Abstract
Streptococcus mutans, the primary causative agent of dental caries, contains two paralogues of the LytR-CpsA-Psr family proteins encoded by brpA and psr, respectively. Previous studies have shown that BrpA plays an important role in cell envelope biogenesis/homeostasis and affects stress responses and biofilm formation by Strep. mutans, traits critical to cariogenicity of this bacterium. In this study, a Psr-deficient mutant, TW251, was constructed. Characterization of TW251 showed that deficiency of Psr did not have any major impact on growth rate. However, when subjected to acid killing at pH 2.8, the survival rate of TW251 was decreased dramatically compared with the parent strain UA159. In addition, TW251 also displayed major defects in biofilm formation, especially during growth with sucrose. When compared to UA159, the biofilms of TW251 were mainly planar and devoid of extracellular glucans. Real-time-PCR and Western blot analyses revealed that deficiency of Psr significantly decreased the expression of glucosyltransferase C, a protein known to play a major role in biofilm formation by Strep. mutans. Transmission electron microscopy analysis showed that deficiency of BrpA caused alterations in cell envelope and cell division, and the most significant defects were observed in TW314, a Psr-deficient and BrpA-down mutant. No such effects were observed with Psr mutant TW251 under similar conditions. These results suggest that while there are similarities in functions between BrpA and Psr, distinctive differences also exist between these two paralogues. Like Bacillus subtilis but different from Staphylococcus aureus, a functional BrpA or Psr is required for viability in Strep. mutans.
Introduction
Streptococcus (Strep.) mutans, the major causative agent of human dental caries, thrives on the tooth surface as tenacious biofilms better known as dental plaque. The environment of the oral cavity has drastic and often unpredictable fluctuations in conditions, such as carbohydrate source and availability, pH, temperature, oxygen tension, toxic metabolites and antimicrobial agents (Burne, 1998). Strep. mutans has evolved multiple mechanisms to survive various environmental insults, persist on the tooth surface and, under certain conditions, become numerically significant, thus causing carious lesions on the tooth surface. The cell envelope plays a fundamental role in environmental sensing and signal transduction, and protection against various environmental insults, including toxic metabolites, antimicrobials and host immune defences. The cell envelope serves as a molecular sieve and is essential in controlled transport of solutes in and out of the bacterial cells. Besides, it is also essential in maintenance of cell shape, cell growth and division.
The LytR-CpsA-Psr (LCP) family of proteins is a group of cell-surface-associated proteins widely distributed in Gram-positive bacteria, which possess a featured LCP extracellular domain anchored by a short N-terminal transmembrane domain and followed by a C terminus that is variable in length and composition between different species (Hübscher et al., 2008; Wen & Burne, 2002). Initially, LytR of Bacillus subtilis has been identified as an autogenous transcriptional attenuator that also regulates the promoter of the divergently transcribed lytABC operon, which includes a lipoprotein (LytA), an autolysin (LytC), and a modifier protein of LytC (LytB) (Lazarevic et al., 1992). CpsA of Streptococcus agalactiae is a transcriptional activator regulating capsule biosynthesis (Cieslewicz et al., 2001). In fact, recent studies by Hanson et al. (2011, 2012) have provided evidence that CpsA of Strep. agalactiae can bind to the promoters of target genes. Recently, evidence has implicated that proteins of the LCP family play a significant role in cell envelope biogenesis and/or homeostasis (Chatfield et al., 2005; Hübscher et al., 2008; Johnsborg & Håvarstein, 2009; Over et al., 2011; Wen & Burne, 2002). LytR in Streptococcus pneumoniae was shown to be essential for normal septum formation (Johnsborg & Håvarstein, 2009). LytR-deficient pneumococci displayed major defects in cell division and cell morphology. Besides LytR (recently renamed as TagU), B. subtilis also possesses TagT and TagV, two additional proteins highly homologous to LytR (Hübscher et al., 2008; Kawai et al., 2011). Deficiency of any two of the LytR homologues causes defects in cell morphology and cell division, but disruption of all three coding sequences led to the eventual loss of viability (Kawai et al., 2011). Staphylococcus (Staph.) aureus also contains three LCP paralogues, including MsrR, a Psr-like protein, and two LytR-like proteins, Sa0908 and Sa2103 (Over et al., 2011). Staph. aureus strains deficient in MsrR, Sa0908 and Sa2103, individually or a combination of any two, were shown to form enlarged cells with irregular septa in various degrees, and were more susceptible to the antimicrobial agents oxacillin and teicoplanin (Hübscher et al., 2009; Over et al., 2011). Staph. aureus deficient in all three proteins was still viable, but had severely impaired septum placement and cell separation (Over et al., 2011). Only recently, studies by Eberhardt et al. (2012) and Kawai et al. (2011) have provided evidence that these widely distributed LCP proteins are enzymes involved in the covalent attachment of anionic cell wall polymers, like teichoic acid and capsular polysaccharides, to peptidoglycan. In B. subtilis, a triple tagTUV mutation carrying an IPTG-inducible tagV greatly reduced wall teichoic acid during growth in the absence of IPTG (Kawai et al., 2011). Similarly, pneumococcal LytR and its two homologues, Cps2A and Psr, contribute to capsule biosynthesis and retention at the cell wall (Eberhardt et al., 2012). Single Cps2A and Psr mutants produced a reduced amount of capsule, and a Cps2A and LytR double mutant showed greatly impaired growth and abnormal cell morphology and lost approximately half of the total capsule material into the cultural supernatant.
Strep. mutans contains two genes encoding proteins with features similar to the LCP family of proteins, including BrpA encoded by SMU.410 and Psr encoded by SMU.787 (www.oralgen.lanl.gov) (Ajdić et al., 2002; Hübscher et al., 2008). BrpA shows better homology to LytR proteins with the highest similarity to LytR of Strep. pneumoniae (44 % identity in amino acid composition, 58.6 % similarity) (Eberhardt et al., 2012; Hübscher et al., 2008; Johnsborg & Håvarstein, 2009) (Table S1, available with the online version of this paper), while the product of SMU.787 is homologous to the penicillin-sensitive repressor proteins (Psr) with the best similarity to the pneumococcal homologue (68 % similarity and 50.1 % identity) (Eberhardt et al., 2012; Hübscher et al., 2008; Rossi et al., 2003) (Table S1). We have shown that BrpA in Strep. mutans plays a major role in regulation of stress responses and biofilm formation (Bitoun et al., 2012; Chatfield et al., 2005; Wen & Burne, 2002; Wen et al., 2006). Deficiency of BrpA caused major defects in acid and oxidative stress tolerance and biofilm formation. Besides, the deficient mutant had an increased autolysis rate, a decreased viability, a weakened tolerance to cell envelope stress, and a substantial alteration in the transcriptional profile (Bitoun et al., 2012; Wen et al., 2006).
This study was designed to investigate the role of Psr in Strep. mutans cellular physiology and determine whether Psr is involved in regulation of stress tolerance and biofilm formation, traits critical to cariogenicity of this human pathogen. Here, a Psr-deficient mutant was constructed by allelic replacement. Characterization of this mutant revealed that like BrpA, Psr deficiency in Strep. mutans also weakens the acid tolerance responses, increases autolysis rate and causes major defects in biofilm formation. Different from BrpA, however, Psr deficiency drastically reduces glucosyltransferase C (GtfC) expression and glucan production. Consistent with our previous studies, transmission electron microscopy analysis showed that deficiency of BrpA caused alterations in cell envelope and defects in cell division, although no major effects were observed with the Psr-deficient mutant. In addition, deficiency of both BrpA and Psr appears to be lethal in Strep. mutans under the conditions studied.
Methods
Plasmids, bacterial strains and growth conditions.
All bacterial strains and plasmids used for this study are listed in Table 1. Unless otherwise stated, Strep. mutans strains were grown in Brain Heart Infusion (BHI, Difico Laboratories) aerobically in a 37 °C chamber containing 5 % CO2 under static conditions. When necessary, kanamycin (Kan, 1 mg ml−1), spectinomycin (Spe, 1 mg ml−1) and/or erythromycin (Erm, 10 µg ml−1) were included in the growth medium. For agar medium, Bacto agar (Difco Laboratories) was added at a level of 1.5 % (w/v). For growth studies, overnight cultures were appropriately diluted in fresh medium and allowed to continue to grow until mid-exponential phase (OD600 0.4–0.5), when they were further diluted by 1 : 100 and growth was continuously monitored using Bioscreen C (Oy Growth Curves) at 37 °C with and without sterile mineral oil overlay (Bitoun et al., 2011; Zeng et al., 2006). For heat stress tolerance response, cultures were grown aerobically in a 42 °C water bath.
Table 1. Bacterial strains and plasmids used in this study.
