Abstract
Genomic and antigenic characterization of the Salehabad virus, a species of the genus Phlebovirus, and four other unclassified phleboviruses (Arbia, Adria, Arumowot and Odrenisrou) demonstrate a serological and genetic relation to one another and are distinct from the eight other recognized species within the genus Phlebovirus. We propose to incorporate these four unclassified viruses as part of the Salehabad species complex within the genus. The known geographical distribution for the members of this species group includes southern Europe, Central Asia and Africa.
The family Bunyaviridae comprises five genera: Hantavirus, Nairovirus, Orthobunyavirus, Phlebovirus and Tospovirus that are differentiated by antigenic and molecular characteristics (Nichol et al., 2005). Viruses in the genus Phlebovirus are further subdivided into two groups: the Sandfly fever and the Uukuniemi groups, based on their antigenic relationships, arthropod vectors and the presence or absence of a non-structural protein in the M segment (Nichol et al., 2005). Phlebotomine sandflies and mosquitoes transmit viruses in the Sandfly fever group, while ticks transmit Uukuniemi group viruses. A number of the phleboviruses are associated with human illness and at least one, Rift Valley fever virus, is a major veterinary pathogen (Peters et al., 2011; Tesh, 1989; Yu et al., 2011).
Because of the paucity of genetic data for most of the named phleboviruses, the International Committee on Taxonomy of Viruses defines species within the genus Phlebovirus by their antigenic relationships (Nichol et al., 2005). Cross-complement fixation (CF) and plaque reduction neutralization tests have generally been used for the classification of viruses in this genus (Tesh et al., 1976, 1982; Travassos da Rosa et al., 1983). The present communication is the fourth report in our effort to develop a more precise classification system for the phleboviruses by sequencing available viruses in the genus and reassessing their antigenic relatedness in order to clarify their phylogenetic relationships (Palacios et al., 2011a, b; 2012). Here, we describe the genetic and antigenic relatedness of another of those recognized species groups, Salehabad, that contains Salehabad (SALV), Arbia (ARBV), Adria (ADRV), Arumowot (AMTV) and Odrenisrou (ODRV) viruses.
The phlebovirus strains used in this study were AMTV strain Ar 1286-64; SALV, strain I-81; ARBV, strain ISS. Phl.18; and ODRV strain ArA 1131/80. ADRV was not available for study, as noted before. Stocks of AMTV, SALV, ARBV and ODRV were obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch, USA.
AMTV was originally isolated in 1963 from Culex antennatus mosquitoes collected in Sudan. Subsequently, it has been isolated from mosquitoes and rodents throughout Africa. Although antibodies to AMTV have been reported in serosurveys in African human populations (Tesh et al., 1976), there is no definitive evidence that it causes human illness.
SALV was isolated from a pool of sandflies (Phlebotomus spp.) collected in a rural village in central Iran in 1959. No subsequent recoveries of this virus have been made and it is unknown whether it is associated with illness in humans or domestic animals.
ARBV was originally isolated from a pool of Phlebotomus perniciosus collected near Toscana, Italy in 1980 (Verani et al., 1988). Subsequent recoveries of the virus have been made in the same region from other sandfly species, but ARBV has not yet been associated with human illness.
ADRV has not yet been isolated. Partial RNA of a novel phlebovirus, related to ARBV was first detected in two pools of phlebotomine sandflies (species unknown) collected on the Adriatic coast of Albania in 2005 (Papa et al., 2011). The same RNA sequence was subsequently detected in the blood of a child hospitalized with a febrile illness and seizure in northern Greece in 2009 (Anagnostou et al., 2011).
ODRV was isolated from a single pool of Culex albiventris mosquitoes collected in the Tai forest, Ivory Coast in 1980 (CDC, 2013; http://wwwn.cdc/gov/arbocat/catalog-listing.asp?VirusID=342&S1=1). It has not yet been associated with human disease.
