Summary
Direct targeting of oncogenic MYC proteins has been an elusive goal of many cancer drug development efforts. In this issue of Cancer Discovery, Stegmaier and colleagues demonstrate that pharmacologically interfering with the bromodomain and extraterminal (BET) class of proteins potently depletes MYCN in neuroblastoma cells, resulting in cellular cytotoxicity and thus providing a novel approach with a potential impact on a previously undruggable major oncogene.
The complexity of the cancer epigenome is being revealed through multiple collaborative experimental approaches such as the ENCODE project (1). Although it is not a novel concept, pharmacologic manipulation of the epigenome is becoming an increasingly valid anticancer therapeutic strategy. In addition to alterations in the DNA methylation, changes in histone modification patterns are commonly seen in malignancies. Many different proteins modify histones, including histone acetyltransferases and deacetylases, methyltransferases and demethylases, kinases and phosphatases, and ubiquitin ligases and deubiquitinases (2). By altering the chromatin structure and accessibility to transcription factors, these histone modifications promote the activation or repression of gene transcription. Bromodomain and extraterminal (BET) proteins contain 2 bromodomain motifs in the amino terminus and an extraterminal protein–protein interaction domain in the carboxy terminus, constituting an additional class of proteins that regulate chromatin structure. These “chromatin readers” bind to acetylated lysines in chromatin, recruiting additional chromatin-modifying proteins and leading to cell context–dependent gene activation or repression.
The BET family consists of BRD2, BRD3, BRD4, and BRDT (2). An oncogenic role for BET proteins was first shown in NUT midline carcinoma (NMC), a rare squamous carcinoma characterized by a recurrent t(15;19) translocation fusing the bromodomain-containing N terminus of BRD4 to the NUT (nuclear protein in testis) protein. Strikingly, treatment with JQ1, a small-molecule inhibitor that competitively binds to bromodomains, displacing the BRD4 fusion oncoprotein from chromatin, results in cell-cycle arrest and differentiation in NMC cells (3). More recently, several groups have shown that BET inhibitors target MYC transcription and show therapeutic efficacy against leukemia, lymphoma, and multiple myeloma models (4, 5).
The MYC family consists of MYC, MYCL, and MYCN. It regulates transcription, metabolism, cell-cycle progression, stemness, and other key cellular processes (6). With this plethora of roles, it is not surprising that the MYC family is frequently altered in multiple cancer histotypes. MYC is translocated in multiple myeloma and Burkitt lymphoma and is amplified in many other human malignancies (6). MYCN is frequently amplified in neuroblastoma, a pediatric malignancy of the developing nervous system, as well as in small-cell lung cancer, medulloblastoma, and neuroendocrine prostate cancer. In neuroblastoma, MYCN amplification is found in approximately 20% of cases and is highly associated with poor outcome, even in patients who have otherwise favorable disease features (7). With the crucial role that the MYC family plays in both solid and liquid malignancies, substantial interest has been shown in therapeutically targeting this pathway. However, due to both the difficulty in drugging transcription factors and the pleiotropic effects of the MYC family, it has been challenging to design therapeutic strategies targeting this family of oncogenes. The discovery that BET proteins are crucial in promoting the transcription of MYC, coupled with the generation of specific inhibitors of these proteins (3), provides fertile ground for designing therapies against MYC-driven malignancies.
