexes formed by PF4 and distinct aptamers. They reacted only very weakly with PF4/15mer–DNA thrombin aptamer complexes (at 40 µg/mL: mean OD: 0.431 ± 0.064; Figure 5D) but strongly with PF4/44mer–DNA protein C aptamer complexes over a broad concentration range (5-30 µg/mL aptamer per 20 µg/mL PF4; mean OD: 1.098 ± 0.140; Figure 5D). Binding was significantly reduced by the addition of high concentrations of heparin (mean OD: 0.593 ± 0.203, P < .0001; Figure 5D). (Marked inhibition of antibody binding against PF4/polyanion complexes at high concentrations of heparin is a characteristic feature of anti-PF4/polyanion antibody binding.)
Less pronounced binding was observed with PF4/57mer–RNA tetracycline aptamer complexes (at 20 µg/mL: mean OD: 0.819 ± 0.115; Figure 5D) and PF4/77mer–RNA FMN aptamer complexes (at 10 µg/mL: mean OD: 0.910 ± 0.190; Figure 5D). Control sera did not react with any PF4/nucleic acid complexes (OD range: 0.02-0.28, n = 11, data not shown in the figure).
Human anti-PF4/heparin antibodies induce platelet activation in the presence of nucleic acids and aptamers
Among 29 human sera that caused platelet activation in the presence of heparin, 18 (62.1%) also caused platelet activation in the presence of nucleic acids. These 18 sera induced platelet aggregation in the presence of all nucleic acid constructs, including aptamers, but not in the presence of buffer. Mean lag time to platelet activation was slightly longer for the nucleic acid constructs (12.69 ± 2.84 minutes) than for heparin (10.73 ± 6.27 minutes), with only minor differences between the single constructs (Figure 5B). Mean reactivity with donor platelets (ie, the platelets of how many donors reacted) was also lower for the nucleic acids (85.31%) than for heparin (93.22%), with the 15mer–DNA thrombin aptamer showing the lowest reactivity (Figure 5C). Platelet aggregation was consistently inhibited by high concentrations of heparin as well as by the monoclonal antibody IV.3.22 In contrast to heparin, the nucleic acid constructs induced platelet aggregation over a wide concentration range (0.005-100 µg/mL). Consistent with what is known from heparin, very high concentrations of nucleic acids exceeding 1000 µg/mL inhibited platelet aggregation. None of the control sera (n = 9) induced platelet activation in the presence of heparin or any nucleic acid construct.
PF4/aptamer complexes induce anti-PF4/polyanion antibodies in vivo
In mice, mPF4/44mer–DNA protein C aptamer complexes induced a strong and robust immune response within 15 days in all animals (median OD: 2.39, range: 1.96-2.61, n = 5; Figure 5E). These antibodies cross-reacted against mPF4/heparin complexes (median OD: 1.73, range 1.10-2.13, n = 5; Figure 5E). Mice immunized with mPF4/heparin complexes (positive control) showed an immune response against mPF4/heparin complexes (median OD: 1.58, range: 0.61-2.76, n = 7). Injection of the aptamer alone did not induce antibody formation with the exception of 1 animal (median OD: 0.20, range: 0.10-1.39; Figure 5E). Immunization with mPF4 alone also caused an immune response (median OD: 0.84, range: 0.32-2.52; Figure 5E), but this was significantly weaker than the immunization induced by mPF4/44mer–DNA protein C aptamer complexes (P = .015). The reason for an immune response to mPF4 alone is most likely the formation of some mPF4 autoaggregates resembling mPF4/aptamer complexes, as mPF4 tends to form spontaneous complexes in buffers with low salt concentrations (unpublished observations of our group). Antibody binding was always inhibited by the addition of high concentrations of heparin.
Discussion
In this study, we demonstrated a charge- and structure-related interaction between PF4 and nucleic acids and we characterized the functional properties of these multimolecular complexes. Nucleic acids induced structural changes in the PF4 molecule similar to those typically observed in PF4/heparin complexes. These structural changes resulted in the exposure of neoepitopes that were recognized by a subset of platelet-activating anti-PF4/heparin antibodies. Moreover, PF4/aptamer complexes induced antibodies in mice, which cross-reacted with PF4/heparin complexes.