Methods used to prepare antigens and antibodies (immune ascitic fluids) have been described previously (Beaty et al., 1989; Travassos da Rosa et al., 1983; Xu et al., 2007). All immunizing antigens and antibodies were prepared in mice. Antigens for serological tests were prepared from infected newborn mouse brain (AMTV and ODRV), or spent medium from infected Vero cell cultures (SALV and ARBV) was used. CF tests were performed by the microtitre technique (Beaty et al., 1989; Xu et al., 2007), using two units of guinea pig complement and overnight incubation of the antigen and antibody at 4 °C. CF titres were recorded as the highest dilutions giving 3+ or 4+ fixation of complement. Haemagglutination-inhibition (HI) tests were done in microtitre plates, as described previously (Travassos da Rosa et al., 1983). HI tests were performed with 4 U haemagglutination of virus at the optimal pH (5.75) against serial twofold antiserum dilutions starting at 1 : 20. HI titres of 1 : 20 or greater were considered positive. All serological tests were performed at the University of Texas Medical Branch, Galveston, USA.
Results obtained in HI and CF tests with SALV, ARBV, AMTV and ODRV are summarized in Table 1. Since ADRV has not yet been isolated and is known only from a partial sequence of its large (L) RNA segment, it was not included in the serological tests. The antigens of SALV, ARBV, AMTV and ODRV were reactive with their homologous antisera. In CF tests, SALV and ARBV antigens and antisera were highly cross-active, as were AMTV and ODRV; in fact, the four viruses formed two distinct serological groups by this method. In HI tests, the two serological groupings were again observed, although ARBV antigen reacted with AMTV and ODRV antibody and ARBV antigen reacted with AMTV and ODRV antigens. On the basis of these cross-reactions, we feel that the four viruses constitute a single (Salehabad) antigenic complex.
Table 1. Salehabad antigenic complex: results of CF and HI tests.
| Antibody | CF test | HI test | ||||||
| Antigen | Antigen 4u. | |||||||
| SALV | ARBV | AMTV | ODRV | SALV | ARBV | AMTV | ODRV | |
| Salehabad | 256/≥Φ | ≥256/≥Φ | 0 | 0 | 1 : 640 | 1 : 80 | 0 | 0 |
| Arbia | 128/≥Φ | 512/≥Φ | 0 | 0 | 1 : 640 | 1 : 1280 | 1 : 80 | 1 : 80 |
| Arumowot | 0 | 0 | 64/128 | 32/8 | 0 | 1 : 80 | ≥1 : 640 | 1 : 320 |
| Odrenisrou | 0 | 0 | 16/16 | 128/16 | – | 1 : 80 | 1 : 160 | 1 : 640 |
CF titres are expressed as highest antibody/highest antigen dilution. 0 = <8/Φ.
Undiluted.
+HI titres are expressed as highest antibody dilution. 0 = <1 : 20.
For genome sequencing, viral stocks were extracted using TRIzol LS (Invitrogen). Total RNA extracts were treated with DNase I (DNA-Free; Ambion). cDNA was generated using the Superscript II system (Invitrogen) employing random hexamers linked to an arbitrary 17-mer primer sequence (Palacios et al., 2007). Resulting cDNA was treated with RNase H and then randomly amplified by PCR with a 9 : 1 mixture of primer corresponding to the 17-mer sequence and the random hexamer linked 17-mer primer (Palacios et al., 2007). Products greater than 70 bp were selected by column chromatography (MinElute; Qiagen) and ligated to specific adapters for sequencing on the 454 Genome Sequencer FLX (454 Life Sciences) without fragmentation (Cox-Foster et al., 2007; Margulies et al., 2005; Palacios et al., 2008). Software programs accessible through the analysis applications at the GreenePortal website (https://tako.cpmc.columbia.edu/Tools/) were used for the removal of primer sequences, redundancy filtering and sequence assembly. Sequence gaps were completed by PCR, using primers based on pyrosequencing data. Amplification products were size-fractionated on 1 % agarose gels, purified (MiniElute; Qiagen) and directly sequenced in both directions with ABI Prism Big Dye Terminator 1.1 Cycle Sequencing kits on ABI Prism 3700 DNA Analysers (Perkin-Elmer Applied Biosystems). For the termini of each segment, a primer with the 8 nt conserved sequence was used for a specific reverse transcription with additional arbitrary nucleotide on the 5′ end (5′-AAGCAGTGGTATCAACGCAGAGTACACACAAAG-3′; the bold portion is the conserved nucleotides). This primer is designed to bind to the 3′ end of the genomic RNA and the 3′ end of the mRNA. Sequences of the genomes were verified by classical dideoxy sequencing using primers designed to create products of 1000 bp with 500 bp overlap from the draft sequence.