In this issue of Cancer Discovery, Puissant and colleagues (8) report on their screen for cellular cytotoxicity and biomarkers of response with the BET inhibitor JQ1. Using 673 human cancer-derived cell lines from the recently described Cancer Cell Line Encyclopedia (CCLE; ref. 9), they showed a wide range of sensitivity to JQ1, indicating that the compound is not broadly cytotoxic. A substantial subset of the cell lines showed exquisite sensitivity to JQ1, displaying cell-cycle arrest followed by programmed cell death. Using the extensive genomic annotation available through the CCLE, the investigators discovered that the most robust biomarker of JQ1 cytotoxicity was MYCN amplification status. Turning to neuroblastoma, the investigators then showed that JQ1 activity was highly correlated with MYCN protein depletion in sensitive cells and directly showed displacement of BRD4 at the MYCN canonical promoter as the likely mediator of this MYCN protein depletion (Fig. 1). In addition, to begin to understand the alterations in cellular networks influenced by BET inhibition, the investigators developed an mRNA expression signature for JQ1 treatment, showing that it influences proliferation, apoptosis, neural crest stemness, neuritogenesis, and cellular differentiation. Finally, the investigators showed that JQ1 monotherapy could slow tumor growth in both xenotransplantation and transgenic mouse model systems, a significant finding because at least one JQ1 derivative molecule has recently entered clinical trials (http://clinicaltrials.gov/ct2/show/NCT01587703).
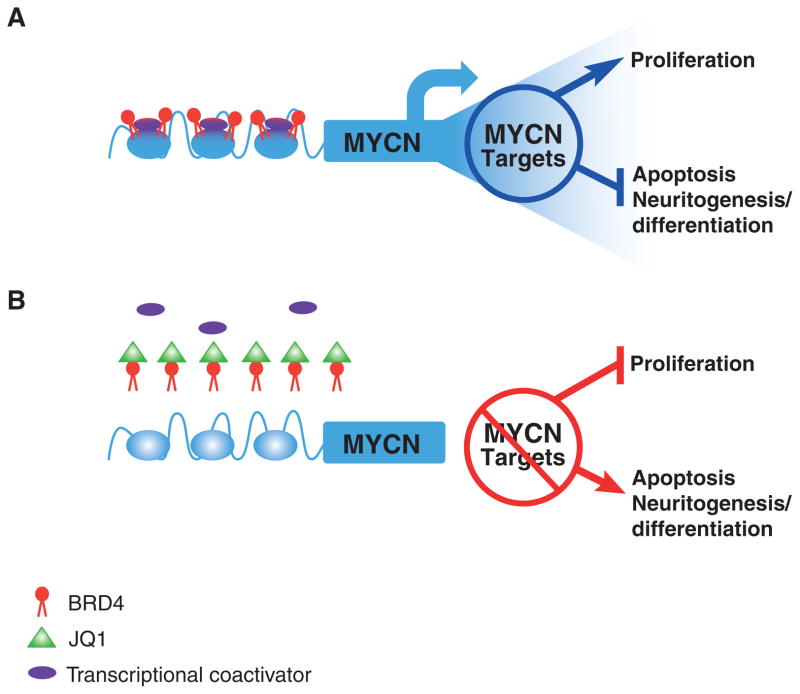
Figure 1.
Inhibition of BRD4 complex acetyl-lysine recognition by JQ1. A, BRD4 binds to the MYCN promoter at acetyl-lysine residues, recruiting additional proteins (such as the histone methyltransferase NSD3) that mediate chromatin density and allow for active MYCN transcription. MYCN target genes subsequently increase cellular proliferation by fostering cell-cycle progression as well as inhibiting apoptosis and blocking neuronal differentiation. B, JQ1 displaces BRD4 from the MYCN promoter, thus inhibiting MYCN transcription and leading to cell-cycle arrest, increased apoptosis, and increased neuritogenesis required for terminal differentiation.
This article further shows the power of pharmacologic screens in highly characterized human cancer-derived cell lines. Despite lingering concerns about the artificial conditions in which cancer cell lines are established and maintained, it is now clear that major oncogenic pathways are typically faithfully maintained in vitro, and thus clinically relevant associations regarding sensitive histotypes and genomic subsets can be generated from these screens. Here, the investigators show that BET inhibition is a valid therapeutic strategy in a broad range of cancers, extending the potential impact beyond hematologic malignancies and the rare NUT midline carcinomas that have been the focus of prior investigations.