Kanr, Sper and Ermr: kanamycin, spectinomycin and erythromycin resistance, respectively.
| Strain or plasmid | Relevant characteristic | Source or reference |
| Strains | ||
| Strep. mutans UA159 | Wild-type, serotype c | Ajdić et al. (2002) |
| Strep. mutans OMZ175 | Wild-type, serotype f | Abranches et al. (2009) |
| Strep. mutans TW14D | Derivative of UA159, ΔbrpA (SMU.410), Ermr | Wen et al. (2006) |
| Strep. mutans TW14DC | TW14D carrying pDL278:brpA, Sper Ermr | Wen et al. (2006) |
| Strep. mutans TW251 | Derivative of UA159, Δpsr (SMU.787), Kanr | This study |
| Strep. mutans TW251C | TW251 carrying pDL278:psr, Kanr Sper | This study |
| Strep. mutans TW314 | Derivative of UA159, Δpsr, brpA-down, Kanr Ermr | This study |
| Strep. mutans TW316 | Derivative of OMZ175, Δpsr, Kanr | This study |
| Strep. mutans TW322 | Derivative of UA159, ΔrgpG, Sper | This study |
| E. coli DH10B | Cloning host, ΔmcrA ΔmcrBC Δmrr Δhsd | Invitrogen |
| Plasmids | ||
| pDL278 | Shuttle vector, Sper | LeBanc & Lee (1991) |
| pDL278:psr | Shuttle carrying psr, Sper | This study |
Construction of mutants and complemented strains.
To construct the Psr-deficient mutant, a PCR–ligation–mutagenesis strategy was used as described previously (Lau et al., 2002; Wen & Burne, 2004). Briefly, the 5′ and 3′ regions flanking psr were amplified by PCR using high fidelity DNA polymerase (New England Biolabs) with gene-specific primers shown in Table 2. Following appropriate restriction enzyme digestions, the flanking regions were ligated to a non-polar kanamycin-resistant element that was digested with the same enzymes (Zeng et al., 2006). The resulting ligation mixtures were used to directly transform Strep. mutans UA159 in the presence of competence stimulating peptide (CSP) (Li et al., 2002). Allelic replacement mutant TW251, with psr deficiency, was isolated on BHI-Kan plates and further confirmed by colony PCR with gene-specific 55 and 33 primers and sequencing. For complementation of TW251, the psr-coding sequence plus its cognate promoter region were amplified by PCR; the sequence was verified by sequencing and cloned into shuttle vector pDL278 (LeBanc & Lee, 1991). The resulting construct, pDL278:psr, was then used to transform TW251, generating the complemented strain TW251C. For the brpA and psr double mutant, PCRs were performed using genomic DNA from TW14D, a BrpA-deficient mutant with its coding sequence deleted and replaced by an erythromycin resistance element (ermr) (Wen et al., 2006), or TW251 as the template with gene-specific 55 and 33 primers (Table 2). With TW14D serving as the template, the resulting amplicon that contains ermr and regions flanking brpA element (Wen et al., 2006) was used to transform the Psr-deficient mutant, TW251. Similarly, when TW251 was used as the template, the resulting amplicon containing elements flanking psr and the non-polar kanamycin-resistance element was used to transform TW14D. The transformation mixtures were then plated on BHI with Erm and Kan. To enhance the transformation efficiency, CSP was added to the transformation mix 30 min prior to the addition of DNA (Li et al., 2002). The resulting double mutants were further examined to verify the deficiency of the respective genes using colony PCR as described above.
Table 2. Primers used in this study.
Underlined sequences are restriction sites engineered for cloning.
| Primer | Sequence (5′ to 3′) | Application | |
| Forward | Reverse | ||
| 787 55 and 53 | (55) tcataatgtcgctttgcatgcatttcc | (53) actggtaagcttcctgataatcatcttc | 5′ fragment for psr mutation |
| 787 35 and 33 | (35) agatccaaccaagctttttacaggttc | (33) tcaacttctacaatatcattc | 3′ fragment for psr mutation |
| 787C | ttctttagctttaacgcttcctg | cgcaggtcattctcgcttgg | psr complementation |
| RgpG 55 and 53 | (55) actactcgtatgaggtaaagactatg | (53) tctcgcatttgaattctcaactgcc | 5′ fragment for rgpG mutation |
| RgpG 35 and 33 | (35) tctcgcatttgaattctcaactgcc | (33) accatccatcataacatgaac | 3′ fragment for rgpG mutation |
| FtsW | gttatcagcaatcttctt | accattactcatagcata | 152 bp, qPCR of ftsW |
| 787 | gtgaagccgttgattcagtagagg | tgagacatgattgccgacataacc | 199 bp, qPCR of psr |
| GtfB | agcaatgcagccatctacaaat | acgaactttgccgttattgtca | 98 bp, qPCR of gtfB |
| RodA | tatcgtatgctgcgtgtca | ccaactgctgctccaatg | 116 bp, qPCR of rodA |
| GidA | taatcttcttgctcctac | ttactgcttcttcatctt | 155 bp, qPCR of gidA |
| GtfC | atggcgacaatatgatta | cggatgaaggaataagaa | 172 bp, qPCR of gtfC |
| GtfD | tgacttctgttcgttatg | ggttattgctggtaatga | 99 bp, qPCR of gtfD |
| FtsX | ggttgttgtaggttacttat | cttcaagaatcgtctcatt | 181 bp, qPCR of ftsX |
| FtsQ | ttacagacggcagtattg | ttcagcattagaggagttc | 128 bp, qPCR of ftsQ |
| Ldh | ttggcgacgctcttgatcttag | gtcagcatccgcacagtcttc | 92 bp, qPCR of ldh |
Biofilm analysis.
Strep. mutans biofilms were grown in modified biofilm medium with glucose (20 mM, BMG), sucrose (10 mM, BMS) or glucose (18 mM) plus sucrose (2 mM) (BMGS) as the supplementary carbon and energy sources as described previously (Bitoun et al., 2012; Li et al., 2002; Loo et al., 2000). When grown on 96-well plates, biofilms were stained using 0.1 % crystal violet after 24 h, and the quantity was measured using a spectrophotometer as described previously (Wen & Burne, 2002). For structural analysis, hydroxylapatite discs were used as substratum (Wen et al., 2006). After 24, 48 or 72 h, biofilms were stained with the BacLight live/dead bacterial viability kit (Invitrogen) or Alexa-488 Concanavalin A (Con A, Invitrogen) and SYTO 59 (Invitrogen). Optical dissections were carried out using a laser scanning confocal microscope (Olympus Fluoview BX61). At least three independent image stacks were acquired at 600× magnification using a 60× water immersion objective lens for each sample at random positions on the hydroxylapatite surface. Post-acquisition image analysis was carried out using SLIDEBOOK 5.0 (Olympus). COMSTAT 2.0 was used to quantify average thickness, biovolume, and surface area of the biofilms as detailed previously (Heydorn et al., 2000; Wen et al., 2006). The biovolume is defined as the volume (μm3) of the biomass per μm2 of substratum area (Heydorn et al., 2000). For scanning electron microscopy (SEM), biofilms were grown on hydroxylapatite discs that were deposited in the wells of 24-well plates for 24 h, and SEM analysis was carried out as described previously (Wen et al., 2005, 2006).
Cell envelope antimicrobial susceptibility assays.
The susceptibility of Strep. mutans strains to cell envelope antimicrobial agents was examined using microtitre plate-based assays as described previously (Bitoun et al., 2012; McBain et al., 2004; Suntharalingam et al., 2009). Cell envelope antimicrobial agents tested included the antibiotics vancomycin, bacitracin and the β-lactam antibiotic pencillin G, the bacteriocin nisin, and the cell envelope active compounds SDS and chlorhexidine (all from Sigma).
Acid and hydrogen peroxide killing assays.
The impact of psr deficiency on the ability of Strep. mutans to withstand acid and hydrogen peroxide challenges was determined by using procedures described elsewhere (Wen & Burne, 2004; Wen et al., 2006). For the assays, planktonic cultures were grown in BHI until early exponential phase (OD600~0.3), and biofilms cells were cultivated in BMGS on sterile glass slides deposited in 50 ml Falcon tubes (Wen et al., 2006).
RNA extraction and qPCR analysis.
For quantitative PCR, total RNA was extracted from early exponential phase (OD600~0.3) cultures, as detailed elsewhere (Wen et al., 2006). Confirmation of selected genes previously identified by DNA microarray analysis was carried out by real-time PCR with gene-specific primers designed using Beacon Designer 7.6 (PREMIER Biosoft) (Table 2) (Ahn et al., 2006; Wen et al., 2006).
Western blot analysis.
