Abstract
Enteroaggregative Escherichia coli (EAEC) is an important agent that causes endemic and epidemic diarrhoeal diseases worldwide. Several EAEC virulence-related genes (VRGs) have been described but their role in the clinical outcome of infection is not completely defined. This study investigated the prevalence of EAEC and potential associations of its VRGs with risk of or protection from diarrhoeal diseases in children from urban communities in north-eastern Brazil. The case–control study included 166 children, who had their stools evaluated for the EAEC diagnostic genes (aaiC and aatA) using PCR. Positive samples were further analysed by multiplex PCR and identified 18 VRGs. EAEC was found in the same proportion in both groups (41 %). The plasmid-borne gene encoding a hexosyltransferase homologue (capU) was the most frequently detected (89.6 %), followed by dispersin protein (aap, 58.2 %) and EAEC HilA homologue (eilA, 57.8 %). The AAF/III fimbrial subunit (agg3A) gene was observed at lower frequency (1.5 %). Plasmid-encoded toxin (pet) or AAF/II fimbrial subunit (aafA) was associated significantly with disease. AAF/IV fimbrial subunit (agg4A) or hypothetical plasmid-encoded haemolysin (orf61) was detected significantly more in controls than in children with diarrhoea. In addition, one set of genes in combination, aaiC and agg3/4C but lacking agg4A and orf61, was associated with diarrhoea cases; and another one, orf61 in the absence of pet and aafA, was correlated with control children. These data confirm a high prevalence, endemicity and heterogeneity of EAEC strains in the developing urban areas of north-eastern Brazil. Statistical correlation between cases and controls was seen with either isolated or combined sets of genes, suggesting that the pathophysiology of EAEC infection involves a complex and dynamic modulation of several VRGs.
Introduction
Since its first description by Nataro et al. (1987) in a prospective study of paediatric diarrhoea in Santiago, Chile, enteroaggregative Escherichia coli (EAEC) has been increasingly recognized as an agent of diarrhoea. EAEC is probably best known for its role in persistent diarrhoea in children living in developing countries, the context in which its pathogenesis was originally defined (Okeke & Nataro, 2001). However, it has emerged as an important pathogen in outbreaks of acute diarrhoea in developed (Huang et al., 2006; Scavia et al., 2008) and developing countries (Bueris et al., 2007; Kermani et al., 2010; Meng et al., 2011; Pereira et al., 2007), international traveller’s diarrhoea (Paschke et al., 2011) and diarrhoea among patients with human immunodeficiency virus-related infection (Samie et al., 2007).
The pathogenesis of EAEC infection is not fully understood. The major obstacle in identifying the mechanism of pathogenesis for this bacterium is the heterogeneity of strains. While some investigations confirm the clear association of EAEC with diarrhoea in some individuals (Meng et al., 2011; Okeke et al., 2000; Opintan et al., 2010), in many others, it appears to cause subclinical infection or only intestinal colonization (Bueris et al., 2007; Piva et al., 2003). EAEC has also been associated with chronic intestinal inflammation, leading to childhood malnutrition and growth impairment (Steiner et al., 1998).
In recent years several advances have been made in the determination of EAEC pathogenesis, including the development of in vivo models (Harrington et al., 2009; Roche et al., 2010), and the use of previously characterized in vitro models to investigate the effects of EAEC infection on intestinal cells (Strauman et al., 2010) and the repair of cell damage (Carvalho et al., 2012). The complete genome of the prototypical strain EAEC 042 has recently been published (Chaudhuri et al., 2010). Despite this progress, the disease pathophysiology remains obscure, as several candidate virulence-related genes (VRGs) have been described but are not present in all EAEC strains. The prevalence and association of these genes with diarrhoeal diseases are not well established, as they may vary by geographical location (Estrada-Garcia & Navarro-Garcia, 2012; Nataro & Kaper, 1998). A recent large-scale outbreak of diarrhoea and haemolytic uraemic syndrome infecting 4137 individuals and resulting in 54 deaths was caused by a Shiga toxin-producing EAEC O104 : H4 with a distinct set of virulence factors (Frank et al., 2011; Muniesa et al., 2012; Scheutz et al., 2011). The importance of combinations of these virulence factors warrants further investigation.
Aggregative adherence fimbriae (AAFs) are the main mucosal adhesins of EAEC, of which at least four variants are known (Bernier et al., 2002; Boisen et al., 2008; Czeczulin et al., 1997; Nataro et al., 1992). The four structural subunits are respectively encoded by aggA (AAF/I), aafA (AAF/II), agg3A (AAF/III) and agg4A (AAF/IV), and they have been shown to be regulated by the transcriptional AraC/XylS activator aggregative adherence regulator (aggR). Pathogenesis and molecular studies suggest the presence of a package of aggR-regulated VRGs (<50 in number) encoded on either chromosomal islands or virulence plasmids (Dudley et al., 2006; Morin et al., 2010). Under the control of aggR is the anti-aggregation protein gene (aap), formerly known as the EAEC secreted protein U gene (aspU), which is present on the pAA-plasmid. The aap gene encodes a secreted low-molecular-mass protein, called dispersin, that promotes dispersal of EAEC on the intestinal mucosa to establish new foci of infection (Sheikh et al., 2002). Dispersin secretion is translocated via a system called the EAEC ABC transporter (aat), encoded by aatPABCD (Nishi et al., 2003). Also regulated by aggR is the chromosomal cluster termed the aggR-activated island (aai), aaiA-P, encoding a type VI secretion system (Dudley et al., 2006). Other putative EAEC virulence factors not regulated by aggR are the EAEC heat-stable toxin 1, encoded by the aggregative heat-stable toxin A gene (astA) (Savarino et al., 1993), and a set of toxins termed serine protease autotransporters of Enterobacteriaceae (SPATEs).
SPATEs can be organized phylogenetically into two classes. Members of class I are cytotoxic and include the plasmid-encoded toxin gene (pet) (Navarro-García et al., 1998) and its homologues, secreted autotransporter toxin gene (sat) and Shigella IgA-like protease homologue gene (sigA) (Guyer et al., 2000; Rajakumar et al., 1997). Members of class II SPATEs are non-cytotoxic and include the protein involved in colonization (pic), a mucinase that promotes intestinal colonization (Henderson et al., 1999) and Shigella extracellular protease (sepA), which appears to be involved in tissue invasion (Benjelloun-Touimi et al., 1995). Recently, sepA was associated with clinical illness in a case–control study from Mali (Boisen et al., 2012).
Other EAEC candidate VRGs include capU (cap locus that encodes a protein 50 % identical to an rfbU-related lipopolysaccharide biosynthetic gene of E. coli O157 : H7) (Czeczulin et al., 1999; Fujiyama et al., 2008), the regulator eilA (EAEC HilA homologue) (Sheikh et al., 2006) and hypotheticals orf3 (cryptic protein) and orf61 (plasmid-encoded haemolysin) (Boisen et al., 2012).