It is well known that PF4 forms complexes with polyanions other than heparin, such as dextran sulfate, pentosan polysulfate, polyvinyl sulfonate, hypersulfated chondroitin sulfate, and PI-88 (investigational antiangiogenic drug).25-27 The formation of epitopes on these complexes that are recognized by anti-PF4/heparin antibodies is dependent on critical structural features of the polyanions, for example, charge density,26,28,29 length,26,28 and charge clustering, such as those that occur on branched polysaccharides.26 Nucleic acids express charged oxygens (O−) of phosphate groups that are spaced 0.5 to 0.7 nm apart.30,31 These dimensions are similar to the known critical charge density of sulfated polysaccharides, in which sulfates are spaced about 0.5 nm apart along the carbohydrate backbone.27 Thus, the surface-exposed phosphate residues in RNA and DNA compounds most likely serve as binding partners for PF4. In fact, polysaccharides in which sulfate groups were substituted by phosphates were demonstrated to bind to PF4, indicating strong PF4-phosphate interaction.27 Moreover, the phosphate groups of lipid A as a component of lipopolysaccharide also interact with PF4.7
Besides charge density, the size of nucleic acids appears to be critical for enhancing the binding of PF4 on the platelet surface. Longer constructs augmented PF4 binding to platelets (Figure 3B), while single nucleotides added at the same concentration did not. Similarly, disaccharides do not bind to PF4, and the synthetic pentasaccharide, fondaparinux, binds only weakly to PF4.32,33
Although nucleic acids seem to share some similarities with heparins in regard to their polyanion character, the structure-function relationships for interactions between PF4 and nucleic acids appear to be more complicated. Other than heparins, nucleic acids can fold into highly organized structures, for example, those exhibiting spatial clustering of negative charges. While several RNA and DNA constructs formed complexes with PF4, the linear homopolymers 45mer–polyA and 45mer–polyC did not (Figure 3), although they were above the critical length and showed a high charge density. Thus, double-stranded domains within nucleic acids seem to be important for binding to PF4. This is in line with our recent observations that double-stranded hairpin-forming, but not single-stranded, nucleic acid oligomers are also potent activators of the intrinsic coagulation pathway by interacting with the RNA-binding cofactor high molecular weight kininogen.34
Figure 6 depicts a simplified model of the interaction between PF4 and nucleic acids: the negatively charged phosphate residues along the backbone of double-stranded nucleic acids bind to PF4, just as PF4 binds to the sulfate groups of heparin (Figure 6A-B). In contrast, due to their higher flexibility and the loose spatial arrangement of negative charges, single-stranded nucleic acids are more likely to wrap around 1 PF4 tetramer instead of bridging several PF4 tetramers (Figure 6C). This is mediated by interactions of their phosphate groups with positively charged amino acid residues of PF4, as it has already been described for polysaccharides comprising >34 sugar units.35
Figure 6.
A schematic model of complex formation between PF4 and nucleic acids is shown. (A) The basic amino acid residues (+) in the PF4 tetramer interact with the negatively charged phosphate groups (−) in double-stranded nucleic acids, thereby forming large multimolecular complexes. (B) PF4 binds to the negatively charged sulfate groups (−) on both sides of the heparin molecule, resulting in the formation of large multimolecular complexes. (C) Due to their high flexibility, single-stranded nucleic acids potentially wrap around PF4 tetramers, thereby inhibiting the formation of large, multimolecular complexes with PF4.
However, the molecular interactions are likely more complex, and certain conformations of nucleic acids appear to be more prone than others to forming multimolecular complexes with PF4. One example is the reactivity of the 44mer–DNA protein C aptamer vs the 77mer–RNA FMN aptamer. Although the latter is much longer and has more double-stranded segments (Figure 1A), anti-PF4/heparin antibodies bound somewhat more strongly to PF4/44mer–protein C aptamer complexes (Figure 5D).