SALV, ARBV, AMTV and ODRV all have the genomic organization characteristic of phleboviruses: three RNA segments including a L segment encoding the RNA polymerase, a medium (M) segment encoding a polyprotein that includes the non-structural protein (NSm) and both glycoproteins (GN and GC), and a small (S) segment encoding the N protein (NP) and, in ambisense orientation, a non-structural protein (NSs) (GenBank accession nos HM566143–HM566145, HM566173–HM566173, JX472400–JX472405).
All phlebovirus sequences available from GenBank (March 2012) was used to determine the phylogenic relationships of SALV, ARBV, AMTV and ODRV. All sequences were aligned using the clustal algorithm (as implemented in the mega package version 3) at the amino acid level with additional manual editing to ensure the highest possible quality of alignment. Neighbour-joining analysis at the amino acid level was performed due to the observed high variability of the underlying nucleotide sequences. Nucleotide trees were also achieved by using neighbour-joining analysis and the Kimura 2-paramater model. The statistical significance of tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Phylogenetic analysis was performed by using the mega software (Kumar et al., 2004).
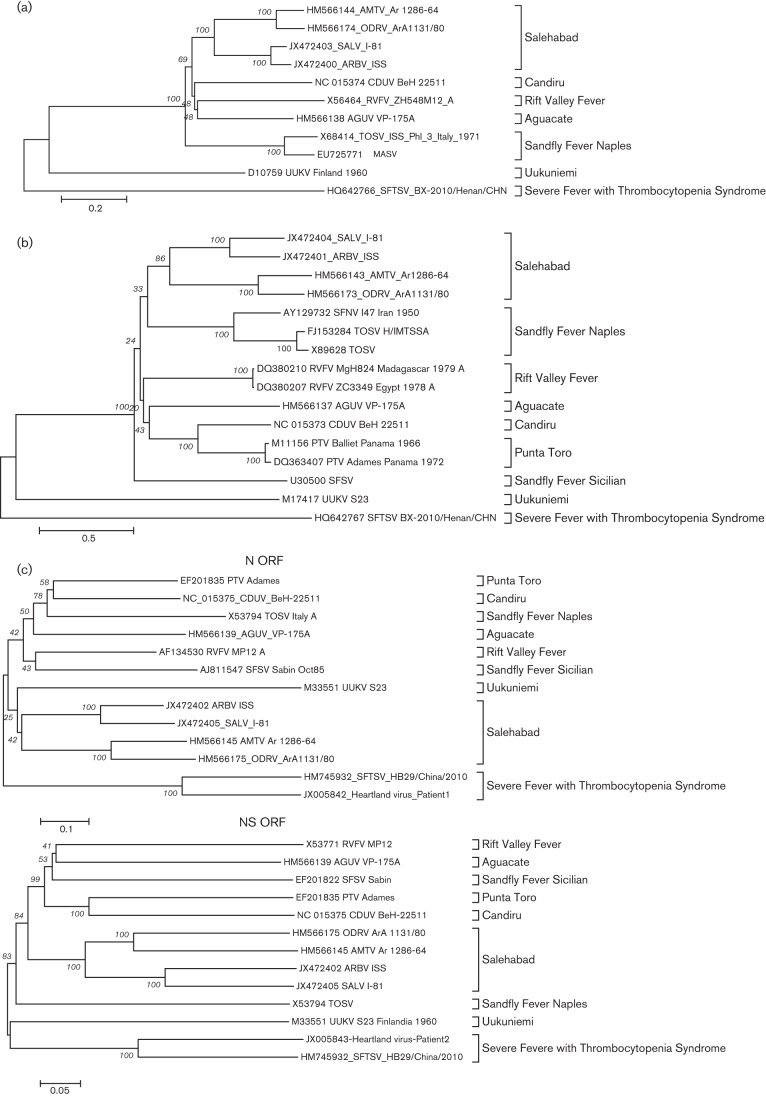
In phylogenetic analysis, major nodes that represented viruses belonging to the same species or antigenic complex were clearly distinct and confirmed previously reported topologies (Charrel et al., 2009; Collao et al., 2009). Although the four viruses reported here clustered together defining the Salehabad virus antigenic complex node (Figs 1 and 2, full trees in Figs S1, S2 and S3, available in JGV Online), only the NS ORF showed a strong bootstrapping value. Interestingly, the serological pairings (SALV and ARBV; AMTV and ODRV) were strongly supported by the phylogenetic analysis. Similar topologies were observed in phylogenetic trees at the nucleotide level (data not shown). No branching inconsistencies were observed suggesting no reassortment among these viruses.
