Abstract
Autophagy, a highly conserved cellular mechanism wherein various cellular components are broken down and recycled through lysosomes, has been implicated in the development of heart failure. However, tools to measure autophagic flux in vivo have been limited. Here, we tested whether monodansylcadaverine (MDC) and the lysosomotropic drug chloroquine could be used to measure autophagic flux in both in vitro and in vivo model systems. Using HL-1 cardiac-derived myocytes transfected with GFP-tagged LC3 to track changes in autophagosome formation, autophagy was stimulated by mTOR inhibitor rapamycin. Administration of chloroquine to inhibit lysosomal activity enhanced the rapamycin-induced increase in the number of cells with numerous GFP-LC3-positive autophagosomes. The chloroquine-induced increase of autophagosomes occurred in a dose-dependent manner between 1 μM and 8 μM, and reached a maximum 2 hour after treatment. Chloroquine also enhanced the accumulation of autophagosomes in cells stimulated with hydrogen peroxide, while it attenuated that induced by Bafilomycin A1, an inhibitor of V-ATPase that interferes with fusion of autophagosomes with lysosomes. The accumulation of autophagosomes was inhibited by 3-methyladenine, which is known to inhibit the early phase of the autophagic process. Using transgenic mice expressing mCherry-LC3 exposed to rapamycin for 4 hr, we observed an increase in mCherry-LC3-labeled autophagosomes in myocardium, which was further increased by concurrent administration of chloroquine, thus allowing determination of flux as a more precise measure of autophagic activity in vivo. MDC injected 1 hr before sacrifice colocalized with mCherry-LC3 puncta, validating its use as a marker of autophagosomes. This study describes a method to measure autophagic flux in vivo even in non-transgenic animals, using MDC and chloroquine.
Keywords: chloroquine, autophagy, cardiac myocytes, LC3, monodansylcadaverine
Introduction
Autophagy is a highly conserved cellular mechanism that plays a key role in the turnover of long-lived proteins, RNA, and other macromolecules.1 In certain contexts including starvation or stress,2,3 autophagy represents an adaptive strategy by which cells clear damaged organelles and survive nutritional bioenergetic stress. Morphological and biochemical studies have shown that autophagy involves formation of a cup-shaped double-membrane structure, followed by sequestration or engulfment of cytoplasmic portions, protein aggregates, or intracellular organelles in double-membrane vacuoles called autophagosomes (AVs).1 Maturation occurs upon fusion with lysosomes to generate acidic single-membrane autophagolysosomes, where lysosomal proteases degrade the inner autophagosomal membrane and cargo. Autophagy is considered a physiological adaptive strategy required for living organisms. In other settings, however, deregulated autophagy may contribute to the pathogenesis of disease, such as cancer,4-6 neurodegenerative disorders,7,8 infectious disease,9 or cardiac disease.10,11 Little is known about the pathological roles of autophagy, due to the limitation of measuring autophagic flux. Flux reflects the dynamic process of autophagosome formation, engulfment, and lysosomal fusion. Static images of cells with numerous AVs could reflect increased autophagy, but could also reflect low autophagic flux due to impaired fusion with lysosomes. We recently reported a method to measure flux in cell culture,12 but methods to assess flux in vivo are lacking.
Accumulating evidence from patients indicates that autophagy occurs in the failing heart.13-15 Patients with cardiac hypertrophy who progress to heart failure have ultrastructural changes consistent with augmented lysosomal activity.13 Cardiac tissue of patients in heart failure demonstrates that ubiquitin-containing autophagic vacuoles appear in cardiomyocytes.14 However, these pathological studies cannot determine whether autophagy is a protective response to the initial cardiac insult or the cause of injury. It is also still under discussion using experimental models whether cardiac autophagy is a cardioprotective16,17 or maladaptive18-20 response to cardiac injury. Recently, pressure overload stress induced by aortic banding was reported to induce heart failure with increased cardiac autophagy, and suppression of autophagy ameliorated the development of heart failure.19 Conversely, another group reported that up-regulation of autophagy is a protective adaptation to hemodynamic stress.16 The role of autophagy in ischemia/reperfusion injury is also controversial. 17,20 Given this, there is a need to clarify the role of autophagy in the development of cardiac disease, a leading cause of mortality in developed countries.21 However, due to limitations in our ability to measure autophagic flux in heart, our understanding of the significance of increased numbers of AVs has been limited and in some cases may have led to incorrect conclusions. Here we present a method by which chloroquine can be used to verify that increased numbers of AVs reflect increased flux.