Our data raise further questions beyond the scope of the current study. It remains unclear why only about 60% of the anti-PF4/heparin antibodies that induced platelet activation in the presence of heparin also caused platelet activation in the presence of nucleic acids. Even more puzzling is our observation that those sera that induced platelet activation in the presence of nucleic acids did so with most nucleic acids, independent of whether these strongly enhanced PF4 binding to the platelet surface or not. In the absence of a 3-dimensional model of the structural changes of PF4 induced by different nucleic acids, we can only speculate that the nucleic acids may expose different epitopes than heparins do.
Another hypothetical explanation could be based on the recent studies of Sachais et al,36 showing that the monoclonal antibody KKO, but not another anti-PF4 monoclonal antibody, is also able to induce oligomerization of PF4 in the absence of heparin, suggesting that some anti-PF4/heparin antibodies are capable of promoting further formation of their own antigen.37 Following this concept, sera causing platelet activation in the presence of nucleic acids might contain antibodies that, like KKO, can cross-link PF4 molecules. In this case, the negative charge of nucleic acids would initiate antigen formation by allowing close approximation of PF4 molecules, which would be sufficient for further oligomerization of PF4 by such anti-PF4/heparin antibodies. Unfortunately, we cannot readily test these hypotheses experimentally due to the polyclonal nature of human antibodies.
Despite these unresolved issues, our study provides an explanation for why patients with major tissue damage develop increased levels of anti-PF4/heparin antibodies compared with patients with minor tissue damage.38 Increased concentrations of cell-free plasma DNA (1-2 µg/mL) have been found in various clinical conditions, including trauma,39 cancer,40 and infections,41 correlating with the severity of trauma or infection.11,41 However, due to their short plasma half-life, elevated RNA/DNA plasma levels return to baseline within a few hours after tissue damage. Yet this might be sufficient to induce an immune response, as also single and minimal applications of heparin have been shown to be sufficient for the induction of HIT.42 Cellular RNA and DNA released by the tissue trauma might complex with PF4, potentially enhancing the immune response against PF4/heparin complexes. In vivo formation of PF4/nucleic acid complexes may also explain the formation of anti-PF4/heparin antibodies in patients after major orthopedic surgery even in the absence of heparin,43-45 as well as the relatively high frequency of antibodies in critically ill patients.46 In addition, formation of PF4/nucleic acid complexes might be another explanation for the reported cases of “spontaneous HIT,” because all patients who suffered from a HITlike thromboembolic disorder, despite no previous heparin administration, suffered from infections45 or had undergone major orthopedic surgery.44
Finally, our findings might be of relevance for the recognition of unwanted effects of therapeutic oligonucleotides such as aptamers or antisense RNA. The PF4/44mer-DNA protein C aptamer complexes induced a strong immune response in mice, and these complexes are recognized by anti-PF4/heparin antibodies. The resulting antibodies cross-reacted with PF4/heparin complexes, indicating that therapeutic aptamers (with a long plasma half-life) could potentially induce an immune response toward PF4/polyanion complexes. To date, there are no reported cases of HITlike adverse effects under treatment with therapeutic aptamers, and we are not aware of thrombotic complications in the ongoing trials using aptamers. Yet clinical data are still sparse, as the only approved therapeutic aptamer, pegaptanib, an RNA aptamer targeting vascular endothelial growth factor, is administered intravitreously and systemic concentrations are very low.47 Moreover, thromboembolic complications occurring under pegaptanib are considered to be a class effect of antivascular endothelial growth factor drugs.48 Beyond this, it should be considered that heparin is much more immunogenic when given to patients with major proinflammatory states, such as following surgery49 or major trauma.38 Thus, studies of aptamer administration in healthy volunteers could be expected to underestimate the anti-PF4/aptamer immune response that could occur in patients. The evaluation of the anti-PF4/polyanion immune response within clinical studies is a far more sensitive marker of evaluating the potential of a given aptamer for inducing serious adverse effects than is monitoring for thrombocytopenia and thrombosis.