The association of JQ1 sensitivity with MYCN amplification status is compelling for 2 distinct reasons. The obvious one is that it nominates MYCN amplification as a predictive biomarker for BET inhibitory activity in patients, suggesting that diseases such as neuroblastoma, small-cell lung cancer, and medulloblastoma should be emphasized in the clinical development of these compounds. Indeed, this may be an instance to design a trial agnostic to histology, and enrichment strategies (to maximize the potential of seeing clinical benefit) could be as simple as MYCN amplification being the major eligibility criterion for solid tumor phase I and II trials. The other reason this observation is compelling is that the vast majority of cell lines that were sensitive to JQ1 did not have MYCN amplification. Only 4 of the 99 sensitive cell lines, all neuroblastoma derived, harbored MYCN amplification. The CCLE includes other cell line models with MYCN amplification and/or very high expression of MYCN, and whether or not MYCN expression across the cell line panel was correlated with JQ1 sensitivity is unclear. Perhaps, more importantly, these data emphasize the fact that MYCN (or MYC) expression is not the sole predictor for sensitivity to BET inhibition, and clearly other biomarkers of sensitivity to BET inhibition remain to be discovered. The preliminary evidence that NOTCH pathway activation may also be enriched in JQ1-sensitive models is of interest but needs confirmation as well as mechanistic explanation.
Puissant and colleagues (8) very nicely show that the correlation of JQ1 cytotoxicity with MYCN has a direct mechanistic link by demonstrating displacement of BRD4 from the MYCN promoter and depletion of MYCN mRNA and protein following JQ1 administration. However, this must represent only one mechanism of action for a drug that likely has pleiotropic effects. It would have been predicted that JQ1 would be an active drug in mice xenotransplanted with human neuroblastoma-derived cell lines harboring MYCN amplification, and indeed, significant growth delay was shown. However, if MYCN depletion was the major mechanism of action, one would have predicted that the drug would be inactive in a transgenic model in which the MYCN transgene is not driven by a native promoter but is overexpressed in the neural crest and driven from an exogenous (tyrosine hydroxylase) promoter (10). The authors note that tyrosine hydroxylase expression is modulated by JQ1 and suggest this as a possible explanation for the downregulation of MYCN in the transgenic model of neuroblastoma, but this was not shown. Alternatively, JQ1 may be slowing tumor growth in the MYCN transgenic mice via destabilization of MYCN by unknown mechanisms or via other yet to be discovered on-target mechanisms.
One of the major issues to date with epigenetic-based cancer therapies is the relative nonspecificity of the compounds. Although BET inhibition may provide increased specificity, it is also clear that this class of proteins is broadly involved in a wide variety of gene regulatory processes, likely in a highly developmentally regulated and tissue-specific manner. Thus, on-target toxicity from these compounds should be anticipated, and the off-target effects are completely unknown. On-target toxicity can be derived from the disruption of MYC in normal tissues or from the potentially large number of other alterations in cellular networks these drugs likely will cause. As the investigators properly point out, we are only at the beginning of understanding the potential therapeutic use of BET bromodomain inhibition strategies.
Despite these cautionary notes, the potential impact of a drug strategy that targets MYCN cannot be underestimated. Since the original description in 1985 of MYCN amplification as a major predictor of death from neuroblastoma (11), an intense and largely frustrating focus has been placed on exploiting this potential oncogenic vulnerability. The work described by Puissant and colleagues (8) is a very important step in the development of precise therapeutic approaches to an important childhood cancer and may also define novel therapeutic strategies for substantial subsets of patients with other MYC-driven malignancies.
Acknowledgments
Grant Support
R. Schnepp is supported on the Abramson Cancer Center T32 training grant (T32 CA009615). J.M. Maris’ effort on this article was supported by the Giulio D’Angio Endowed Chair at the Children’s Hospital of Philadelphia.
Footnotes
Disclosure of Potential Conflicts of Interest
J.M. Maris has commercial research grants from GlaxoSmithKline and Novartis. No potential conflicts of interest were disclosed by the other author.
References
- 1.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–6. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]