Fig. 1.
Phylogenetic analysis of select phlebovirus (a) L, (b) M, (c) N and NS ORFs. Neighbour-joining analysis at the amino acid level was performed due to the high variability of the underlying nucleotide sequences. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times.
Fig. 2.
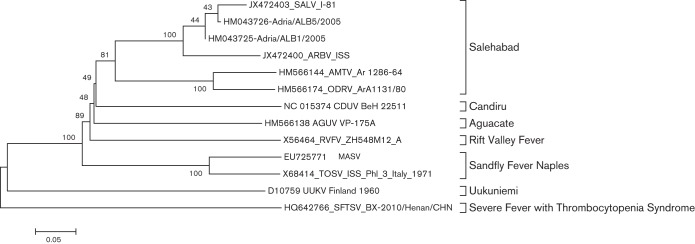
Phylogenetic analysis of the Adria virus L sequence. Neighbour-joining analysis at the amino acid level was performed due to the high variability of the underlying nucleotide sequences. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times.
For sequence assembly and analysis, Geneious 4.8.3 (Biomatters Inc.) was used. Topology and targeting predictions were generated by employing SignalP, NetNGlyc, TMHMM (http://www.cbs.dtu.dk/services), TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html), and integrated predictions in Geneious (Bendtsen et al., 2004; Claros & von Heijne, 1994; Kahsay et al., 2005; Käll et al., 2004; Krogh et al., 2001)
ORF analysis of the RNA-dependent RNA polymerase (L) revealed that the four members of the Salehabad virus antigenic complex show high conservation with previously described conserved phlebovirus functional domains (Aquino et al., 2003; Palacios et al., 2011b; Poch et al., 1989; Xiong & Eickbush, 1990). The polyprotein is co-translationally cleaved into three protein products, NSm, Gn and Gc. The topology of the polyprotein of the Salehabad virus antigenic complex is similar to the predicted topology for TOSV, PTV, RVFV and CDUV (Gerrard & Nichol, 2002; Grò et al., 1997; Ihara et al., 1985; Matsuoka et al., 1996; Palacios et al., 2011b; Valentini et al., 2008) with signal sequences, transmembrane domains and predicted cleavage sites conserved (data not shown). Similarly, all the functional domains described in the NP (Gerrard & Nichol, 2002; Le May et al., 2005; Palacios et al., 2011b) were conserved (data not shown). No singularities were found in the NSs ORF.
With the advent of high-throughput sequencing, virus taxonomy has begun to rely less on serology and more on genetic characteristics. One of the problems with antigenic or serological classification is the variability in the reactivity and potency of each batch of antigen or antibody. A second problem, in the case of the phleboviruses, is that some of the viruses are not pathogenic in mice, so production of ‘clean’ murine antisera is difficult. Also as more and more investigators use RT-PCR and other RNA detection systems for virus assay, virus isolates are sometimes not available for serological study, as in the case of ADRV. Genetic classification provides a more objective standard from which to classify viruses, with numerical cut-offs to distinguish different taxonomic levels. Furthermore, genome information may be the only information available, as in the case of ADRV.
However, a genetics-only based classification system for the bunyaviruses also has risks. Among phleboviruses, we have demonstrated that serological relatedness can sometimes be detected in cases where no genetic similarity is observed (Palacios et al., 2012). The serological and genetic data among the members of the Salehabad group enhance that view. Taken as a whole, we feel there is evidence to group AMTV, SALV, ARBV, ADRV and ODRV, as a single species group or complex in the genus Phlebovirus that is geographically distributed in Africa, Asia and Europe. Sequence or serological data alone show only a slight association; together the two datasets support this clustering. This data correlates and emphasizes our recent observation on the Uukuniemi antigenic complex, suggesting that a system of classification for the family Bunyaviridae that considers both genetic and serological data, when available, is optimal.