While induction of an immune reaction resembling typical HIT may be rather unlikely due to the usually low plasma concentrations of aptamers, a strong antibody response to PF4/aptamer complexes may result in an immune reaction resembling “delayed onset HIT,”50 an autoimmunelike syndrome in which strongly reactive anti-PF4/polyanion antibodies activate platelets and induce thrombosis even in the absence of heparin. Yet various aptamers differ considerably in their structure and are often chemically modified, which may interfere with their binding to PF4. Using the methods described in this study, it should be possible to screen aptamers for their potential interaction with PF4. We suggest that clinical trials of aptamers or other therapeutic nucleic acid constructs should include screening for the development of anti-PF4/aptamer antibodies and potential clinical features of HIT as part of their safety end points.
Acknowledgments
The authors thank Professor Dr Bernd Pötzsch and Dr Müller (Institut für Experimentelle Hämatologie und Transfusionsmedizin, University of Bonn, Germany) for kindly providing the 44mer–DNA protein C aptamer.
This work was supported by the Gerhard-Domagk-Program of the Medical School, University of Greifswald (M.E.J.) and by the Zentrum für Innovationskompetenz Humorale Immunreaktionen bei kardiovaskuläen Erkrankungen (ZIK HIKE, Federal Ministry of Education and Research, BMBF FKZ 03Z2CN12 and FKZ 03Z2CN11) (K.K., S. Brandt, and S. Block). Part of the work was supported by the Excellence-cluster Cardio-pulmonary system (ECCPS) and grant FI 543/2-2 from the Deutsche Forschungsgemeinschaft (Bonn, Germany) (K.T.P. and S.F.).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.E.J. designed and performed the in vitro experiments and wrote the manuscript; K.K. designed the study, supervised the experiments, reviewed and interpreted the results, and revised the manuscript; B.F. performed and supervised the mouse experiments and interpreted the results; T.M. and B.A. designed and synthesized the nucleic acid constructs and reviewed the results; S. Brandt, S. Block, and C.A.H. performed the CD spectroscopy experiments and reviewed and interpreted the results; S.F. and J.G. isolated the cellular RNA, performed the preliminary experiments on PF4-RNA interaction, and reviewed the manuscript; S.M. designed the nucleic acid constructs and reviewed the results and the manuscript; K.T.P. created the hypothesis of PF4–nucleic acid interaction, designed the experiments on RNA, and wrote the manuscript; A.G. developed the hypothesis of PF4-aptamer interaction, designed the experiments, reviewed the results, and wrote the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Greinacher, Institut für Immunologie und Transfusionsmedizin, Universitätsmedizin, Ernst-Moritz-Arndt-Universität Greifswald, Sauerbruchstrasse, D-17475 Greifswald, Germany; e-mail: greinach@uni-greifswald.de.