Molecular approaches, especially DNA-based tests involving PCR, have increasingly been applied to identify diarrhoeagenic E. coli, which was among the first pathogens for which molecular diagnostic techniques were developed (Nataro & Kaper, 1998). In this case–control study, we determined the prevalence of EAEC strains in children, with and without diarrhoea, living in a poor urban area from Fortaleza, Ceara, north-eastern Brazil. We used PCR assays to amplify aggR-activated island C (aaiC) and EAEC ABC transporter A (aatA) genes. In addition, samples positive for EAEC were further analysed for the presence of homologous sequences to the putative VRGs associated with the EAEC pathotype.
Methods
Study site and ethical clearance.
The study was conducted in two urban communities in Fortaleza. Gonçalves Dias is a five-block area that houses 1826 people, and Parque Universitario is a community of 11 018 inhabitants.
The research protocol was reviewed and approved by the Research Ethics Committee of the Federal University of Ceara, the National Commission on Ethics in Research of Brazil and the Institutional Review Board of the University of Virginia. A consent form was read and signed by all parents or guardians.
Study design.
This was a case–control study in children 2–36 months old, as previously described by da Silva Quetz et al. (2010). Briefly, the study included children with (cases) and without (controls) diarrhoea in the last 2 weeks. Diarrhoea was defined as three or more liquid stools in a 24 h period. Stool samples were collected between March and July 2007, during the rainy season, the period which accounts for the higher incidence of diarrhoeal diseases in Fortaleza (Façanha & Pinheiro, 2005). A questionnaire with information regarding the socioeconomic and clinical conditions and the occurrence of diarrhoea was completed with the guardians. Stool samples were collected from all children at each household and they were transported within 4 h in ice boxes to the Infectious Diseases Laboratory, Institute of Biomedicine for Brazilian Semi-Arid & Clinical Research Unit/Center for Global Health, Federal University of Ceara. All the samples were aliquoted without dilution and stored at −80 °C until molecular tests were performed.
DNA extraction.
Bacterial DNA was extracted from all stool samples using the QIAamp DNA Stool Mini kit (Qiagen) according to the manufacturer’s instructions. The options of incubating the stools with lysis solution at 95 °C and removing the second washing solution in two steps were used for enhancing pathogen DNA extraction. DNA quality and quantity were checked by a spectrophotometric method (BioPhotometer; Eppendorf). Extracted DNA was stored at −20 °C until further use.
PCR assays.
Molecular diagnosis of EAEC was performed by gene amplification of aaiC (chromosomal gene) and aatA (plasmid gene), which had no apparent homologues within GenBank. Single PCR was performed using AmpliTaq Gold PCR Master Mix (Applied Biosystems), which contains Taq polymerase, dNTPs, MgCl2 and the appropriate buffer. Each PCR tube contained 25 µl reaction mixture composed of 12.5 µl of the master mix, 2.5 µl of each forward and reverse primer solution (in a final concentration of 200 nM), 1–2.5 µl of faecal DNA (this volume was DNA concentration-dependent) and nuclease-free water to complete the final volume. The PCR conditions were one cycle for 5 min at 95 °C; 35 cycles for 20 s at 95 °C, 20 s at 57 °C and 1 min at 72 °C; and a final extension step for 10 min at 72 °C in a thermal cycler (Bio-Rad Laboratories). Bands were visualized and photographed (ChemiDoc XRS; Bio-Rad Laboratories) after electrophoresis of an ethidium bromide-stained 1.2 % agarose gel in 1× Tris-acetate-EDTA-buffer. The primers used are described in Table 1. Only the presence of the correctly sized PCR product was interpreted as a positive result. One sample was considered positive for EAEC when it presented one or both researched genes.
Table 1. Description of genes, GenBank accession numbers, primer sequences, size of the obtained products, PCR conditions of the genes used for diagnosis of EAEC and its VRGs.
For all PCR conditions, an initial denaturation step (5 min for single PCR, 15 min for multiplex PCR 1, and 2 min for multiplexes 2–4 at 95 °C) and a final extension (10 min at 72 °C) were performed.
| Target gene (GenBank accession no.) | Type of PCR | Primer sequence (5′→3′) | Amplicon size (bp) | PCR conditions (35 cycles) | Reference |
| Diagnostic genes | |||||
| aaiC – aggR-activated island (FN554766.1) | Single | ATTGTCCTCAGGCATTTCACACGACACCCCTGATAAACAA | 215 | 20 s at 95 °C, 20 s at 57 °C, 1 min at 72 °C | Designed for this study |
| aatA – anti-aggregation protein transporter (AY351860) | Single | CTGGCGAAAGACTGTATCATCAATGTATAGAAATCCGCTGTT | 630 | 20 s at 95 °C, 20 s at 57 °C, 1 min at 72 °C | Schmidt et al. (1995) |
| Virulence genes | |||||
| astA – aggregative heat-stable toxin A, EAST1 (L11241) | Multiplex 1 | ATGCCATCAACACAGTATATGCGAGTGACGGCTTTGTAGT | 110 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Mohamed et al. (2007) |
| pet – plasmid-encoded toxin (AF056581) | Multiplex 1 | GGCACAGAATAAAGGGGTGTTTCCTCTTGTTTCCACGACATAC | 302 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Restieri et al. (2007) |
| sigA – Shigella IgA-like protease homologue (NC_004337) | Multiplex 1 | CCGACTTCTCACTTTCTCCCGCCATCCAGCTGCATAGTGTTTG | 430 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Boisen et al. (2009) |
| pic – protein involved in colonization (AF097644) | Multiplex 1 | ACTGGATCTTAAGGCTCAGGATGACTTAATGTCACTGTTCAGCG | 572 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Restieri et al. (2007) |
| sepA – Shigella extracellular protease (Z48219) | Multiplex 1 | GCAGTGGAAATATGATGCGGCTTGTTCAGATCGGAGAAGAACG | 794 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Restieri et al. (2007) |
| sat – secreted autotransporter toxin (AE014075) | Multiplex 1 | TCAGAAGCTCAGCGAATCATTGCCATTATCACCAGTAAAACGCACC | 932 | 60 s at 94 °C, 1.5 min at 58 °C, 1.5 min at 72 °C | Boisen et al. (2009) |
| orf3 – cryptic protein (FN554767.1) | Multiplex 2 | CAGCAACCATCGCATTTCTACGCATCTTTCAATACCTCCA | 121 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| aap – anti-aggregation protein, dispersin (Z32523) | Multiplex 2 | GGACCCGTCCCAATGTATAACCATTCGGTTAGAGCACGAT | 250 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| aggR – aggregative adherence regulator (Z18751) | Multiplex 2 | GCAATCAGATTAARCAGCGATACACATTCTTGATTGCATAAGGATCTGG | 426 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| agg4A – AAF/IV fimbrial subunit (EU637023) | Multiplex 3 | TGAGTTGTGGGGCTAYCTGGACACCATAAGCCGCCAAATAAGC | 169 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| aggA – AAF/I fimbrial subunit (Y18149, AY344586) | Multiplex 3 | TCTATCTRGGGGGGCTAACGCTACCTGTTCCCCATAACCAGACC | 220 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| aafA – AAF/II fimbrial subunit (AF012835) | Multiplex 3 | CTACTTTATTATCAAGTGGAGCCGCTAGGAGAGGCCAGAGTGAATCCTG | 289 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| agg3A – AAF/III fimbrial subunit (AF411067) | Multiplex 3 | CCAGTTATTACAGGGTAACAAGGGAATTGGTCTGGAATAACAACTTGAACG | 370 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| agg3/4C* – usher, AAF/III-IV assembly unit (AF411067, AB255435, EU637023) | Multiplex 3 | TTCTCAGTTAACTGGACACGCAATTTAATTGGTTACGCAATCGCAATTCTGACCAAATGTTATACCTTCAYTATG | 409 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| aafC – usher, AAF/II assembly unit (AF114828) | Multiplex 3 | ACAGCCTGCGGTCAAAAGCGCTTACGGGTACGAGTTTTACGG | 491 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| orf61 – plasmid-encoded haemolysin (FN554767.1) | Multiplex 4 | AGCTCTGGAAACTGGCCTCTAACCGTCCTGATTTCTGCTT | 108 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| eilA – Salmonella HilA homologue (FN554766.1) | Multiplex 4 | AGGTCTGGAGCGCGAGTGTTGTAAAACGGTATCCACGACC | 248 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
| capU – hexosyltransferase homologue (AF134403) | Multiplex 4 | CAGGCTGTTGCTCAAATGAAGTTCGACATCCTTCCTGCTC | 395 | 50 s at 94 °C, 1.5 min at 57 °C, 1.5 min at 72 °C | Boisen et al. (2012) |
Two forward primers and one reverse primer were used for the amplification of agg3/4C. This primer set was designed to amplify the usher gene from both AAF/III and IV.