Salehabad has been detected in the Mediterranean region in clinically ill persons and sandflies. In the same geographical region, human phlebovirus seroprevalance is high. Moreover, a Sicilian-like virus (Cyprus virus) was associated with a major outbreak of febrile illness in 2002. Characterization of the genetic diversity of the members of this group might be crucial to enhance molecular diagnostic methods in the area.
Acknowledgements
This work was supported by Google.org, National Institutes of Health award AI57158 (North-east Biodefence Center - Lipkin), and USAID Predict funding source code 07-301-7119-52258 (Center for Infection and Immunity) and the Department of Defense. A. T. R., R. T. and H. G. were supported by NIH contract HHSN2722010000ROI/HHSN27200004/DO4.
Footnotes
Supplementary figures are available with the online version of this paper.
References
- Anagnostou V., Pardalos G., Athanasiou-Metaxa M., Papa A. (2011). Novel phlebovirus in febrile child, Greece. Emerg Infect Dis 17, 940–941 10.3201/eid1705.101958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino V. H., Moreli M. L., Moraes Figueiredo L. T. (2003). Analysis of oropouche virus L protein amino acid sequence showed the presence of an additional conserved region that could harbour an important role for the polymerase activity. Arch Virol 148, 19–28 10.1007/s00705-002-0913-4 [DOI] [PubMed] [Google Scholar]
- Beaty B. J., Calisher C. H., Shope R. E. (1989). Arboviruses. In Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, pp. 797–855 Edited by Schmidt N. J., Emmons R. W. Washington, DC: American Public Health Association [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Charrel R. N., Moureau G., Temmam S., Izri A., Marty P., Parola P., da Rosa A. T., Tesh R. B., de Lamballerie X. (2009). Massilia virus, a novel Phlebovirus (Bunyaviridae) isolated from sandflies in the Mediterranean. Vector Borne Zoonotic Dis 9, 519–530 10.1089/vbz.2008.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., von Heijne G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10, 685–686 [DOI] [PubMed] [Google Scholar]
- Collao X., Palacios G., Sanbonmatsu-Gámez S., Pérez-Ruiz M., Negredo A. I., Navarro-Marí J. M., Grandadam M., Aransay A. M., Lipkin W. I. & other authors (2009). Genetic diversity of Toscana virus. Emerg Infect Dis 15, 574–577 10.3201/eid1504.081111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster D. L., Conlan S., Holmes E. C., Palacios G., Evans J. D., Moran N. A., Quan P. L., Briese T., Hornig M. & other authors (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 10.1126/science.1146498 [DOI] [PubMed] [Google Scholar]
- Gerrard S. R., Nichol S. T. (2002). Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J Virol 76, 12200–12210 10.1128/JVI.76.23.12200-12210.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grò M. C., Di Bonito P., Fortini D., Mochi S., Giorgi C. (1997). Completion of molecular characterization of Toscana phlebovirus genome: nucleotide sequence, coding strategy of M genomic segment and its amino acid sequence comparison to other phleboviruses. Virus Res 51, 81–91 10.1016/S0168-1702(97)00076-2 [DOI] [PubMed] [Google Scholar]
- Ihara T., Smith J., Dalrymple J. M., Bishop D. H. (1985). Complete sequences of the glycoproteins and M RNA of Punta Toro phlebovirus compared to those of Rift Valley fever virus. Virology 144, 246–259 10.1016/0042-6822(85)90321-6 [DOI] [PubMed] [Google Scholar]
- Kahsay R. Y., Gao G., Liao L. (2005). An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21, 1853–1858 10.1093/bioinformatics/bti303 [DOI] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5, 150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Le May N., Gauliard N., Billecocq A., Bouloy M. (2005). The N terminus of Rift Valley fever virus nucleoprotein is essential for dimerization. J Virol 79, 11974–11980 10.1128/JVI.79.18.11974-11980.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J. & other authors (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Chen S. Y., Holland C. E., Compans R. W. (1996). Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch Biochem Biophys 336, 184–189 10.1006/abbi.1996.0547 [DOI] [PubMed] [Google Scholar]