References
- 1.Kaplan KL, Broekman MJ, Chernoff A, Lesznik GR, Drillings M. Platelet alpha-granule proteins: studies on release and subcellular localization. Blood. 1979;53(4):604–618. [PubMed] [Google Scholar]
- 2.Greinacher A, Gopinadhan M, Günther JU, et al. Close approximation of two platelet factor 4 tetramers by charge neutralization forms the antigens recognized by HIT antibodies. Arterioscler Thromb Vasc Biol. 2006;26(10):2386–2393. doi: 10.1161/01.ATV.0000238350.89477.88. [DOI] [PubMed] [Google Scholar]
- 3.Rauova L, Poncz M, McKenzie SE, et al. Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia. Blood. 2005;105(1):131–138. doi: 10.1182/blood-2004-04-1544. [DOI] [PubMed] [Google Scholar]
- 4.Arepally GM, Ortel TL. Clinical practice. Heparin-induced thrombocytopenia. N Engl J Med. 2006;355(8):809–817. doi: 10.1056/NEJMcp052967. [DOI] [PubMed] [Google Scholar]
- 5.Warkentin TE, Chong BH, Greinacher A. Heparin-induced thrombocytopenia: towards consensus. Thromb Haemost. 1998;79(1):1–7. [PubMed] [Google Scholar]
- 6.Warkentin TE, Greinacher A, Koster A, Lincoff AM. Treatment and prevention of heparin-induced thrombocytopenia: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition). Chest. 2008;133(6 Suppl):340S-380S. [DOI] [PubMed] [Google Scholar]
- 7.Krauel K, Weber C, Brandt S, Zähringer U, Mamat U, Greinacher A, Hammerschmidt S. Platelet factor 4 binding to lipid A of Gram-negative bacteria exposes PF4/heparin-like epitopes. Blood. 2012;120(16):3345–3352. doi: 10.1182/blood-2012-06-434985. [DOI] [PubMed] [Google Scholar]
- 8.Anderson GP. Insights into heparin-induced thrombocytopenia. Br J Haematol. 1992;80(4):504–508. doi: 10.1111/j.1365-2141.1992.tb04564.x. [DOI] [PubMed] [Google Scholar]
- 9.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittra I, Nair NK, Mishra PK. Nucleic acids in circulation: are they harmful to the host? J Biosci. 2012;37(2):301–312. doi: 10.1007/s12038-012-9192-8. [DOI] [PubMed] [Google Scholar]
- 11.Lam NY, Rainer TH, Chan LY, Joynt GM, Lo YM. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003;49(8):1286–1291. doi: 10.1373/49.8.1286. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi Y, Rykova EY, editors. Extracellular Nucleic Acids. In: Gross HJ, Bujnicki JM, eds. Nucleic Acids and Molecular Biology. Vol 25. Berlin: Springler-Verlag; 2010. [Google Scholar]
- 13.Ni X, Castanares M, Mukherjee A, Lupold SE. Nucleic acid aptamers: clinical applications and promising new horizons. Curr Med Chem. 2011;18(27):4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krauel K, Fürll B, Warkentin TE, Weitschies W, Kohlmann T, Sheppard JI, Greinacher A. Heparin-induced thrombocytopenia—therapeutic concentrations of danaparoid, unlike fondaparinux and direct thrombin inhibitors, inhibit formation of platelet factor 4-heparin complexes. J Thromb Haemost. 2008;6(12):2160–2167. doi: 10.1111/j.1538-7836.2008.03171.x. [DOI] [PubMed] [Google Scholar]
- 15.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 16.Müller J, Isermann B, Dücker C, et al. An exosite-specific ssDNA aptamer inhibits the anticoagulant functions of activated protein C and enhances inhibition by protein C inhibitor. Chem Biol. 2009;16(4):442–451. doi: 10.1016/j.chembiol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31(7):1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strohbach D, Novak N, Müller S. Redox-active riboswitching: allosteric regulation of ribozyme activity by ligand-shape control. Angew Chem Int Ed Engl. 2006;45(13):2127–2129. doi: 10.1002/anie.200503820. [DOI] [PubMed] [Google Scholar]