All EAEC-positive samples were analysed by additional PCR with the primers and conditions described in Table 1 to amplify fragments of 18 genes encoding putative virulence factors, which were divided into four multiplex reactions. All multiplexes were performed as described by Boisen et al. (2012). PCR products were separated on 2 % pre-cast agarose gels (Life Technologies).
The following EAEC strains were used as positive controls: JM221 (aggA, sat) (Mathewson et al., 1987), 042 (aatA, aggR, aaiC, aap, orf3, pic, pet, astA, aafA, aafC, capU, eilA) (Nataro et al., 1985), 55989 (agg3A, agg3/4C) (Bernier et al., 2002), H223-1 (sigA) (Czeczulin et al., 1999) and C1010-00 (agg4A, agg3/4C, sat, sepA) (Olesen et al., 2005). E. coli MC1061 (Clermont et al., 2000) and HS (Levine et al., 1978), and water were used as negative controls.
Data analysis.
Data were typed in duplicate and analysed by two independent investigators using Microsoft Office Access software (Microsoft Corporation). Classification and regression tree (CART software, pro version 6.0; Salford Systems) was used inputting 18 factors of interest as binary (present/absent) independent predictive variables. Case/control status was the binary dependent outcome variable. Chi-squared, Fisher’s exact, Mann–Whitney and odds ratio (OR) tests in addition to logistic regression analysis were used to compare data derived from case and control children depending on data requirements. Statistical analyses were performed using Statistical Package for Social Sciences (SPSS software, version 11.0) and GraphPad Prism (GraphPad software, version 5.01). P-values of ≤0.05 were considered statistically significant.
Results
Characteristics of the study population
A total of 325 children were screened for enteric pathogens, 43 inhabitants of Gonçalves Dias and 282 of Parque Universitario. Children were from 295 houses, and 268 of those (90.8 %) had one child per house. Only two houses (0.7 %, one in each community) had more than three children. Study households had a median of 5.5 persons who slept in 1.9 rooms and lived in four compartments. Most dwellings were permanent structures built of brick and adobe (98.3 %), supplied with piped water (85.5 %), with an interior flush toilet (83.0 %). About 42.1 % of these families received up to US$ 175.00 per month.
Of the 325 children, 172 (52.9 %) were male and 219 (67.4 %) were more than 12 months of age [median 18 months; only 37 (11.4 %) were between 2 and 6 months of age]. Criteria for inclusion in the case group were met in 83 children (25.5 %). Age-matched controls were randomly selected among the other 242 children of the study. Thus, we analysed data from 83 case children and 83 controls.
Prevalence of EAEC
At least one of the two EAEC diagnostic genes analysed in this study was found in 34 (41.0 %) children of the case group. A similar prevalence of EAEC was obtained among children without diarrhoea (34/83, 41.0 %, P = 1.000, by Pearson’s chi-squared). In total, 68 samples were positive for EAEC and three different patterns of diagnostic genes were observed: (i) only positive for aaiC, (ii) only positive for aatA or (iii) positive for both aaiC and aatA. Of the two EAEC diagnostic genes, aaiC was the most common. It was detected in 33 (39.8 %) of 83 cases, of which 18 (21.7 %) children presented only this gene and 15 (18.1 %) were positive for both aaiC and aatA genes. Only one (1.2 %) case was positive only for the aatA gene. Fifteen (18.1 %) and three (3.6 %) children from the control group had positive PCR for aaiC or aatA, respectively, and 16 (19.3 %) children were positive for the two diagnostic genes. Neither was associated with the case or control groups, as shown in Table 2.
Table 2. Prevalence of enteroaggregative E. coli (EAEC) and its virulence-related genes in stool samples from case and control children.
The aaiC and aatA genes were used for EAEC diagnosis. Samples were considered positive for EAEC when they presented one or both genes. Comparison of gene frequencies between case and control groups was performed by chi-squared using 2×2 contingency tables. Fisher’s exact test was applied when the observed frequency was <5. The total number of samples (83 cases and 83 controls) was considered for analysis of diagnostic genes. For statistical tests of the virulence genes, only EAEC-positive samples were eligible (33 cases and 34 controls). One positive case did not contain a sufficient amount of DNA sample to perform the tests.