For Western blot analysis, Strep. mutans cultures were grown in BHI until early exponential phase (OD600~0.3), and whole-cell lysates, surface-associated fractions and cell-free supernates were prepared as described previously (Ahn & Burne, 2006; Bitoun et al., 2012; Wen et al., 2005). Briefly, whole-cell lysates were obtained by glass bead-beating in SDS-boiling buffer (60 mM Tris, pH 6.8, 10 % glycerol and 5 % SDS). For surface-associated fractions, cells from 500 ml cultures were suspended in 25 ml 0.2 % N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (Zwittergent, Sigma) and incubated at 28 °C with shaking at 80 r.p.m. for 1 h. Following centrifugation at 4000 r.p.m. at 4 °C for 10 min, the supernatants were further concentrated using Amicon Ultra-15 centrifugal filters (10 kDa NMWL) (Millipore) (Bitoun et al., 2012). For extracellular proteins, cell-free supernatants from these early exponential phase cultures were concentrated 300-fold by passage through a Centriprep filter with 50 kDa MW cut-off (Millipore). Total proteins (10 µg for whole-cell lysates, 15 µg for surface-associated fractions and 100 µg for cell-free supernatants, respectively) were separated by 7.5 % SDS-PAGE, blotted to Immobilon-FL membranes, and then probed for BrpA, GtfB, GtfC and GtfD using polyclonal antibodies against Strep. mutans BrpA, GtfB, GtfC or GtfD (kind gift from Dr Bowen), respectively (Bowen & Koo, 2011; Wen et al., 2005).
Autolysis assay.
Strep. mutans strains were grown in BHI overnight. Bacterial cells were harvested by centrifugation at 4000 r.p.m., at 4 °C for 10 min, washed once with PBS, and then resuspended in PBS containing 0.2 % (v/v) Triton X-100 (Sigma) (Wen & Burne, 2002). The OD600 of the cell suspension was then monitored automatically using a Bioscreen C at 37 °C every 30 min with moderate shaking (10 s) before measurement.
Wax worm infection model.
Galleria mellonella killing assays were performed as described previously (Kajfasz et al., 2010). Briefly, aliquots (5 µl each) of appropriately diluted mid-exponential phase (OD600~0.5) cultures of Strep. mutans wild-type or the Psr-deficient mutant were injected into the haemocoel. Groups receiving heat-inactivated (10 min at 75 °C) Strep. mutans wild-type or sterile saline were used as controls. After injection, larvae were incubated at 37 °C, and the appearance of melanization and survival were recorded and further analysed.
TEM analysis.
For TEM analysis of Strep. mutans strains, early exponential phase cultures grown in BHI were harvested by centrifugation at 4000 r.p.m., 4 °C for 10 min, washed once with PBS and then fixed in 2 % paraformaldehyde/2.5 % glutaraldehyde (Polysciences) in PBS for 1 h at room temperature. Cells were washed in PBS and post-fixed in 1 % osmium tetroxide (Polysciences) for 1 h. Samples were then rinsed extensively in dH2O prior to en bloc staining with 1 % aqueous uranyl acetate (Ted Pella) for 1 h. The cells were then washed with dH2O and dehydrated in a graded ethanol series and embedded in Eponate 12 resin (Ted Pella). Sections of 90–100 nm were prepared, stained with uranyl acetate and lead citrate, and viewed under a JEOL 1200 EX transmission electron microscope (JEOL USA).
Statistical analysis.
All quantitative data were further analysed using paired Student’s t-test.
Results
BrpA and Psr double deficiency is not viable
A Psr-deficient Strep. mutans strain, TW251, was constructed; this has 925 bp of its coding sequence (1386 bp) from nucleotides 230 to 1155 relative to the start codon ATG deleted and replaced with a non-polar kanamycin-resistance element (Zeng et al., 2006). To assess the impact of BrpA and Psr double deficiency, repeated efforts were made to generate a mutant deficient in both BrpA and Psr by using the PCR amplicon generated with TW251 genomic DNA as the template to transform the BrpA-deficient mutant, TW14D, and the PCR amplicon generated from TW14D to transform the Psr-deficient mutant, TW251 (Lau et al., 2002). While thousands of transformants were readily obtained with UA159 serving as a transformation control, no colonies were obtained with TW14D as the recipient. However, a few colonies were obtained on selective plates when TW251 was transformed with the PCR amplicon generated from TW14D. Colony PCR was carried out using the 5′ and 3′ end primers BrpA55 and BrpA33 (Wen et al., 2006), and all yielded two bands. As confirmed by sequencing, the smaller band contained an intact copy of the wild-type brpA, and the bigger band possessed the ermr element that replaces the brpA-coding sequence in TW14D (Wen et al., 2006), suggesting that these mutants were likely resulting from Campbell-like insertion and duplication, although the exact underlying mechanism awaits further investigation. One mutant, designated TW314, was selected for further study. When analysed by Western blotting, the level of BrpA expression in TW314 was found to be decreased by more than threefold compared with the wild-type, UA159, and the parent strain, TW251 (Fig. S1). Therefore, TW314 is a Psr-deficient and BrpA-down mutant, although the mechanism that underlies the regulation awaits further investigation.
To confirm whether BrpA and Psr double deficiency is lethal, the brpA- and psr-complemented strains, TW14DC and TW251C, which carry a wild-type copy of brpA and psr plus the cognate promoter region in shuttle vector pDL278, respectively, were used as the recipients in the transformation assays. As expected and further confirmed by colony PCR, mutants with allelic replacement of the chromosomal copy of both brpA and psr were readily achieved in the presence of a wild-type copy of the brpA or psr in the shuttle vector, suggesting double deficiency of BrpA and Psr is not viable under the conditions studied.
Psr deficiency and BrpA-down expression in TW314 causes major defects in growth
Deficiency of Psr did not have any major effects on colony morphology and growth rate during planktonic growth in BHI at 37 °C (Fig. 1). Similarly, no major differences were observed between the Psr-deficient mutant, TW251, and the wild-type, UA159, when incubated aerobically in a 42 °C water bath (Fig. 1). Unlike TW251, however, the BrpA-deficient mutant, TW14D, was found to display a significant delay in growth rate during incubation at 42 °C, with an average doubling time of 132 (±6) min compared with 84 (±3) (P<0.001) min for UA159. As expected, complementation of the deficient mutant, TW14D, with a wild-type copy of brpA plus the cognate promoter region in trans was able to restore the phenotype to a level similar to UA159 (data not shown).
Fig. 1.
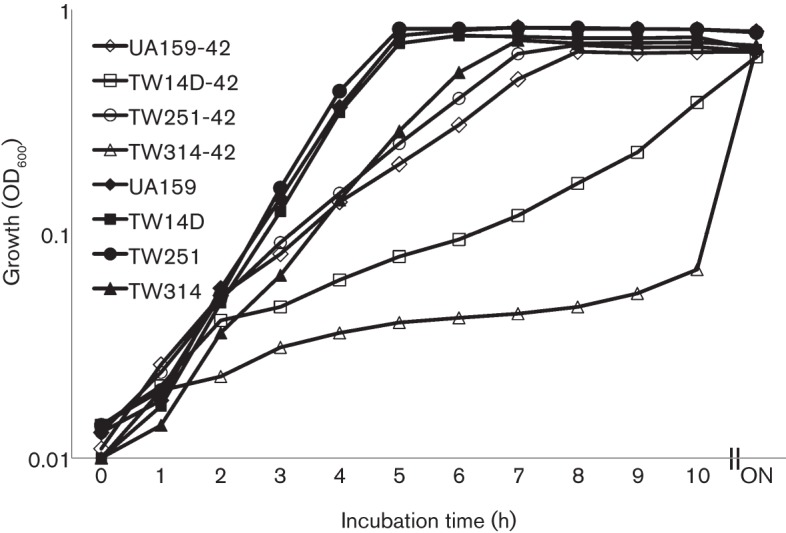
Growth study of Strep. mutans strains UA159, TW14D, TW251 and TW314 in BHI broth in a 42 °C water bath (open symbols) and a 37 °C aerobic chamber with 5 % CO2 (closed symbols). While no significant differences were observed between the strains at 37 °C, severe defects in growth rate of TW14D (□) and especially TW314 (▵) were measured during growth at 42 °C, when compared to UA159 (◊). The data presented here are representative of three independent experiments. ON, Overnight.
The Psr-deficient and BrpA-down mutant, TW314, was shown to have severe defects in growth under similar conditions (Fig. 1). It formed severe aggregates as it grew, precipitating to the bottom of the culture tube, although the aggregates were easily dispersed and resuspended with a brief shaking or vortexing. When grown in Bioscreen C at 37 °C, it took more than 174 (±6) (P<0.001) min to double its optical density, compared to 90 (±5) min for the wild-type, UA159. When incubated in a 42 °C water bath, TW314 had a doubling time of more than 6 h (P<0.001) (Fig. 1).