Chloroquine is an anti-inflammatory drug that has been used for over 60 years in the treatment of patients with malaria22 and inflammatory disorders.23 Chloroquine is thought to suppress inflammation by raising lysosomal pH24,25 and thereby inhibiting lysosomal activity.26,27 Chronic intoxication with this drug is known to result in myopathic changes with high lysosomal enzyme activities and the formation of numerous rimmed vacuoles.28,29 We previously used Bafilomycin A1, to inhibit lysosomal function to measure autophagic flux in cell culture.17 However, Bafilomycin A1 is not suitable for in vivo studies. We therefore investigated the use of chloroquine as a tool to measure autophagic flux in vitro and in vivo, in cardiomyocytes exposed to rapamycin or hydrogen peroxide (H2O2), and in the hearts of mice exposed to rapamycin.
Results
Chloroquine enhanced AV accumulation in cardiomyocytes stimulated with rapamycin
To examine the autophagic changes in cells, we transfected the cardiomyocyte-derived HL-1 cell line12,30-32 with GFP-tagged LC3. The 16-kDa processed isoform of LC3 is recruited from the cytoplasm to autophagosomal membranes. Thus, punctate GFP-LC3-labeled structures represent autophagosomes, referred to herein as AVs. Importantly, previous studies reported that overexpression of GFP-LC3 did not affect autophagic activity.35,36 The transfection efficiency achieved at least 40–60%, which was confirmed by control transfection with the plasmid expressing pEGFP-C1 (data not shown). We used rapamycin to stimulate autophagy. Rapamycin is known to induce autophagy via mTOR, which suppresses the initiation of autophagy and stimulates protein synthesis.37,38
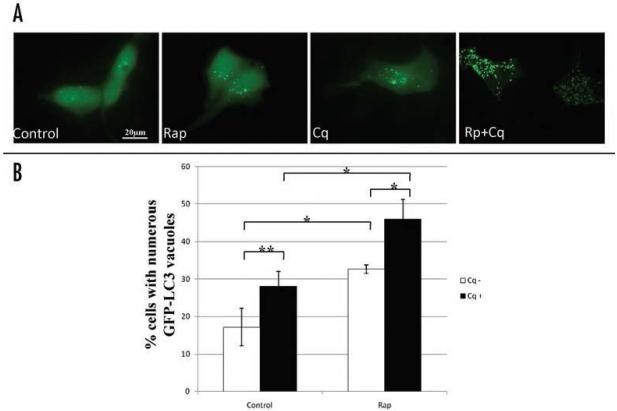
To avoid the influence of serum on the experiment, the serum-depleted medium was prepared as a control. Compared to the cells cultured with serum-containing medium, serum deprivation slightly increased the percentage of cardiomyocytes with numerous punctate GFP-LC3-labeled structures from 10.6 ± 1.5% to 17.2 ± 5.0%, consistent with previous reports of autophagic induction by serum deprivation.39-41 Rapamycin treatment greatly increased the percentage of cells with numerous punctate GFP-LC3-labeled structures. We previously used Bafilomycin A1 to inhibit lysosome-autophagosome fusion and therefore to measure flux.12 To quantify autophagic flux, activation of autophagy, indicated by the conversion of cytosolic diffuse GFP-LC3 to membrane associated GFP-LC3 puncta, was examined by fluorescence microscopy. Cells showing few or many GFP-LC3 puncta were classified (as previously described in ref. 12). In this study, to test the effect of chloroquine on rapamycin-activated autophagy, cells were treated with 5 μM rapamycin in the absence or presence of 3 μM chloroquine for 2 hours. Administration of 3 μM chloroquine increased the percentage of cells with high numbers of GFP-LC3 puncta relative to control. Concurrent administration of chloroquine with rapamycin induced a further increase in the percentage of cells with high numbers of puncta. Representative maximal Z-projection images of cells showing the pattern of distribution of LC3-GFP are shown in Figure 1A. Figure 1B shows the quantitative analysis of the percentage of cells with high numbers of GFP-LC3 vacuoles. These results suggested that chloroquine, like Bafilomycin A1, can be used as a tool to measure autophagic flux.