| EAEC genes | No. of cases (%) | No. of controls (%) | Total no. (%) | OR | Risk estimate (95 % CI) | χ2 | P-value |
| aaiC* | 33 (39.8) | 31 (37.3) | 64 (38.5) | 1.11 | 0.59–2.07 | 0.10 | 0.750 |
| aatA | 16 (19.3) | 19 (22.9) | 35 (21.1) | 0.80 | 0.38–1.70 | 0.33 | 0.568 |
| aggR | 17 (51.5) | 21 (61.8) | 38 (56.7) | 0.66 | 0.25–1.74 | 0.72 | 0.397 |
| aap | 18 (54.5) | 21 (61.8) | 39 (58.2) | 0.75 | 0.28–1.97 | 0.36 | 0.549 |
| orf3 | 18 (54.5) | 16 (47.1) | 34 (50.7) | 1.35 | 0.52–3.53 | 0.38 | 0.540 |
| sat | 10 (30.3) | 16 (47.1) | 26 (38.8) | 0.49 | 0.18–1.33 | 1.98 | 0.159 |
| sepA | 9 (27.3) | 7 (20.6) | 16 (23.9) | 1.45 | 0.47–4.48 | 0.42 | 0.521 |
| pic* | 7 (21.2) | 9 (26.5) | 16 (23.9) | 0.75 | 0.25–2.32 | 0.26 | 0.614 |
| sigA* | 1 (3.0) | 3 (8.8) | 4 (6.0) | 0.32 | 0.32–3.28 | − | 0.613 |
| pet | 6 (18.2) | 1 (2.9) | 7 (10.5) | 7.33 | 0.83–64.73 | − | 0.050† |
| astA | 15 (45.5) | 17 (50.0) | 32 (47.8) | 0.83 | 0.32–2.18 | 0.14 | 0.710 |
| aafC | 4 (12.1) | 1 (2.9) | 5 (7.5) | 4.55 | 0.48–43.10 | − | 0.197 |
| agg3/4C | 14 (42.4) | 13 (38.2) | 27 (40.3) | 1.19 | 0.45–3.16 | 0.12 | 0.727 |
| agg3A | 1 (3.0) | 0 (0.0) | 1 (1.5) | 3.19 | 0.13–81.08 | − | 0.492 |
| aafA | 4 (12.1) | 0 (0.0) | 4 (6.0) | 10.53 | 0.54–203.80 | − | 0.050† |
| aggA | 3 (9.1) | 8 (23.5) | 11 (16.4) | 0.33 | 0.78–1.36 | − | 0.186 |
| agg4A | 2 (6.1) | 9 (26.5) | 11 (16.4) | 0.18 | 0.03–0.91 | − | 0.045† |
| capU | 30 (90.9) | 30 (88.2) | 60 (89.6) | 1.33 | 0.27–6.48 | 0.13 | 0.721 |
| eilA* | 20 (60.6) | 17 (50.0) | 37 (57.8) | 1.54 | 0.58–4.06 | 0.76 | 0.383 |
| orf61 | 7 (21.2) | 18 (52.9) | 25 (37.3) | 0.24 | 0.08–0.70 | 7.21 | 0.007† |
EAEC chromosomal genes. All other genes are pAA-plasmid encoded.
P≤0.05.
Individual frequencies of VRGs
To investigate if other EAEC VRGs could be associated with disease, we performed four multiplex PCR assays targeting 18 genes of 33 cases and 34 controls positive for this bacterium. Individual frequencies of each gene are given in Table 2. Results from amplification reactions of chromosomal and pAA-encoded genes showed that all samples carried at least one of the 18 assayed VRGs. Several genotypes were found in this population. The plasmid-borne gene encoding the hexosyltransferase homologue capU was the most frequently detected (89.6 %), followed by aap (58.2 %), eilA (57.8 %), aggR (56.7 %) and hypothetical orf3 (50.7 %).
Regarding the AAF pilin, AAF/I and AAF/IV, encoded by aggA and agg4A genes, respectively, were the most frequent at 16.4 % each, followed by AAF/II (aafA, 6.0 %) and AAF/III (agg3A, 1.5 %), which was the least frequently detected among all 18 genes studied. The closely related usher for AAF/III and AAF/IV variants (agg3/4C) was detected in 40.3 % of samples. A total of 59.7 % of samples were negative for any described AAF and none was positive for more than one variant.
Among the genes encoding SPATEs, the frequencies of detection were: sat, 38.8 %; pic, 23.9 %; sepA, 23.9 %; pet, 10.5 %; and sigA, 6.0 %.
Considering the frequencies of VRGs between case and control groups, pet and aafA showed significant associations with diarrhoea (P = 0.054 and P = 0.053, respectively). In addition, two potential protective genes were identified: agg4A and hypothetical orf61. The first was detected in 6.1 % of cases versus 26.5 % of controls [P = 0.045, OR = 0.179 and 95 % confidence interval (CI) = 0.035–0.906] and orf61 was identified in 21.2 % of cases and 52.9 % of controls (P = 0.007, OR = 0.239, and 95 % CI = 0.082–0.699). In the logistic regression analysis, orf61 kept its protective role (P = 0.009, OR = 0.239 and 95 % CI = 0.082–0.699) (Table 2).
Combinations of VRGs
To investigate if a specific combination of VRGs could be correlated with disease, we employed CART analysis, which constructs a model in stepwise fashion and the outcome is a combination of factors most strongly associated with cases or controls.
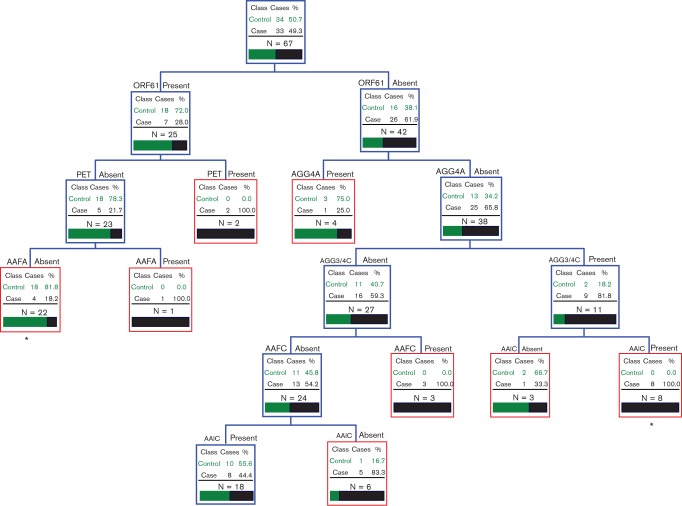
We analysed all genotypic assays of 34 controls and 33 cases. The best CART fit for the dataset is shown in Fig. 1. The analysis suggested two important trait clusters, which were associated with each studied group. Samples harbouring aaiC and agg3/4C but lacking agg4A and orf61 were associated with children with diarrhoea (P = 0.002, OR = 23.00 and 95 % CI = 1.267–417.4). In addition, the presence of orf61 in the absence of pet and aafA was associated with the control group (P = 0.0006, OR = 0.123 and 95 % CI = 0.0353–0.425).
Fig. 1.
CART model for VRGs in stool samples from case and control children positive for EAEC. The presence or absence of each gene between case and control groups is given inside each node. Terminal nodes are indicated by the red boxes. The tree is hierarchical in nature. *P≤0.05.
Characterization of diarrhoeal diseases and its correlation with EAEC
The median duration of diarrhoea in 83 cases was 4.2 days (range 1–18 days). Sick children with EAEC infection did not show differences in the median durations of diarrhoea in comparison with sick children without EAEC infection (4.3 versus 4.2 days, P = 0.752, by Mann–Whitney U test, data not shown). Similarly, no differences were seen in the peak number of liquid stools per day (4.0 versus 4.1, P = 0.635, by Mann–Whitney U test, data not shown).
Only three of 83 children (3.6 %) had diarrhoea lasting longer than 14 days (classified as persistent diarrhoea). Of these, 2/3 (66.7 %) were positive for EAEC. Of 80 cases of acute diarrhoea found in this study, 32 (40.0 %) were positive for this bacterium. The frequency of persistent diarrhoea did not differ between cases positive and negative for EAEC (P = 0.565, by Fisher’s exact test, data not shown).