Psr deficiency affects acid tolerance responses
When subjected to acid killing at pH 2.8 for periods of 30, 45 and 60 min, the survival rate of TW251 was 2-log lower (P<0.001) than UA159 after 30 min (Fig. 2a), which is similar to TW14D and an indicator of weakened tolerance to acid stress as a result of Psr deficiency. Strep. mutans is known to possess an adaptive acid tolerance response, it has an enhanced tolerance to lower pH after initial exposure to a low pH environment (Burne, 1998). To assess whether Psr deficiency affects the ability of the mutant to launch an adaptive acid tolerance response, TW251 was incubated in BHI adjusted to pH of 5.0 for 1 h before being subjected to acid killing at pH 2.8. Like UA159, TW251 significantly increased its survival rate after initial incubation at pH 5.0 (Fig. 2b). Relative to UA159, however, the rate was still significantly lower, which is again similar to TW14D. Similar results were also obtained with 48 h biofilm cells of UA159 and TW251 grown in BMGS (Fig. S2). As expected, complementation with a wild-type copy of psr plus its cognate promoter in multiple copy number shuttle vector pDL278 (LeBanc & Lee, 1991) was able to restore the acid-sensitive phenotype of TW251 to a level similar to the wild-type, UA159. When compared to the wild-type, UA159, and its parent strain, TW251, the BrpA-down and Psr-deficient mutant, TW314, displayed the most severe acid-sensitive phenotype in cells that were both low pH adapted and non-adapted (Fig. 2). Unlike the BrpA-deficient mutant, TW14D (Wen et al., 2006), no significant differences in oxidative stress responses were observed between UA159 and TW251 when challenged with paraquat and hydrogen peroxide (data not shown).
Fig. 2.
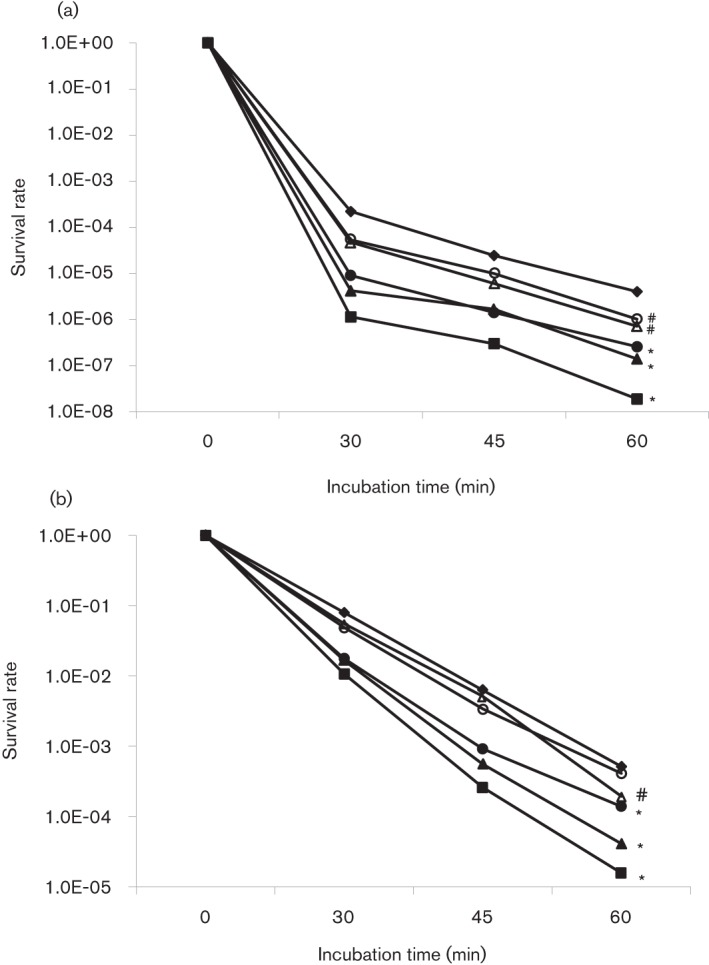
Acid killing assay of non-adapted (a) and adapted (b) cells. Strep. mutans UA159 (⧫), TW14D (•), TW14DC (○), TW251 (▴), TW251C (▵) and TW314 (▪) were grown in BHI until mid-exponential phase (OD~0.3). Bacterial cells were then harvested by centrifugation and washed once in 0.1 M glycine buffer, pH 7.0. Acid killing was performed by incubating the cells in 0.1 M glycine buffer, pH 2.8, for 30, 45 and 60 min. For adapted conditions, bacterial cells were incubated in BHI adjusted to pH 5.0 for 1 h prior to the killing assay. The surviving cells were plated in triplicate on BHI agar. Data presented here are the average of three independent experiments; significant difference is indicated by *, P<0.001, and #, P<0.01, compared with the wild-type under similar conditions.
Psr deficiency had no effect on cell envelope stress responses
Recently, we have shown that deficiency of BrpA weakens the ability of the deficient mutants to withstand several cell-wall-active antimicrobial agents, including nisin, bacitracin, chlorhexidine and SDS (Bitoun et al., 2012). To investigate whether Psr plays a similar role in cell envelope stress responses, the Psr-deficient mutant TW251 was subjected to antimicrobial susceptibility assays (Bitoun et al., 2012). Unlike TW14D, TW251 showed no significant differences in susceptibility against the antimicrobial agents evaluated (data not shown).
Psr deficiency has a limited effect on infectivity and virulence in the wax worm
Recent studies showed that BrpA deficiency in Strep. mutans OMZ175, a highly invasive strain (Abranches et al., 2009), significantly decreases its virulence in wax worm infection and toxicity (Bitoun et al., 2012). To investigate whether Psr has similar effects in regulation of virulence and infectivity, a Psr-deficient mutant of OMZ175 was created using the PCR amplicon generated with genomic DNA of TW251 to transform OMZ175 by following the strategy described in Methods. When the Psr-deficient mutant, TW316, was analysed using a wax worm infection model, a slight increase in survival rate was also observed, although the differences between the wild-type, OMZ175, and the Psr-deficient mutant, TW316, were not statistically significant (P = 0.0599) (data not shown).
Psr protects against autolysis
Cell division and cycle are cell-envelope-associated functions tightly regulated by autolysins. Previously, BrpA-deficiency was shown to cause increased autolysis rate, when compared to the wild-type (Chatfield et al., 2005; Wen & Burne, 2002). The Psr-deficient mutant, TW251, also displayed an increased autolysis rate (Fig. 3). Unlike the observation with acid tolerance responses, however, complementation of the mutant with a wild-type psr plus its cognate promoter region in trans was only able to moderately restore its phenotype. As expected, the Psr-deficient and BrpA-down mutant, TW314, appeared to have the most severe effect on autolysis, when compared with UA159 and TW251. Efforts were also made to analyse the expression of known autolysins, including AtlA, by Western blotting (Ahn & Burne, 2007). Consistent with previous DNA microarray analysis (Bitoun et al., 2012; Wen et al., 2006), no major differences in AtlA expression were detected between the wild-type and the Psr- and the BrpA-deficient mutants.
Fig. 3.
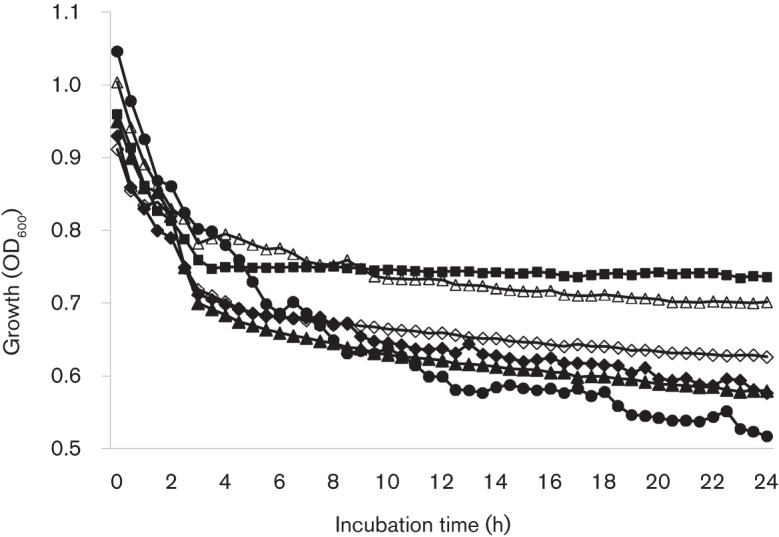
Autolysis assay. Overnight cultures of Strep. mutans UA159 (▪), TW14D (▴), TW14DC (▵), TW251 (⧫), TW251C (◊) and TW314 (•) were washed and resuspended in PBS, pH 7.2, to similar optical density with inclusion of Triton X-100 (0.2 %, w/v), and the optical density of these cells was then monitored using Bioscreen C. Similar to TW14D, the OD600 of TW251 and especially TW314 was significantly reduced after 24 h, suggesting that the autolysis rates were increased. Unlike TW14DC, however, complementation of psr in TW251C showed only slight effects. Data presented here are representative of three separate experiments.