- 19.Juhl D, Eichler P, Lubenow N, Strobel U, Wessel A, Greinacher A. Incidence and clinical significance of anti-PF4/heparin antibodies of the IgG, IgM, and IgA class in 755 consecutive patient samples referred for diagnostic testing for heparin-induced thrombocytopenia. Eur J Haematol. 2006;76(5):420–426. doi: 10.1111/j.1600-0609.2005.00621.x. [DOI] [PubMed] [Google Scholar]
- 20.Warkentin TE, Greinacher A. Laboratory Testing for Heparin-Induced Thrombocytopenia. In: Warkentin TE, Greinacher A, editors. Heparin-Induced Thrombocytopenia. New York, NY: Informa Healthcare; 2007. pp. 227–238. [Google Scholar]
- 21.Greinacher A, Pötzsch B, Amiral J, Dummel V, Eichner A, Mueller-Eckhardt C. Heparin-associated thrombocytopenia: isolation of the antibody and characterization of a multimolecular PF4-heparin complex as the major antigen. Thromb Haemost. 1994;71(2):247–251. [PubMed] [Google Scholar]
- 22.Kelton JG, Sheridan D, Santos A, et al. Heparin-induced thrombocytopenia: laboratory studies. Blood. 1988;72(3):925–930. [PubMed] [Google Scholar]
- 23.Böhm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5(3):191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 24.Suvarna S, Qi R, Arepally GM. Optimization of a murine immunization model for study of PF4/heparin antibodies. J Thromb Haemost. 2009;7(5):857–864. doi: 10.1111/j.1538-7836.2009.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alban S, Franz G. Gas-liquid chromatography-mass spectrometry analysis of anticoagulant active curdlan sulfates. Semin Thromb Hemost. 1994;20(2):152–158. doi: 10.1055/s-2007-1001898. [DOI] [PubMed] [Google Scholar]
- 26.Greinacher A, Alban S, Dummel V, Franz G, Mueller-Eckhardt C. Characterization of the structural requirements for a carbohydrate based anticoagulant with a reduced risk of inducing the immunological type of heparin-associated thrombocytopenia. Thromb Haemost. 1995;74(3):886–892. [PubMed] [Google Scholar]
- 27.Visentin GP, Moghaddam M, Beery SE, McFarland JG, Aster RH. Heparin is not required for detection of antibodies associated with heparin-induced thrombocytopenia/thrombosis. J Lab Clin Med. 2001;138(1):22–31. doi: 10.1067/mlc.2001.115525. [DOI] [PubMed] [Google Scholar]
- 28.Leroux D, Canépa S, Viskov C, et al. Binding of heparin-dependent antibodies to PF4 modified by enoxaparin oligosaccharides: evaluation by surface plasmon resonance and serotonin release assay. J Thromb Haemost. 2012;10(3):430–436. doi: 10.1111/j.1538-7836.2012.04618.x. [DOI] [PubMed] [Google Scholar]
- 29.Petitou M, Hérault JP, Bernat A, Driguez PA, Duchaussoy P, Lormeau JC, Herbert JM. Synthesis of thrombin-inhibiting heparin mimetics without side effects. Nature. 1999;398(6726):417–422. doi: 10.1038/18877. [DOI] [PubMed] [Google Scholar]
- 30.Schneider B, Patel K, Berman HM. Hydration of the phosphate group in double-helical DNA. Biophys J. 1998;75(5):2422–2434. doi: 10.1016/S0006-3495(98)77686-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saenger W. Principles of Nucleic Acid Structure. Berlin: Springer-Verlag; 1984. [Google Scholar]
- 32.Savi P, Chong BH, Greinacher A, et al. Effect of fondaparinux on platelet activation in the presence of heparin-dependent antibodies: a blinded comparative multicenter study with unfractionated heparin. Blood. 2005;105(1):139–144. doi: 10.1182/blood-2004-05-2010. [DOI] [PubMed] [Google Scholar]
- 33.Weimann G, Lubenow N, Selleng K, Eichler P, Albrecht D, Greinacher A. Glucosamine sulfate does not crossreact with the antibodies of patients with heparin-induced thrombocytopenia. Eur J Haematol. 2001;66(3):195–199. doi: 10.1034/j.1600-0609.2001.00353.x. [DOI] [PubMed] [Google Scholar]