Figure 1.
Effect of chloroquine on autophagosome accumulation in cardiac myocytes. GFP-LC3-expressing HL-1 cardiac myocytes were treated with saline (Control), rapamycin (Rap, 5 μM), chloroquine (Cq, 3 μM), or rapamycin plus chloroquine (Rap+Cq) in serum free media for 2 hr. (A) Shown are maximum projection images of Z-stacks taken of GFP-LC3 fluorescence in HL-1 cardiac myocytes treated with the indicated conditions. Scale bar, 25 mm. (B) Autophagic flux was determined via fluorescent imaging of GFP-LC3 without and with chloroquine. Represented is the percentage of cells with numerous punctate GFP-LC3 labeled vacuoles per total cells scored. Results are mean ± SE of three independent experiments. *p< 0.01. **p< 0.05.
Dose-response and time-dependent effects of chloroquine on autophagy in cardiac myocytes
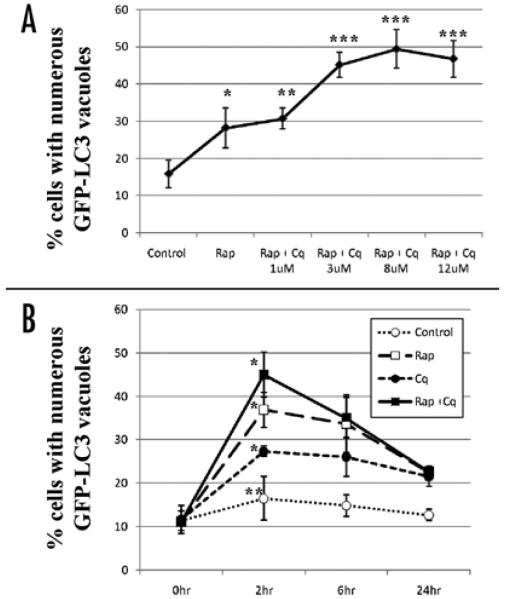
We examined the concentration of chloroquine required to block autophagosome-lysosome fusion. As shown in Figure 2A, in HL-1 cardiac myocytes, chloroquine increased the accumulation of autophagosomes stimulated by rapamycin in a dose-dependent manner, with the optimal effect at 3 μM. Importantly, even 12 μM chloroquine was not toxic to the cells during the 2 hr incubation, based on nuclear staining with Hoechst dye. Incubation of HL-1 cells with 3 μM chloroquine for 24 hour did not result in a significant increase in cell death, although cytotoxicity was observed with concentrations of 12 μM and higher (data not shown).
Figure 2.
Dose-response and time-course dependency of chloroquine effects on autophagy in cardiac myocytes. GFP-LC3-expressing HL-1 cardiac myocytes were incubated for 2 hr in the indicated conditions in serum free media. Represented is the percentage of cells with numerous punctate GFP-LC3 labeled vacuoles per total cells scored. Results are mean ± SE of 3 independent experiments. rapamycin (Rp), chloroquine (Cq), saline (control). (A) Dose-response curve of chloroquine effects on AV accumulation. Cardiac myocytes stimulated with rapamycin were treated with indicated dose of chloroquine. *p < 0.01 vs Control. **p = N.S. vs Rp. ***p < 0.01 vs Rp (B) Time-course dependency for maximal effect of chloroquine under the indicated conditions. *p < 0.001 vs 0 hour. **p < N.S. vs 0 hour
Figure 2B shows the time-course changes in the number of cells with numerous puncta after exposure to rapamycin and/or chloroquine. The maximal effect of chloroquine was observed 2 hr after addition, under all conditions.