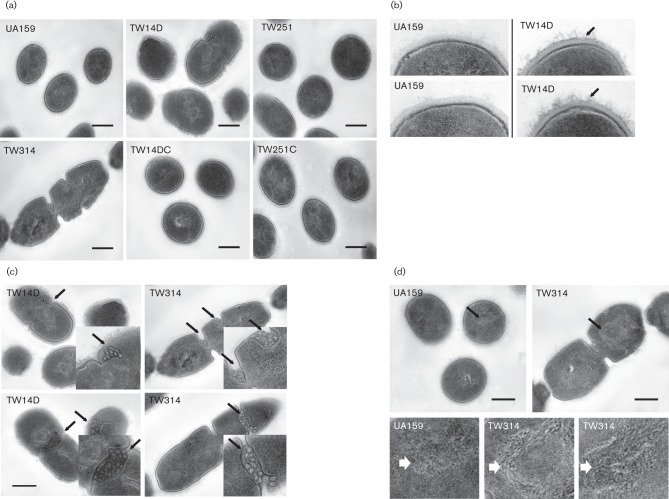
BrpA and Psr deficiency affect cell division and cell envelope
When analysed using TEM, UA159 generally appeared coccoid with some dividing cells (Fig. 4a). Relative to UA159, TW14D had more dividing cells and more long chains, and no significant differences were observed between TW251 and UA159. In contrast, TW314 generally appeared in long chains of dividing, giant cells with multiple septa. When compared to UA159, TW14D also appeared to have a rough and fibrous cell outer surface (peptidoglycan) (Fig. 4b), a feature associated with increased autolytic activity, but no such defects were observed with TW251 and TW314. Relative to UA159, the amount of mesosomes as well as their frequency of presence in TW14D and especially in TW314 were significantly increased (Fig. 4c). In addition, the nucleoid of TW314, and to some extent that of TW14D, was looser in structure of DNA fibre, as compared to the tight structure in the wild-type UA159 (Fig. 4d).
Fig. 4.
TEM analysis of Strep. mutans strains grown in BHI to mid-exponential phase (OD600 0.4). (a) Shows defects in cell division in TW14D and especially, in TW314; (b) highlights rougher, fibrous outer surface (peptidoglycan) in TW14D (indicated by arrows); (c) shows increased presence of mesosomes in TW14D and TW314 (marked by arrows), with inserts showing magnification of the regions indicated); and (d) shows looser nucleoid structure of TW314 (indicated by arrows), as compared to UA159, with magnified images of the selected regions shown below. Images were taken at magnifications of 25K and 60K. Scale bars (all images), 250 nm.
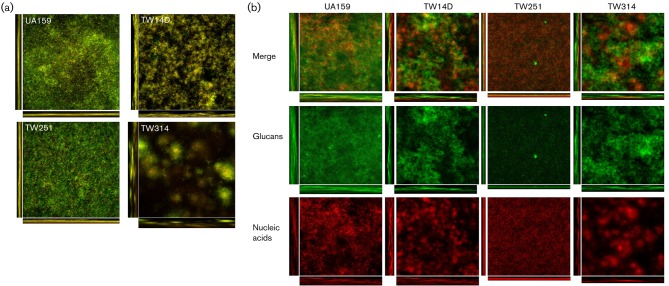
Psr deficiency significantly affects biofilm formation
BrpA deficiency causes major defects in biofilm formation by Strep. mutans, especially during growth under continuous flow conditions (Wen & Burne, 2002; Wen et al., 2006; Yoshida & Kuramitsu, 2002). To assess whether Psr-deficiency also affects biofilm formation, TW251 was grown in BMG, BMS and BMGS on glass slides and hydroxylapatite discs. Relative to UA159, biofilm formation (measured by c.f.u. count) by TW251 was decreased by more than 25-fold, especially during growth in BMS. After 24 h of biofilm growth in BMS, UA159 accumulated 5.17E7 (±1.42E7) c.f.u. while TW251 accumulated only 2.09E6 (±7.89E5) c.f.u. (P<0.001). Interestingly, the biofilms of TW251 were flat and tight (Fig. 5a), a phenotype drastically different from that observed for UA159. When Alexa 488-conjugated Concanavalin A was used to stain the glucans, followed by treatment with SYTO 59, which stains nucleic acids with red fluorescence (Bitoun et al., 2011), it was found that TW251 biofilms contained significantly less glucans than UA159 (Fig. 5b). COMSTAT calculations concluded that the total biovolume of TW251 when grown in BMS was 9.25 (±4.48) µm3 µm−2, while the wild-type contained a biovolume of 23.46 (±3.68) µm3 µm−2 (P<0.001). Glucan biovolume comparisons suggested that Psr deficiency also affects the ability of the cells to adhere to the surface since TW251 was nearly devoid of glucans, with glucan biovolume of 2.06 (±1.05) µm3 µm−2 versus 14.21 (±3.73) µm3 µm−2 (P<0.001) for the wild-type. In this static biofilm model system, BrpA deficiency in TW14D also increased the total biovolume by 1.87-fold (P<0.01) compared with the wild-type. Consistent with previous findings, however, TW14D was shown to have a 29-fold (P<0.001) decrease in cell volume when the cell populations were stained with SYTO 59. Interestingly, a 2.75-fold (P<0.001) increase in glucans was also measured, when compared with the wild-type. Similarly, the biofilm formation by the Psr-deficient and BrpA-down mutant, TW314, was further significantly decreased compared with UA159 and TW251 and again, the cell biovolume of SYTO 59-stained TW314 was decreased by 185-fold compared to the wild-type UA159, with an average of 7.5E−4 (±9.0E−5) µm3 µm−2 for TW314 versus 1.4E−1 (±8.9E−2) µm3 µm−2 for UA159 (P<0.001). Characteristically, TW314 biofilms were loose, patchy and contained what appeared to be plumes of microcolonies, with drastic increases in glucan production (Fig. 5). As revealed by thickness analysis using COMSTAT, both TW14D and TW314 biofilms were significantly thicker than UA159, with an average thickness of 162.6 (±5.2) µm for TW14D and 140.9 (±7.9) µm for TW314 compared with 105.1 (±11.3) µm (P<0.001) for UA159. Lastly, COMSTAT analysis also revealed that the total surface area of Psr-deficient TW251 was substantially decreased (P<0.001), with average area of 3.13E6 (±1.02E5) µm2 versus 7.68E6 (±1.53E5) µm2 for UA159.
Fig. 5.
Biofilm analysis. Strep. mutans biofilms were grown on hydroxylapatite discs in BM medium supplemented with glucose, sucrose, and glucose plus sucrose for 48 h. Following proper staining using BacLight live/dead fluorescent dye, biofilms were optically dissected using an Olympus laser scanning confocal microscope, and post-acquisition analyses were performed using SLIDEBOOK 5.0 and COMSTAT 2.0. As shown in (a), biofilm formation by TW14D, TW251 and especially TW314 during growth in BMGS was reduced significantly, when compared to the UA159. Relative to UA159, alterations in biofilm structure were also apparent, especially with TW314. (b) To examine glucan production by Strep. mutans, biofilms were treated with Alexa 488-conjugated Concanavalin A and SYTO 59, prior to optical dissections. The green fluorescent glucans (glucans) and red fluorescent cells (nucleic acids) and the merged (merge) images of UA159, TW14D, TW251 and TW314. In comparison, TW251 biofilms had significantly less glucans than the parent strain UA159. Images presented here are representatives composed of xyz, xz and yz images (512×512).
When analysed by SEM, UA159 formed robust biofilms (Fig. S3). In comparison, biofilm formation by the deficient mutants, especially the BrpA-deficient mutant, TW14D, and the Psr-deficient and BrpA-down mutant, TW314, was decreased significantly, similar to what has been described previously and above (Wen & Burne, 2002; Wen et al., 2006). Unlike UA159 and TW251, TW14D and TW314 existed primarily in chains, like Buddha-beads, with asymmetrical, giant cells widely distributed in TW314, which is consistent with the morphological characteristics shown by TEM (Fig. 4). Besides, TW14D and TW314 also possessed more wool-like glucans during growth in BMGS, as compared to UA159.