Discussion
In this case–control study, we examined children 2–36 months of age living in resource-poor urban communities in Fortaleza, individuals who are particularly susceptible to infectious diarrhoea (Keusch et al., 2006). We employed PCR assays to diagnose EAEC directly from stool samples. This molecular technique enables the differentiation of micro-organisms that otherwise appear to be identical by identification of minor differences among the strains. It also identifies VRGs and their products, providing new perspectives for studying the epidemiology of diarrhoeal diseases (Keusch et al., 2006; Nataro & Kaper, 1998; Wright & Wynford-Thomas, 1990). The highly heterogeneous characteristics of EAEC are major obstacles for its efficient molecular diagnosis (Nataro & Kaper, 1998). A recently described conserved chromosomal locus, the aaiC gene, was chosen for the development of a specific bacterial primer pair as well as the pAA plasmid-encoded aatA. The aatA gene has been applied in the molecular identification of EAEC since 1990, when Baudry et al. (1990) published the first specific probe (CVD432) for EAEC. Neither gene has been identified among non-EAEC genomes deposited in GenBank (Baudry et al., 1990; Dudley et al., 2006).
The prevalence of EAEC in this population was high (41 %). In previous studies performed in industrialized countries, the prevalence of EAEC varied between 2 and 12 % (Chan et al., 1994; Cohen et al., 2005; Huppertz et al., 1997; Nataro et al., 2006; Usein et al., 2009). Although diarrhoeal disease is a less important cause of morbidity and mortality in industrialized countries than in developing countries, sporadic diarrhoeal diseases constitute the second most common infectious disease in industrialized areas, with 1–2 episodes per person per year. Besides, they are frequently associated with costly hospital admissions (Guerrant et al., 2001; Steiner et al., 2006). Studies carried out in England, Germany, USA and Romania implicated EAEC as a common bacterial cause of diarrhoea (Chan et al., 1994; Cohen et al., 2005; Huppertz et al., 1997; Nataro et al., 2006; Usein et al., 2009). In developing regions, the percentage of EAEC among diarrhoea cases ranged from 4.5 to 39 % (Meng et al., 2011; Ochoa et al., 2011; Okeke et al., 2000; Rúgeles et al., 2010). In Brazil, reports found a prevalence of EAEC between 11 and 45 % (Bueris et al., 2007; Pereira et al., 2007; Piva et al., 2003; Scaletsky et al., 2002), consistent with the percentage observed in this work.
In the present study, we detected similar EAEC distribution between diarrhoea and non-diarrhoea groups. Similar findings have been made by other researchers (Bueris et al., 2007; Piva et al., 2003). In previous work by our group, studying children from an urban community in Fortaleza, we found a prevalence of EAEC in 38 and 34.8 % of children with acute and persistent diarrhoea, respectively (≥14 days of duration), and in 34.8 % of children representing controls (Lima et al., 2000). We therefore hypothesized that some VRGs or a specific combination of them might be found in EAEC from Fortaleza and possibly be associated with enteric infection symptoms and their risks for dehydration and/or malnutrition, consequences of the EAEC infection previously described (Steiner et al., 1998).
In the last two decades, several bacterial putative VRGs have been suggested to influence the course of EAEC enteric infection (Nataro & Kaper, 1998). This work explored 18 candidate EAEC VRGs in a childhood population living in a poor urban community with a very high prevalence for EAEC colonization and diarrhoeal diseases (Lima et al., 2000). In agreement with previous reports, our samples harboured a diverse range and combination of VRGs. Genes encoding Pet and AAF/II were individually associated with diarrhoea; agg4A (AAF/IV) and the hypothetical orf61 genes were detected significantly more often in children without diarrhoea. Other reports have not found a correlation between the pet gene and the occurrence of diarrhoeal disease (Huang et al., 2007; Pereira et al., 2007; Piva et al., 2003; Regua-Mangia et al., 2009). However, studies in vitro showed that this cytotoxin induces modification in the cytoskeleton followed by cell rounding and detachment of cell monolayers in culture (Betancourt-Sanchez & Navarro-Garcia, 2009). The importance of AAF/II in EAEC pathogenesis was reported by Okeke et al. (2000), which suggested the possibility of using it as a reference marker to identify potentially pathogenic EAEC. Nevertheless, results obtained by Regua-Mangia et al. (2009), analysing EAEC strains isolated from Brazilian children, did not support this suggestion. Surprisingly, the newly described AAF/IV (Boisen et al., 2008) was the most frequently detected AAF, with a similar percentage to that of AAF/I. Furthermore, AAF/IV was associated with control children, data so far not described. The increased number of samples negative for any known AAF strengthens the wide diversity of EAEC adhesive structures, which include non-fimbrial and uncharacterized fimbrial adhesins. The recently identified orf61 was first investigated by Boisen et al. (2012). Similar to our findings, the authors detected its significant association with control children.
The terms ‘typical’ and ‘atypical’ EAEC have been suggested to classify EAEC strains harbouring or lacking the aggR regulon, respectively (Kaper et al., 2004). In this study the aggR gene was found only in 56.7 % of the EAEC-positive samples, and it was not associated with disease. Intriguingly, this regulatory gene controls a number of plasmid genes encoding virulence factors, in addition to pathogenicity islands in the EAEC chromosome involved in several steps of its pathogenesis (Nataro & Kaper, 1998; Okeke & Nataro, 2001). Correlations between aggR and diarrhoea are not consistent. While some authors found significant differences in aggR alone or in combination with cases compared with controls (Huang et al., 2007; Sarantuya et al., 2004), others did not observe any correlation (Boisen et al., 2012; Regua-Mangia et al., 2009). The lack of aggR in the majority of EAEC-positive samples also showed that this gene may not be a good marker to diagnose EAEC samples as suggested by several reports (Cerna et al., 2003; Samie et al., 2007; Sarantuya et al., 2004). Furthermore, we found samples that did not have the aggR gene but were positive for some genes under its control and vice versa. Similar findings were described by Bouzari et al. (2005) and Boisen et al. (2012). These data could be explained by the presence of mutated plasmids in our wild-type strains, and/or by the mosaic nature of EAEC genomes.
CART analysis suggested a trait cluster that indicates virulent EAEC, and another one that is associated with less-pathogenic EAEC. In a recent study by Boisen et al. (2012), characterizing EAEC strains isolated as part of a case–control study of diarrhoea among children in Mali, the investigators identified two gene combinations correlated with diarrhoea: a group harbouring the EAEC heat-stable toxin 1 and the flagellar type H33, and a group carrying several of the typical EAEC VRGs. However, this report did not detect any set of EAEC genes associated with controls.
The variation of EAEC VRGs found in our samples, as well as from different populations of distinct geographical regions of the world, confirmed the genetic heterogeneity of this organism. As most of the VRGs are encoded on the plasmid, this may help to explain the dynamic horizontal acquisition and loss of these VRGs, which favour the variety of genetic profiles found in these EAEC strains (Czeczulin et al., 1999).