- Nichol S. T., Beaty B. J., Elliott R. M., Goldbach R., Plyusnin A., Schmaljohn C. S., Tesh R. B. (2005). Family Bunyaviridae. In Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses, pp. 695–716 Edited by Fauquet C. M., Mayo M. A., Desselberger J., Ball L. A. San Diego: Elsevier Academic Press [Google Scholar]
- Palacios G., Quan P. L., Jabado O. J., Conlan S., Hirschberg D. L., Liu Y., Zhai J., Renwick N., Hui J. & other authors (2007). Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 13, 73–81 10.3201/eid1301.060837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P. L., Hui J. & other authors (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358, 991–998 10.1056/NEJMoa073785 [DOI] [PubMed] [Google Scholar]
- Palacios G., da Rosa A. T., Savji N., Sze W., Wick I., Guzman H., Hutchison S., Tesh R., Lipkin W. I. (2011a). Aguacate virus, a new antigenic complex of the genus Phlebovirus (family Bunyaviridae). J Gen Virol 92, 1445–1453 10.1099/vir.0.029389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Tesh R., Travassos da Rosa A., Savji N., Sze W., Jain K., Serge R., Guzman H., Guevara C. & other authors (2011b). Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol 85, 3811–3820 10.1128/JVI.02275-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G., Savji N., Rosa A. T. d., Guzman H., Yu X., Desai A., Rosen G. E., Hutchison S., Lipkin W. I., Tesh R. (2012) Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): demonstration that Severe fever with thrombocytopenia syndrome virus and Heartland virus are related members. J Virol (in press). 10.1099/vir.0.048850-0 [DOI] [Google Scholar]
- Papa A., Velo E., Bino S. (2011). A novel phlebovirus in Albanian sandflies. Clin microbiol infect 17, 585–587 10.1111/j.1469-0691.2010.03371.x [DOI] [PubMed] [Google Scholar]
- Peters C. J., Makino S., Morrill J. C. (2011). Rift Valley Fever. In Tropical Infecitous Diseases Principals, Pathogens and Practice, 3rd edn, pp. 462–465 Edited by Guerrant R. L., Walker D. H., Weller P. F. Philadelphia: Suanders Elsevier; 10.1016/B978-0-7020-3935-5.00069-0 [DOI] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B. (1989). Phlebotomus fevers. In The Arboviruses: Epidemiology and Ecology, pp. 15–27 Edited by Monath T. P. Boca Raton, FL: CRC Press, Inc [Google Scholar]
- Tesh R. B., Saidi S., Gajdamovic S. J., Rodhain F., Vesenjak-Hirjan J. (1976). Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ 54, 663–674 [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Peters C. J., Meegan J. M. (1982). Studies on the antigenic relationship among phleboviruses. Am J Trop Med Hyg 31, 149–155 [DOI] [PubMed] [Google Scholar]
- Travassos da Rosa A. P., Tesh R. B., Pinheiro F. P., Travassos da Rosa J. F., Peterson N. E. (1983). Characterization of eight new phlebotomus fever serogroup arboviruses (Bunyaviridae: Phlebovirus) from the Amazon region of Brazil. Am J Trop Med Hyg 32, 1164–1171 [DOI] [PubMed] [Google Scholar]
- Valentini M., Valassina M., Savellini G. G., Cusi M. G. (2008). Nucleotide variability of Toscana virus M segment in strains isolated from clinical cases. Virus Res 135, 187–190 10.1016/j.virusres.2008.01.016 [DOI] [PubMed] [Google Scholar]
- Verani P., Ciufolini M. G., Caciolli S., Renzi A., Nicoletti L., Sabatinelli G., Bartolozzi D., Volpi G., Amaducci L. & other authors (1988). Ecology of viruses isolated from sand flies in Italy and characterized of a new Phlebovirus (Arabia virus). Am J Trop Med Hyg 38, 433–439 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. (1990). Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9, 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Liu D., Nunes M. R., DA Rosa A. P., Tesh R. B., Xiao S. Y. (2007). Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am J Trop Med Hyg 76, 1194–1200 [PubMed] [Google Scholar]
- Yu X. J., Liang M. F., Zhang S. Y., Liu Y., Li J. D., Sun Y. L., Zhang L., Zhang Q. F., Popov V. L. & other authors (2011). Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364, 1523–1532 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]