- 34.Gansler J, Jaax M, Leiting S, Appel B, Greinacher A, Fischer S, Preissner KT. Structural requirements for the procoagulant activity of nucleic acids. PLoS ONE. 2012;7(11):e50399. doi: 10.1371/journal.pone.0050399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuckey JA, St Charles R, Edwards BF. A model of the platelet factor 4 complex with heparin. Proteins. 1992;14(2):277–287. doi: 10.1002/prot.340140213. [DOI] [PubMed] [Google Scholar]
- 36.Sachais BS, Litvinov RI, Yarovoi SV, et al. Dynamic antibody-binding properties in the pathogenesis of HIT. Blood. 2012;120(5):1137–1142. doi: 10.1182/blood-2012-01-407262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greinacher A, Krauel K, Jensch I. HIT-antibodies promote their own antigen. Blood. 2012;120(5):930–931. doi: 10.1182/blood-2012-06-430694. [DOI] [PubMed] [Google Scholar]
- 38.Lubenow N, Hinz P, Thomaschewski S, et al. The severity of trauma determines the immune response to PF4/heparin and the frequency of heparin-induced thrombocytopenia. Blood. 2010;115(9):1797–1803. doi: 10.1182/blood-2009-07-231506. [DOI] [PubMed] [Google Scholar]
- 39.Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000;46(3):319–323. [PubMed] [Google Scholar]
- 40.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 41.Huttunen R, Kuparinen T, Jylhävä J, et al. Fatal outcome in bacteremia is characterized by high plasma cell free DNA concentration and apoptotic DNA fragmentation: a prospective cohort study. PLoS ONE. 2011;6(7):e21700. doi: 10.1371/journal.pone.0021700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gettings EM, Brush KA, Van Cott EM, Hurford WE. Outcome of postoperative critically ill patients with heparin-induced thrombocytopenia: an observational retrospective case-control study. Crit Care. 2006;10(6):R161. doi: 10.1186/cc5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jay RM, Warkentin TE. Fatal heparin-induced thrombocytopenia (HIT) during warfarin thromboprophylaxis following orthopedic surgery: another example of ‘spontaneous’ HIT? J Thromb Haemost. 2008;6(9):1598–1600. doi: 10.1111/j.1538-7836.2008.03040.x. [DOI] [PubMed] [Google Scholar]
- 44.Pruthi RK, Daniels PR, Nambudiri GS, Warkentin TE. Heparin-induced thrombocytopenia (HIT) during postoperative warfarin thromboprophylaxis: a second example of postorthopedic surgery ‘spontaneous’ HIT. J Thromb Haemost. 2009;7(3):499–501. doi: 10.1111/j.1538-7836.2008.03263.x. [DOI] [PubMed] [Google Scholar]
- 45.Warkentin TE, Makris M, Jay RM, Kelton JG. A spontaneous prothrombotic disorder resembling heparin-induced thrombocytopenia. Am J Med. 2008;121(7):632–636. doi: 10.1016/j.amjmed.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Levine RL, Hergenroeder GW, Francis JL, Miller CC, Hursting MJ. Heparin-platelet factor 4 antibodies in intensive care patients: an observational seroprevalence study. J Thromb Thrombolysis. 2010;30(2):142–148. doi: 10.1007/s11239-009-0425-0. [DOI] [PubMed] [Google Scholar]
- 47.Drolet DW, Nelson J, Tucker CE, et al. Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm Res. 2000;17(12):1503–1510. doi: 10.1023/a:1007657109012. [DOI] [PubMed] [Google Scholar]
- 48.Costagliola C, Agnifili L, Arcidiacono B, et al. Systemic thromboembolic adverse events in patients treated with intravitreal anti-VEGF drugs for neovascular age-related macular degeneration. Expert Opin Biol Ther. 2012;12(10):1299–1313. doi: 10.1517/14712598.2012.707176. [DOI] [PubMed] [Google Scholar]
- 49.Warkentin TE, Sheppard JA, Sigouin CS, Kohlmann T, Eichler P, Greinacher A. Gender imbalance and risk factor interactions in heparin-induced thrombocytopenia. Blood. 2006;108(9):2937–2941. doi: 10.1182/blood-2005-11-012450. [DOI] [PubMed] [Google Scholar]
- 50.Rice L, Attisha WK, Drexler A, Francis JL. Delayed-onset heparin-induced thrombocytopenia. Ann Intern Med. 2002;136(3):210–215. doi: 10.7326/0003-4819-136-3-200202050-00009. [DOI] [PubMed] [Google Scholar]