In conclusion, this study has demonstrated a high prevalence of EAEC in children living in a resource-poor urban community of north-eastern Brazil. The overall prevalence of EAEC-positive samples in the diarrhoea group was similar to that of the non-diarrhoea control group. The frequency of 18 different VRGs among these children confirmed the genetic heterogeneity of EAEC. Identification of trait clusters (isolated genes or in combination) correlated with both sick (pet and aafA) and healthy children (agg4A and orf61) suggests that the pathophysiology of this enteric infection involves a complex and dynamic modulation of several VRGs. Further efforts are being directed to elucidate if the expressed proteins encoded by the studied VRGs play a role in EAEC infection outcome and pathogenesis.
Acknowledgements
This study was supported by the Fogarty International Center (grants 3D43TW006578-04S1, 3D43TW006578-05S1 and 5R24TW007988) and the National Institute of Allergy and Infectious Diseases (NIAID grants 2U01AI026512 and AI-033096), US National Institutes of Health (NIH); and the National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grants 152098/2008-9 and 140242/2009-0) and the Foundation for Support to Scientific and Technological Development from the Ceara government (Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico, FUNCAP grant BCP-0055-00042.01.01/11) of Brazil. We thank field workers Sayonara Alencar, Maria Luzia Melo, Rosânia Silva and Lucia de Fátima Alves; laboratory technicians Maria do Carmo Pinho, Verônica Oliveira, Conceição Nogueira and Leah Barrett; the data management team, José Quirino da Silva Filho, Francisco Sousa Júnior and Charles Melo; and ethics coordinator Fabiana Nascimento. We also thank the participating children and their families.
Abbreviations:
- AAF
aggregative adherence fimbria
- aai
aggR-activated island
- aap
anti-aggregation protein gene
- aatA
EAEC ABC transporter A gene
- aggR
activator aggregative adherence regulator gene
- CART
classification and regression tree
- CI
confidence interval
- EAEC
enteroaggregative Escherichia coli
- OR
odds ratio
- pet
plasmid-encoded toxin gene
- sat
secreted autotransporter toxin gene
- SPATE
serine protease autotransporter of Enterobacteriaceae
- sigA
Shigella IgA-like protease homologue gene
- VRG
virulence-related gene.
References
- Baudry B., Savarino S. J., Vial P., Kaper J. B., Levine M. M. (1990). A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J Infect Dis 161, 1249–1251 10.1093/infdis/161.6.1249 [DOI] [PubMed] [Google Scholar]
- Benjelloun-Touimi Z., Sansonetti P. J., Parsot C. (1995). SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol 17, 123–135 10.1111/j.1365-2958.1995.mmi_17010123.x [DOI] [PubMed] [Google Scholar]
- Bernier C., Gounon P., Le Bouguénec C. (2002). Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun 70, 4302–4311 10.1128/IAI.70.8.4302-4311.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt-Sanchez M., Navarro-Garcia F. (2009). Pet secretion, internalization and induction of cell death during infection of epithelial cells by enteroaggregative Escherichia coli. Microbiology 155, 2895–2906 10.1099/mic.0.029116-0 [DOI] [PubMed] [Google Scholar]
- Boisen N., Struve C., Scheutz F., Krogfelt K. A., Nataro J. P. (2008). New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun 76, 3281–3292 10.1128/IAI.01646-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisen N., Ruiz-Perez F., Scheutz F., Krogfelt K. A., Nataro J. P. (2009). Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg 80, 294–301 [PMC free article] [PubMed] [Google Scholar]
- Boisen N., Scheutz F., Rasko D. A., Redman J. C., Persson S., Simon J., Kotloff K. L., Levine M. M., Sow S., et al. (2012). Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 205, 431–444 10.1093/infdis/jir757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzari S., Jafari A., Zarepour M. (2005). Distribution of virulence related genes among enteroaggregative Escherichia coli isolates: using multiplex PCR and hybridization. Infect Genet Evol 5, 79–83 10.1016/j.meegid.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Bueris V., Sircili M. P., Taddei C. R., dos Santos M. F., Franzolin M. R., Martinez M. B., Ferrer S. R., Barreto M. L., Trabulsi L. R. (2007). Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem Inst Oswaldo Cruz 102, 839–844 10.1590/S0074-02762007005000116 [DOI] [PubMed] [Google Scholar]
- Carvalho E. B., Maga E. A., Quetz J. S., Lima I. F. N., Magalhães H. Y. F., Rodrigues F. A. R., Silva A. V. A., Prata M. M. G., Cavalcante P. A., et al. (2012). Goat milk with and without increased concentrations of lysozyme improves repair of intestinal cell damage induced by enteroaggregative Escherichia coli. BMC Gastroenterol 12, 106 10.1186/1471-230X-12-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerna J. F., Nataro J. P., Estrada-Garcia T. (2003). Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol 41, 2138–2140 10.1128/JCM.41.5.2138-2140.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. N., Phillips A. D., Knutton S., Smith H. R., Walker-Smith J. A. (1994). Enteroaggregative Escherichia coli: another cause of acute and chronic diarrhoea in England? J Pediatr Gastroenterol Nutr 18, 87–91 10.1097/00005176-199401000-00015 [DOI] [PubMed] [Google Scholar]
- Chaudhuri R. R., Sebaihia M., Hobman J. L., Webber M. A., Leyton D. L., Goldberg M. D., Cunningham A. F., Scott-Tucker A., Ferguson P. R., et al. (2010). Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS ONE 5, e8801 10.1371/journal.pone.0008801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66, 4555–4558 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. B., Nataro J. P., Bernstein D. I., Hawkins J., Roberts N., Staat M. A. (2005). Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J Pediatr 146, 54–61 10.1016/j.jpeds.2004.08.059 [DOI] [PubMed] [Google Scholar]
- Czeczulin J. R., Balepur S., Hicks S., Phillips A., Hall R., Kothary M. H., Navarro-Garcia F., Nataro J. P. (1997). Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun 65, 4135–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeczulin J. R., Whittam T. S., Henderson I. R., Navarro-Garcia F., Nataro J. P. (1999). Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun 67, 2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Quetz J., Lima I. F. N., Havt A., de Carvalho E. B., Lima N. L., Soares A. M., Mota R. M. S., Guerrant R. L., Lima A. A. M. (2010). Campylobacter jejuni and Campylobacter coli in children from communities in Northeastern Brazil: molecular detection and relation to nutritional status. Diagn Microbiol Infect Dis 67, 220–227 10.1016/j.diagmicrobio.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E. G., Thomson N. R., Parkhill J., Morin N. P., Nataro J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol 61, 1267–1282 10.1111/j.1365-2958.2006.05281.x [DOI] [PubMed] [Google Scholar]
- Estrada-Garcia T., Navarro-Garcia F. (2012). Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol 66, 281–298 10.1111/j.1574-695X.2012.01008.x [DOI] [PubMed] [Google Scholar]