Deficiency of Psr significantly decreases expression of glucosyltransferase C
To uncover the mechanism that underlies the defects in glucan production and biofilm formation as a result of BrpA and Psr deficiency, the deficient mutants were analysed by Western blotting with polyclonal antibodies against the respective GtfB, GtfC and GtfD proteins as the probes. As shown in Fig. S4(a), GtfB was detected at a similar level in the whole-cell lysates of UA159, TW14D and TW251. In TW314, however, it was increased by 2.1-fold and two major bands were detected. In cell-free supernates, a 3- and 8.7-fold increase was measured in TW14D and TW314, respectively, compared with UA159, while no major differences were detected between UA159 and TW251. Little or no significant amount of GtfB was detected in the surface-associated fractions under the conditions studied. Like GtfB, GtfC was detected primarily as a secreted protein in cell-free supernatants, and multiple bands reactive to the probe existed, especially in the whole-cell lysates likely as a result of degradation (Fig. S4b). When compared with UA159, GtfC in TW251 was decreased by more than 70 % in whole-cell lysates and more than 40 % in cell-free supernatants, while a reduction of 30 % was also measured in TW14D whole-cell lysate. Relative to UA159, TW314 had a 2.1- and 1.5-fold increase in whole-cell lysate and cell-free supernatants, respectively, which is similar to GtfB. Unlike GtfB and GtfC, no detectable GtfD was measured in the whole-cell lysates (Fig. S4c), indicating a low level of expression under the conditions studied. In the cell-free supernatants, no significant differences were measured between TW14D, TW251 and UA159, but a 60 % reduction was apparent in the cell-free supernatants of TW314, which is opposite of both GtfB and C (Fig. S4c).
Consistently, real-time-PCR analysis also showed that relative to the wild-type, UA159, transcription of gtfC was reduced by 3.96-fold (P<0.001) in the Psr-deficient mutant, TW251 (Table 3), and was increased by more than 6-fold (P<0.001) in TW314, but no major differences were measured between UA159 and TW14D. The amount of gtfB transcripts was increased slightly in TW14D, but the most significant differences were detected in TW314 with an increase of more than 16-fold (P<0.001) relative to UA159. The transcripts of gtfD were moderately decreased in the deficient mutants, including TW314, when compared to the wild-type (Table 3).
Table 3. Real-time-PCR analysis of selected genes.
| Unique ID | Description/putative function* | Fold-difference relative to UA159† | ||
| TW14D | TW251 | TW314 | ||
| ldh | Lactate dehydrogenase, Ldh | 1.08 | 1.21 | −1.01 |
| SMU.787 | Transcriptional regulator, Psr-like protein | 1.37 | nd | nd |
| SMU.246 | Glycosyltransferase N-acetylglucosaminyltransferase, RgpG | −4.0a | −2.74a | 1.1 |
| gtfB | Glucosyltransferase B | 2.36a | −1.43 | 16.0a |
| gtfC | Glucosyltransferase C | 1.17 | −3.96a | 6.13a |
| gtfD | Glucosyltransferase D | −1.35 | −3.0a | −1.52 |
| SMU.713 | Cell division protein, FtsW | −4.75 a | −2.77a | −1.77b |
| SMU.550 | Cell division protein, FtsQ | −2.08a | −4.80a | 4.56a |
| SMU.1324 | Cell division ABC transporter, FtsX | −1.94a | −1.78a | −1.08 |
| SMU.1003 | Glucose-inhibited division protein, GidA | −2.29a | −2.9a | 1.8b |
| SMU.1279 | Cell division protein; shape-determining protein, RodA | 1.09 | −3.23a | 1.69b |
Description and putative function of the selected genes are based upon those in the published Strep. mutans database.
Data presented here are ratios of the levels of expression in the BrpA- and Psr-deficient mutants, TW14D and TW251, and the Psr-deficient and BrpA-down mutant, TW314, relative to those of the wild-type, UA159, with ‘−’ representing downregulation. Superscript a and b indicate statistical difference between the wild-type and the respective mutant at significance level of P<0.001 and 0.01, respectively. nd, Not detectable.
Discussion
Cell envelope is known to be crucial in cell growth, cell division, maintenance of structural integrity as well as the characteristic shape of bacterial cells. Consistent with previous findings of BrpA influencing cell envelope stress responses (Bitoun et al., 2012), results presented here further support a major role for BrpA in Strep. mutans cell envelope biogenesis/homeostasis (Bitoun et al., 2012; Chatfield et al., 2005; Wen & Burne, 2002; Wen et al., 2006). As shown by TEM, BrpA deficiency altered the cell surface structure, resulting in an apparently rough and fibrous outer surface (peptidoglycan) in TW14D, when compared to the parent strain, UA159. Besides, TW14D also had more dividing cells and long chains, indicative of defects in cell envelope biogenesis and morphogenesis as a result of BrpA deficiency. While Psr deficiency alone had little or no major impact on cell division and cell morphology, Psr deficiency and BrpA-down expression in TW314 was shown to cause severe defects in cell division and morphology, which is probably the major factor contributing to the defects in biofilm formation. Consistent with the severe aggregation during growth in broth, TW314 existed predominantly in long chains of dividing cells and cells with multiple septa, a likely result of delayed and incomplete cell division in response to Psr deficiency and compromised BrpA expression. Moreover, the looser DNA fibres in TW314 and TW14D are also a likely consequence of delayed or disrupted cell division.
Mesosomes are rosette-like clusters of folds in plasma membrane, protruding toward the interior of the cell. They are not independent structures, rather elaborations of the plasma membrane, and may be artefacts of fixation. However, mesosomes have been linked to functionality (Li et al., 2008; Santhana Raj et al., 2007). For example, recent studies in Staph. aureus showed that mesosomes were present only in cells treated with antibiotics and that in comparison, cells treated with oxacillin and vancomycin seemed to have more and deeper mesosomes than those treated with amikacin, gentamicin and ciprofloxacin (Santhana Raj et al., 2007). While their nature and functionality are somewhat controversial, such a significant increase in frequency and quantity in TW14D and TW314 also implicates defects and/or alterations in the plasma membrane, particularly at sites of septum formation. To the best of our knowledge, such effects on cell plasma membrane have not yet been reported for any other members of the LCP proteins.
Optimal cell growth and division require highly coordinated actions of enzymes in cell envelope synthesis, hydrolysis and maintenance. Recently, we have shown by DNA microarray analysis that BrpA deficiency in Strep. mutans resulted in an alteration in expression of several genes with a potential role in biosynthesis of peptidoglycan precursors (Bitoun et al., 2012; Boyd et al., 2000). Additionally, among the genes identified by microarray analysis were those with a potential role in cell division, including SMU.713, SMU.550 and SMU.1003 (Table 3). SMU.713 encodes a protein with homology to FtsW, a transporter of the lipid-linked peptidoglycan precursors across the cytoplasmic membrane (Mohammadi et al., 2011). SMU.550 encodes an FtsQ homologue, a protein essential for the divisome assembly in Escherichia coli (D’Ulisse et al., 2007). SMU.1003 codes for a protein homologous to glucose-inhibited division protein GidA. First described in E. coli (von Meyenburg et al., 1982), GidA has recently been shown to play a role in cell division of other bacterial pathogens (Shippy et al., 2012). Consistent with previous DNA microarray analysis, real-time-PCR analysis in this study further verified that transcription of ftsW, ftsQ and gidA was decreased in TW14D by 4.75-, 2.08- and 2.29-fold (P<0.001), respectively. While the underlying mechanism remains unclear, downregulation of genes involved in cell envelope biogenesis and cell division will have an impact on cell envelope biogenesis and cell morphology, and consequently affect the integrity of the cell envelope, which can in part be attributed to the increased autolysis and the decreased acid tolerance responses of the mutants (Bitoun et al., 2012); (Chatfield et al., 2005; Wen & Burne, 2002; Wen et al., 2006). Interestingly, similar patterns in transcription of these genes were also seen with the Psr-deficient mutant, TW251, although it did not display major morphological defects, as compared to the wild-type, UA159. These results further suggest that factors other than FtsW, FtsQ and GidA are likely involved in BrpA-mediated regulation in cell division and morphology of S. mutans.