- Façanha M. C., Pinheiro A. C. (2005). Acute diarrhea treated by health care services in Fortaleza, Ceará State, Brazil, from 1996 to 2001. Cad Saude Publica 21, 49–54 (in Portuguese). 10.1590/S0102-311X2005000100006 [DOI] [PubMed] [Google Scholar]
- Frank C., Werber D., Cramer J. P., Askar M., Faber M., an der Heiden M., Bernard H., Fruth A., Prager R., et al. (2011). Epidemic profile of Shiga-toxin-producing Escherichia coli O104 : H4 outbreak in Germany. N Engl J Med 365, 1771–1780 10.1056/NEJMoa1106483 [DOI] [PubMed] [Google Scholar]
- Fujiyama R., Nishi J., Imuta N., Tokuda K., Manago K., Kawano Y. (2008). The shf gene of a Shigella flexneri homologue on the virulent plasmid pAA2 of enteroaggregative Escherichia coli 042 is required for firm biofilm formation. Curr Microbiol 56, 474–480 10.1007/s00284-008-9115-y [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., Van Gilder T., Steiner T. S., Thielman N. M., Slutsker L., Tauxe R. V., Hennessy T., Griffin P. M., DuPont H., et al. (2001). Practice guidelines for the management of infectious diarrhea. Clin Infect Dis 32, 331–351 10.1086/318514 [DOI] [PubMed] [Google Scholar]
- Guyer D. M., Henderson I. R., Nataro J. P., Mobley H. L. (2000). Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol 38, 53–66 10.1046/j.1365-2958.2000.02110.x [DOI] [PubMed] [Google Scholar]
- Harrington S. M., Sheikh J., Henderson I. R., Ruiz-Perez F., Cohen P. S., Nataro J. P. (2009). The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun 77, 2465–2473 10.1128/IAI.01494-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., Czeczulin J., Eslava C., Noriega F., Nataro J. P. (1999). Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun 67, 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. B., Nataro J. P., DuPont H. L., Kamat P. P., Mhatre A. D., Okhuysen P. C., Chiang T. (2006). Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin Infect Dis 43, 556–563 10.1086/505869 [DOI] [PubMed] [Google Scholar]
- Huang D. B., Mohamed J. A., Nataro J. P., DuPont H. L., Jiang Z. D., Okhuysen P. C. (2007). Virulence characteristics and the molecular epidemiology of enteroaggregative Escherichia coli isolates from travellers to developing countries. J Med Microbiol 56, 1386–1392 10.1099/jmm.0.47161-0 [DOI] [PubMed] [Google Scholar]
- Huppertz H. I., Rutkowski S., Aleksic S., Karch H. (1997). Acute and chronic diarrhoea and abdominal colic associated with enteroaggregative Escherichia coli in young children living in western Europe. Lancet 349, 1660–1662 10.1016/S0140-6736(96)12485-5 [DOI] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. T. (2004). Pathogenic Escherichia coli. Nat Rev Microbiol 2, 123–140 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Kermani N. A., Jafari F., Mojarad H. N., Hoseinkhan N., Zali R. (2010). Prevalence and associated factors of persistent diarrhoea in Iranian children admitted to a paediatric hospital. East Mediterr Health J 16, 831–836 [PubMed] [Google Scholar]
- Keusch G. T., Fontaine O., Bhargava A., Boschi-Pinto C., Bhutta Z. A., Gotuzzo E., Rivera J., Chow J., Shahid-Salles S. A., Laxminarayan R. (2006). Diarrheal diseases. In Disease Control Priorities in Developing Countries, 2nd edn, pp. 371–387 Edited by Jamison D. T., Breman J. G., Measham A. R., Alleyne G., Claeson M., Evans D. B., Jha P., Mills A., Musgrov P. New York: Oxford University Press [Google Scholar]
- Levine M. M., Bergquist E. J., Nalin D. R., Waterman D. H., Hornick R. B., Young C. R., Sotman S. (1978). Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1, 1119–1122 10.1016/S0140-6736(78)90299-4 [DOI] [PubMed] [Google Scholar]
- Lima A. A. M., Moore S. R., Barboza M. S., Jr, Soares A. M., Schleupner M. A., Newman R. D., Sears C. L., Nataro J. P., Fedorko D. P., et al. (2000). Persistent diarrhea signals a critical period of increased diarrhea burdens and nutritional shortfalls: a prospective cohort study among children in northeastern Brazil. J Infect Dis 181, 1643–1651 10.1086/315423 [DOI] [PubMed] [Google Scholar]
- Mathewson J. J., Oberhelman R. A., Dupont H. L., Javier de la Cabada F., Garibay E. V. (1987). Enteroadherent Escherichia coli as a cause of diarrhea among children in Mexico. J Clin Microbiol 25, 1917–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C. Y., Smith B. L., Bodhidatta L., Richard S. A., Vansith K., Thy B., Srijan A., Serichantalergs O., Mason C. J. (2011). Etiology of diarrhea in young children and patterns of antibiotic resistance in Cambodia. Pediatr Infect Dis J 30, 331–335 10.1097/INF.0b013e3181fb6f82 [DOI] [PubMed] [Google Scholar]
- Mohamed J. A., Huang D. B., Jiang Z. D., DuPont H. L., Nataro J. P., Belkind-Gerson J., Okhuysen P. C. (2007). Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol 45, 121–126 10.1128/JCM.01128-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin N., Tirling C., Ivison S. M., Kaur A. P., Nataro J. P., Steiner T. S. (2010). Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol Med Microbiol 58, 344–355 [DOI] [PubMed] [Google Scholar]
- Muniesa M., Hammerl J. A., Hertwig S., Appel B., Brüssow H. (2012). Shiga toxin-producing Escherichia coli O104 : H4: a new challenge for microbiology. Appl Environ Microbiol 78, 4065–4073 10.1128/AEM.00217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B. (1998). Diarrheagenic Escherichia coli. Clin Microbiol Rev 11, 142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Baldini M. M., Kaper J. B., Black R. E., Bravo N., Levine M. M. (1985). Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J Infect Dis 152, 560–565 10.1093/infdis/152.3.560 [DOI] [PubMed] [Google Scholar]
- Nataro J. P., Kaper J. B., Robins-Browne R., Prado V., Vial P., Levine M. M. (1987). Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J 6, 829–831 10.1097/00006454-198709000-00008 [DOI] [PubMed] [Google Scholar]
- Nataro J. P., Deng Y., Maneval D. R., German A. L., Martin W. C., Levine M. M. (1992). Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun 60, 2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J. P., Mai V., Johnson J., Blackwelder W. C., Heimer R., Tirrell S., Edberg S. C., Braden C. R., Glenn Morris J., Jr, Hirshon J. M. (2006). Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis 43, 402–407 10.1086/505867 [DOI] [PubMed] [Google Scholar]
- Navarro-García F., Eslava C., Villaseca J. M., López-Revilla R., Czeczulin J. R., Srinivas S., Nataro J. P., Cravioto A. (1998). In vitro effects of a high-molecular-weight heat-labile enterotoxin from enteroaggregative Escherichia coli. Infect Immun 66, 3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi J., Sheikh J., Mizuguchi K., Luisi B., Burland V., Boutin A., Rose D. J., Blattner F. R., Nataro J. P. (2003). The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J Biol Chem 278, 45680–45689 10.1074/jbc.M306413200 [DOI] [PubMed] [Google Scholar]
- Ochoa T. J., Mercado E. H., Durand D., Rivera F. P., Mosquito S., Contreras C., Riveros M., Lluque A., Barletta F., et al. (2011). Frequency and pathotypes of diarrheagenic Escherichia coli in Peruvian children with and without diarrhea. Rev Peru Med Exp Salud Publica 28, 13–20 (in Spanish). 10.1590/S1726-46342011000100003 [DOI] [PubMed] [Google Scholar]
- Okeke I. N., Nataro J. P. (2001). Enteroaggregative Escherichia coli. Lancet Infect Dis 1, 304–313 10.1016/S1473-3099(01)00144-X [DOI] [PubMed] [Google Scholar]
- Okeke I. N., Lamikanra A., Czeczulin J., Dubovsky F., Kaper J. B., Nataro J. P. (2000). Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J Infect Dis 181, 252–260 10.1086/315204 [DOI] [PubMed] [Google Scholar]
- Olesen B., Neimann J., Böttiger B., Ethelberg S., Schiellerup P., Jensen C., Helms M., Scheutz F., Olsen K. E. P., et al. (2005). Etiology of diarrhea in young children in Denmark: a case-control study. J Clin Microbiol 43, 3636–3641 10.1128/JCM.43.8.3636-3641.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opintan J. A., Newman M. J., Ayeh-Kumi P. F., Affrim R., Gepi-Attee R., Sevilleja J. E. A. D., Roche J. K., Nataro J. P., Warren C. A., Guerrant R. L. (2010). Pediatric diarrhea in southern Ghana: etiology and association with intestinal inflammation and malnutrition. Am J Trop Med Hyg 83, 936–943 10.4269/ajtmh.2010.09-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschke C., Apelt N., Fleischmann E., Perona P., Walentiny C., Löscher T., Herbinger K.-H. (2011). Controlled study on enteropathogens in travellers returning from the tropics with and without diarrhoea. Clin Microbiol Infect 17, 1194–1200 10.1111/j.1469-0691.2010.03414.x [DOI] [PubMed] [Google Scholar]
- Pereira A. L., Ferraz L. R., Silva R. S., Giugliano L. G. (2007). Enteroaggregative Escherichia coli virulence markers: positive association with distinct clinical characteristics and segregation into 3 enteropathogenic E. coli serogroups. J Infect Dis 195, 366–374 10.1086/510538 [DOI] [PubMed] [Google Scholar]
- Piva I. C., Pereira A. L., Ferraz L. R., Silva R. S. N., Vieira A. C., Blanco J. E., Blanco M., Blanco J., Giugliano L. G. (2003). Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasília, Brazil. J Clin Microbiol 41, 1827–1832 10.1128/JCM.41.5.1827-1832.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar K., Sasakawa C., Adler B. (1997). Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun 65, 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regua-Mangia A. H., Gomes T. A. T., Vieira M. A. M., Irino K., Teixeira L. M. (2009). Molecular typing and virulence of enteroaggregative Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro city, Brazil. J Med Microbiol 58, 414–422 10.1099/jmm.0.006502-0 [DOI] [PubMed] [Google Scholar]
- Restieri C., Garriss G., Locas M. C., Dozois C. M. (2007). Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol 73, 1553–1562 10.1128/AEM.01542-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche J. K., Cabel A., Sevilleja J., Nataro J. P., Guerrant R. L. (2010). Enteroaggregative Escherichia coli (EAEC) impairs growth while malnutrition worsens EAEC infection: a novel murine model of the infection malnutrition cycle. J Infect Dis 202, 506–514 10.1086/654894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rúgeles L. C., Bai J., Martínez A. J., Vanegas M. C., Gómez-Duarte O. G. (2010). Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int J Food Microbiol 138, 282–286 10.1016/j.ijfoodmicro.2010.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie A., Obi C. L., Dillingham R., Pinkerton R. C., Guerrant R. L. (2007). Enteroaggregative Escherichia coli in Venda, South Africa: distribution of virulence-related genes by multiplex polymerase chain reaction in stool samples of human immunodeficiency virus (HIV)-positive and HIV-negative individuals and primary school children. Am J Trop Med Hyg 77, 142–150 [PubMed] [Google Scholar]
- Sarantuya J., Nishi J., Wakimoto N., Erdene S., Nataro J. P., Sheikh J., Iwashita M., Manago K., Tokuda K., et al. (2004). Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol 42, 133–139 10.1128/JCM.42.1.133-139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino S. J., Fasano A., Watson J., Martin B. M., Levine M. M., Guandalini S., Guerry P. (1993). Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci U S A 90, 3093–3097 10.1073/pnas.90.7.3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletsky I. C., Fabbricotti S. H., Silva S. O., Morais M. B., Fagundes-Neto U. (2002). HEp-2-adherent Escherichia coli strains associated with acute infantile diarrhea, São Paulo, Brazil. Emerg Infect Dis 8, 855–858 10.3201/eid0808.010492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavia G., Staffolani M., Fisichella S., Striano G., Colletta S., Ferri G., Escher M., Minelli F., Caprioli A. (2008). Enteroaggregative Escherichia coli associated with a foodborne outbreak of gastroenteritis. J Med Microbiol 57, 1141–1146 10.1099/jmm.0.2008/001362-0 [DOI] [PubMed] [Google Scholar]
- Scheutz F., Nielsen E. M., Frimodt-Møller J., Boisen N., Morabito S., Tozzoli R., Nataro J. P., Caprioli A. (2011). Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104 : H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill 16, 19889 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19889 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Knop C., Franke S., Aleksic S., Heesemann J., Karch H. (1995). Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol 33, 701–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J., Czeczulin J. R., Harrington S., Hicks S., Henderson I. R., Le Bouguénec C., Gounon P., Phillips A., Nataro J. P. (2002). A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest 110, 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J., Dudley E. G., Sui B., Tamboura B., Suleman A., Nataro J. P. (2006). EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol Microbiol 61, 338–350 10.1111/j.1365-2958.2006.05234.x [DOI] [PubMed] [Google Scholar]
- Steiner T. S., Lima A. A. M., Nataro J. P., Guerrant R. L. (1998). Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J Infect Dis 177, 88–96 10.1086/513809 [DOI] [PubMed] [Google Scholar]
- Steiner T. S., Samie A., Guerrant R. L. (2006). Infectious diarrhea: new pathogens and new challenges in developed and developing areas. Clin Infect Dis 43, 408–410 10.1086/505874 [DOI] [PubMed] [Google Scholar]
- Strauman M. C., Harper J. M., Harrington S. M., Boll E. J., Nataro J. P. (2010). Enteroaggregative Escherichia coli disrupts epithelial cell tight junctions. Infect Immun 78, 4958–4964 10.1128/IAI.00580-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usein C. R., Tatu-Chitoiu D., Ciontea S., Condei M., Damian M. (2009). Escherichia coli pathotypes associated with diarrhea in Romanian children younger than 5 years of age. Jpn J Infect Dis 62, 289–293 [PubMed] [Google Scholar]
- Wright P. A., Wynford-Thomas D. (1990). The polymerase chain reaction: miracle or mirage? A critical review of its uses and limitations in diagnosis and research. J Pathol 162, 99–117 10.1002/path.1711620203 [DOI] [PubMed] [Google Scholar]