Strep. mutans produces at least three glucosyltransferases, GtfB, GtfC and GtfD, that synthesize glucose polymers with glucose moiety of sucrose. The adhesive glucans, especially the water-insoluble α1,3-linked glucans of GtfB and GtfC are fundamental in sucrose-induced biofilm formation and cariogenicity of Strep. mutans (Bowen & Koo, 2011; Koo et al., 2010). The product of GtfD, on the other hand, is soluble and readily metabolized to function as a primer for GtfB (Bowen & Koo, 2011). Recent studies by us and some other laboratories have also generated evidence that alteration of the glucan structures and/or the ratio of glucans to glucan-binding proteins could have a significant effect on Strep. mutans adherence and accumulation on a surface (Bitoun et al., 2011; Hazlett et al., 1998, 1999; Wen et al., 2005), although the basis for this phenomenon remains unclear. As revealed by Western blotting and real-time PCR, a major reduction of GtfC in TW251 suggests that the deficiency of glucan production and reduction of biofilm formation by TW251 could at least in part be attributed to the reduced expression of GtfC in response to Psr deficiency. On the other hand, increase in glucan production in TW314 can be attributed to the elevation in expression of GtfB and GtfC as a result of Psr deficiency and BrpA-down expression. It is likely that elevation of GtfB and GtfC and reduction of GtfD in TW314 will cause alterations in glucan composition and/or the ratio of glucans to glucan-binding proteins, and consequently influence surface adherence and biofilm formation by this bacterium.
The gtfB and gtfC genes are linked and can be co-transcribed as an operon, but there is also evidence that these two genes can be differentially transcribed under certain conditions (Goodman & Gao, 2000; Senadheera et al., 2007; Smorawinska & Kuramitsu, 1995; Wen et al., 2005). The results presented here provide further evidence that these two genes are differentially regulated under the conditions studied. Both Western blot and real-time-PCR analyses showed that expression of GtfC, but not GtfB and GtfD, is under the control of Psr; and such regulation is mostly governed at the transcriptional level. Interestingly, however, the opposing effects in GtfC and GtfB expression in the BrpA-down and Psr-deficient mutant, TW314, as compared to the respective single mutants, also suggest that other factors are likely involved in the BrpA- and Psr-mediated regulation, and the details of the underlying mechanisms await further investigation.
Bacterial cell-surface interactions, such as adhesin–receptor interactions, ionic interactions, hydrogen bonding and the hydrophobic effect, are known to play an important role in bacterial adherence and biofilm formation (Nobbs et al., 2009). As recently shown by Kawai et al. (2011) and Eberhardt et al. (2012), the LCP proteins in B. subtilis and Strep. pneuomoniae are involved in attachment of anionic polymers, such as teichoic acid and capsular polysaccharides, to cell wall peptidoglycan. While Strep. mutans is not known to possess capsules (Hamada & Slade, 1980), deficiency of BrpA and Psr will likely cause alterations in teichoic acid and lipoteichoic acid composition, influencing cell-surface interactions and consequently reducing biofilm formation by the deficient mutants. Besides, both BrpA and Psr deficiency was shown to reduce expression of RgpG (SUM.246) (Table 3) (Bitoun et al., 2012), an enzyme involved in the biosynthesis of rhamnose glucose polysaccharide (RGP) (Yamashita et al., 1999), which is a major constituent of streptococcal cell walls. We have also constructed an RgpG-deficient mutant, TW322, and a preliminary study of TW322 showed that deficiency of RgpG had no major effect on growth rate, but slightly decreased biofilm formation by the deficient mutant (data not shown). These results further suggest that the LCP paralogues in Strep. mutans could also be involved in biosynthesis of rhamnose–glucose polysaccharides, which in turn affects biofilm formation.
Data presented here, as well as recent studies in a few other species, also suggest that despite the high similarities in the LCP domains, distinctive differences in structure and function exist between members of this family of proteins (Hübscher et al., 2008, 2009; Johnsborg & Håvarstein, 2009; Kawai et al., 2011; Over et al., 2011; Rossi et al., 2003; Steidl et al., 2008) (Table S1). Staph. aureus strains deficient in all three proteins were viable (Over et al., 2011), while triple disruption of the LytR homologues in B. subtilis resulted in lethality (Kawai et al., 2011). Like B. subtilis but different from Staph. aureus, the failure to isolate a double mutant deficient in both BrpA and Psr in Strep. mutans suggests that one functional LCP paralogue, BrpA or Psr, is required for viability under the conditions studied. The severe defects in growth, autolysis rate, cell morphology and division of TW314, a result of Psr-deficiency and BrpA-down expression, also support this notion. While both BrpA and Psr deficiency led to increased autolysis rates (Chatfield et al., 2005; Wen & Burne, 2002), neither brpA nor psr is directly linked to genes coding for autolysins or proteins with known autolytic activity, which is also different from B. subtilis and Staph. aureus (Lazarevic et al., 1992; Over et al., 2011). Besides, no major differences in expression of known autolysins were detected in strains deficient in BrpA, as shown by Western blotting and DNA microarray analysis (Bitoun et al., 2012).
Unlike other micro-organisms that possess many LCP homologues, Strep. mutans has only two in BrpA and Psr. It is therefore possible to finely dissect the role of these LCP proteins in cell envelope metabolism and other related aspects of Strep. mutans physiology. Psr and BrpA are paralogues of different subfamilies, as shown by similarities in amino acid composition (Table S1), with BrpA belonging to the F2 cluster and Psr the main subfamily of F1 (Hübscher et al., 2008; Wen & Burne, 2002). Different from Psr and other members of the LCP proteins, BrpA possesses a seemingly unique C terminus of approximately 100 amino acids rich in serines and threonines (Wen & Burne, 2002). Like BrpA, deficiency of Psr affects the ability of the deficient mutants to cope with acid tolerance response, influences the autolysis rate, and causes major alterations in biofilm formation by the deficient mutants. Unlike BrpA, however, Psr deficiency showed no significant effect on cell morphology, oxidative stress responses induced by hydrogen peroxide and paraquat, susceptibility to cell-wall-active antimicrobials and detergents tested, and infectivity and virulence in the wax worm model (Bitoun et al., 2012). Both BrpA and Psr deficiency caused major defects in biofilm formation by the respective deficient mutants, but only Psr deficiency drastically decreased GtfC expression. In addition, deficiency of BrpA was previously shown to cause substantial alterations in the transcriptional profile of the deficient mutant, including those related to cell envelope biogenesis (Bitoun et al., 2012; Wen et al., 2006). Discerning the effect Psr deficiency has on the transcriptome in TW251 awaits further investigation, however. These phenotypic characteristics further suggest that while there is some functional redundancy between BrpA and Psr, distinctive differences also exist between these two LCP paralogues in Strep. mutans. How BrpA and Psr modulate gene expression in Strep. mutans also awaits further investigation.
In summary, BrpA and Psr are two paralogues of LCP family proteins in Strep. mutans. Like BrpA, Psr plays an important role in Strep. mutans cellular physiology and affects biofilm formation. It is apparent that of the two paralogues, BrpA plays a major role in cell envelope biogenesis/homeostasis, but neither BrpA nor Psr could completely complement the deficiency of the other. One functional paralogue is required for viability in Strep. mutans. Major efforts are being directed to further investigate the mechanisms underlying BrpA- and Psr-mediated functions in cell envelope biogenesis/homeostasis.
Acknowledgements
We are grateful to Dr William H. Bowen at the University of Rochester School of Medicine and Dentistry for his generous gift of the anti-Gtf antibodies and to Dr Robert A. Burne at the University of Florida College of Dentistry for the gift of anti-autolysin AtlA antibodies and for the sequence information on the LytR-CpsA-Psr proteins in some other Strep. mutans strains. We would also like to thank Drs Robert G. Quivey and Jose Lemos at the University of Rochester Center for Oral Biology for the thoughtful discussions during the preparation of this manuscript; and Mr James H. Miller for his assistance with worm infection assays. This study was supported in part by NIH/NIDCR grant DE19452 to Z. T. W. and in part by Southern Louisiana Institute for Infectious Disease Research.
Abbreviations:
- LCP
LytR-CpsA-Psr
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
Footnotes
A supplementary table and four supplementary figures are available with the online version of this paper.
References
- Abranches J., Zeng L., Bélanger M., Rodrigues P. H., Simpson-Haidaris P. J., Akin D., Dunn W. A., Jr, Progulske-Fox A., Burne R. A. (2009). Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol Immunol 24, 141–145. 10.1111/j.1399-302X.2008.00487.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. J., Burne R. A. (2006). The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol 188, 6877–6888. 10.1128/JB.00536-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. J., Burne R. A. (2007). Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol 189, 6293–6302. 10.1128/JB.00546-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. J., Wen Z. T., Burne R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74, 1631–1642. 10.1128/IAI.74.3.1631-1642.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdić D., McShan W. M., McLaughlin R. E., Savić G., Chang J., Carson M. B., Primeaux C., Tian R., Kenton S. & other authors (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99, 14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Nguyen A. H., Fan Y., Burne R. A., Wen Z. T. (2011). Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol Lett 320, 110–117. 10.1111/j.1574-6968.2011.02293.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Liao S., Yao X., Ahn S. J., Isoda R., Nguyen A. H., Brady L. J., Burne R. A., Abranches J., Wen Z. T. (2012). BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 78, 2914–2922. 10.1128/AEM.07823-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H., Koo H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45, 69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. A., Cvitkovitch D. G., Bleiweis A. S., Kiriukhin M. Y., Debabov D. V., Neuhaus F. C., Hamilton I. R. (2000). Defects in D-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J Bacteriol 182, 6055–6065. 10.1128/JB.182.21.6055-6065.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne R. A. (1998). Oral streptococci... products of their environment. J Dent Res 77, 445–452. 10.1177/00220345980770030301 [DOI] [PubMed] [Google Scholar]
- Chatfield C. H., Koo H., Quivey R. G., Jr (2005). The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151, 625–631. 10.1099/mic.0.27604-0 [DOI] [PubMed] [Google Scholar]
- Cieslewicz M. J., Kasper D. L., Wang Y., Wessels M. R. (2001). Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276, 139–146. 10.1074/jbc.M005702200 [DOI] [PubMed] [Google Scholar]
- D’Ulisse V., Fagioli M., Ghelardini P., Paolozzi L. (2007). Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153, 124–138. 10.1099/mic.0.2006/000265-0 [DOI] [PubMed] [Google Scholar]
- Eberhardt A., Hoyland C. N., Vollmer D., Bisle S., Cleverley R. M., Johnsborg O., Håvarstein L. S., Lewis R. J., Vollmer W. (2012). Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18, 240–255. 10.1089/mdr.2011.0232 [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Gao Q. (2000). Characterization of the gtfB and gtfC promoters from Streptococcus mutans GS-5. Plasmid 43, 85–98. 10.1006/plas.1999.1444 [DOI] [PubMed] [Google Scholar]
- Hamada S., Slade H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev 44, 331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. R., Lowe B. A., Neely M. N. (2011). Membrane topology and DNA-binding ability of the streptococcal CpsA protein. J Bacteriol 193, 411–420. 10.1128/JB.01098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. R., Runft D. L., Streeter C., Kumar A., Carion T. W., Neely M. N. (2012). Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 194, 1668–1678. 10.1128/JB.06373-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett K. R., Michalek S. M., Banas J. A. (1998). Inactivation of the gbpA gene of Streptococcus mutans increases virulence and promotes in vivo accumulation of recombinations between the glucosyltransferase B and C genes. Infect Immun 66, 2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett K. R., Mazurkiewicz J. E., Banas J. A. (1999). Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect Immun 67, 3909–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersbøll B. K., Molin S. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hübscher J., Lüthy L., Berger-Bächi B., Stutzmann Meier P. (2008). Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9, 617. 10.1186/1471-2164-9-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher J., McCallum N., Sifri C. D., Majcherczyk P. A., Entenza J. M., Heusser R., Berger-Bächi B., Stutzmann Meier P. (2009). MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol Lett 295, 251–260. 10.1111/j.1574-6968.2009.01603.x [DOI] [PubMed] [Google Scholar]
- Johnsborg O., Håvarstein L. S. (2009). Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J Bacteriol 191, 5859–5864. 10.1128/JB.00724-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz J. K., Rivera-Ramos I., Abranches J., Martinez A. R., Rosalen P. L., Derr A. M., Quivey R. G., Lemos J. A. (2010). Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192, 2546–2556. 10.1128/JB.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N. & other authors (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J 30, 4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Xiao J., Klein M. I., Jeon J. G. (2010). Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol 192, 3024–3032. 10.1128/JB.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. C. Y., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49, 193–205. 10.1016/S0167-7012(01)00369-4 [DOI] [PubMed] [Google Scholar]
- Lazarevic V., Margot P., Soldo B., Karamata D. (1992). Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol 138, 1949–1961. 10.1099/00221287-138-9-1949 [DOI] [PubMed] [Google Scholar]
- LeBanc D., Lee L. (1991). Replication function of pVA380–1. In Genetics and Molecular Biology of Streptococci, Lactococci, and Enterococci, pp. 235–239. Edited by Dunny G., Cleary P. P., Mckay L. L. Washington, DC: American Society for Microbiology. [Google Scholar]
- Li Y. H., Tang N., Aspiras M. B., Lau P. C., Lee J. H., Ellen R. P., Cvitkovitch D. G. (2002). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184, 2699–2708. 10.1128/JB.184.10.2699-2708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Feng H. Q., Pang X. Y., Li H. Y. (2008). Mesosome formation is accompanied by hydrogen peroxide accumulation in bacteria during the rifampicin effect. Mol Cell Biochem 311, 241–247. 10.1007/s11010-007-9690-4 [DOI] [PubMed] [Google Scholar]
- Loo C. Y., Corliss D. A., Ganeshkumar N. (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182, 1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain A. J., Ledder R. G., Sreenivasan P., Gilbert P. (2004). Selection for high-level resistance by chronic triclosan exposure is not universal. J Antimicrob Chemother 53, 772–777. 10.1093/jac/dkh168 [DOI] [PubMed] [Google Scholar]
- Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Distèche M., de Kruijff B., Breukink E. (2011). Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J 30, 1425–1432. 10.1038/emboj.2011.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs A. H., Lamont R. J., Jenkinson H. F. (2009). Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73, 407–450. 10.1128/MMBR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Over B., Heusser R., McCallum N., Schulthess B., Kupferschmied P., Gaiani J. M., Sifri C. D., Berger-Bächi B., Stutzmann Meier P. (2011). LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol Lett 320, 142–151. 10.1111/j.1574-6968.2011.02303.x [DOI] [PubMed] [Google Scholar]
- Rossi J., Bischoff M., Wada A., Berger-Bächi B. (2003). MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob Agents Chemother 47, 2558–2564. 10.1128/AAC.47.8.2558-2564.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhana Raj L., Hing H. L., Baharudin O., Teh Hamidah Z., Aida Suhana R., Nor Asiha C. P., Vimala B., Paramsarvaran S., Sumarni G., Hanjeet K. (2007). Mesosomes are a definite event in antibiotic-treated Staphylococcus aureus ATCC 25923. Trop Biomed 24, 105–109. [PubMed] [Google Scholar]
- Senadheera M. D., Lee A. W., Hung D. C., Spatafora G. A., Goodman S. D., Cvitkovitch D. G. (2007). The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol 189, 1451–1458. 10.1128/JB.01161-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy D. C., Heintz J. A., Albrecht R. M., Eakley N. M., Chopra A. K., Fadl A. A. (2012). Deletion of glucose-inhibited division (gidA) gene alters the morphological and replication characteristics of Salmonella enterica Serovar Typhimurium. Arch Microbiol 194, 405–412. 10.1007/s00203-011-0769-7 [DOI] [PubMed] [Google Scholar]
- Smorawinska M., Kuramitsu H. K. (1995). Primer extension analysis of Streptococcus mutans promoter structures. Oral Microbiol Immunol 10, 188–192. 10.1111/j.1399-302X.1995.tb00141.x [DOI] [PubMed] [Google Scholar]
- Steidl R., Pearson S., Stephenson R. E., Ledala N., Sitthisak S., Wilkinson B. J., Jayaswal R. K. (2008). Staphylococcus aureus cell wall stress stimulon gene-lacZ fusion strains: potential for use in screening for cell wall-active antimicrobials. Antimicrob Agents Chemother 52, 2923–2925. 10.1128/AAC.00273-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P., Senadheera M. D., Mair R. W., Lévesque C. M., Cvitkovitch D. G. (2009). The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191, 2973–2984. 10.1128/JB.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K., Jørgensen B. B., Nielsen J., Hansen F. G. (1982). Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol Gen Genet 188, 240–248. 10.1007/BF00332682 [DOI] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2002). Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol 68, 1196–1203. 10.1128/AEM.68.3.1196-1203.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2004). LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J Bacteriol 186, 2682–2691. 10.1128/JB.186.9.2682-2691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Suntharaligham P., Cvitkovitch D. G., Burne R. A. (2005). Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun 73, 219–225. 10.1128/IAI.73.1.219-225.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Baker H. V., Burne R. A. (2006). Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol 188, 2983–2992. 10.1128/JB.188.8.2983-2992.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y., Shibata Y., Nakano Y., Tsuda H., Kido N., Ohta M., Koga T. (1999). A novel gene required for rhamnose-glucose polysaccharide synthesis in Streptococcus mutans. J Bacteriol 181, 6556–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Kuramitsu H. K. (2002). Multiple Streptococcus mutans genes are involved in biofilm formation. Appl Environ Microbiol 68, 6283–6291. 10.1128/AEM.68.12.6283-6291.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Wen Z. T., Burne R. A. (2006). A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol 62, 187–200. 10.1111/j.1365-2958.2006.05359.x [DOI] [PubMed] [Google Scholar]