Effect of chloroquine on lysosomal acidification
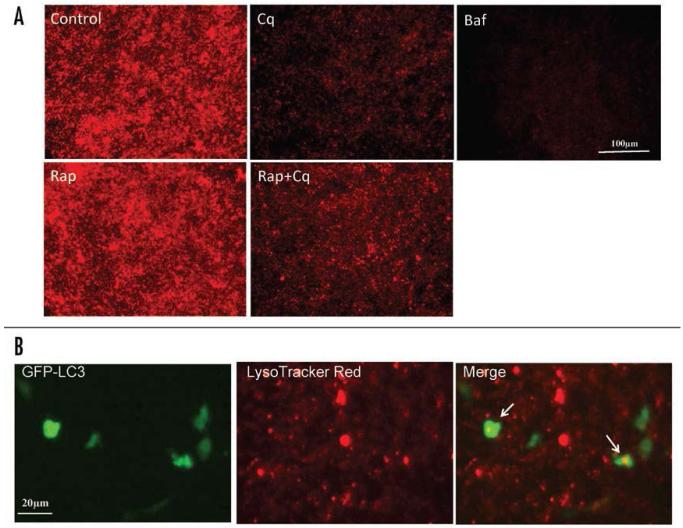
To examine the effects of chloroquine on lysosomal activity, HL-1 cells were incubated in LysoTracker Red, which labels the highly acidic lysosomal vacuoles and thus reports activity of the vacuolar H+-ATPase (v-ATPase). As shown in Figure 3A, lysosomes were numerous and increased further in number in response to rapamycin. However, treatment with 3μM chloroquine for 2 hr significantly reduced the uptake of LysoTracker Red into lysosomes, confirming the alkalinizing effect of chloroquine on lysosomes in HL-1 cells. Partial colocalization of GFP-LC3 and LysoTracker Red is shown in Figure 3B.
Figure 3.
The effect of chloroquine on lysosomal activity in HL-1 cardiac myocytes. (A) HL-1 cardiac myocytes were incubated with saline (Control), rapamycin (Rap, 5 μM), chloroquine (Cq, 3 μM), rapamycin plus chloroquine (Rap+Cq, 5 μM and 3 μM, respectively), or Bafilomycin A1 (Baf, 50 nM), in serum free media for 2 hr. Following treatment, cells were loaded with 50 nM LysoTracker Red for 5 min in the culture medium and analyzed by fluorescence microscopy. LysoTracker Red assessments were performed in three independent experiments; results presented are representative. (B) Higher magnification image of cells transfected with GFP-LC3 and loaded with LysoTracker Red, showing colocalization of GFP-LC3 and LysoTracker Red in a subset of structures (arrows).
The effect of chloroquine on autophagosome accumulation
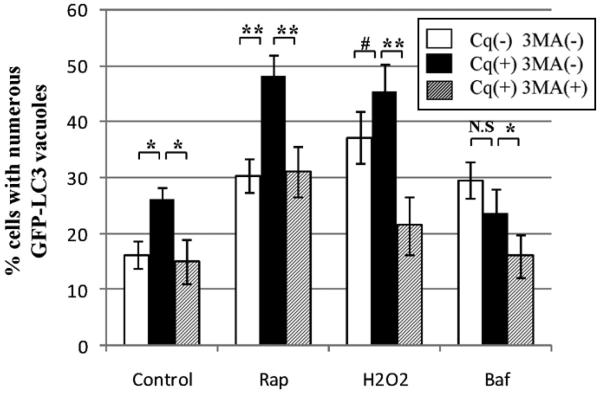
To further validate the use of chloroquine, we exposed HL-1 cardiac myocytes to H2O2, which induces oxidant stress, and rapamycin, which upregulates autophagy through inhibition of the negative regulator mTOR.37,38 As shown in Figure 4, both rapamycin and H2O2 increased the percentage of cells with numerous puncta. Concurrent administration of chloroquine increased the percentage further, verifying that hydrogen peroxide and rapamycin upregulate the formation of autophagosomes rather than impair lysosomal clearance. The formation of the autophagosome is dependent upon a class III PI3-K (VPS34) in mammalian cells,44 which can be inhibited by 3-methyladenine (3-MA).45,46 As shown in Figure 4, 3-MA completely inhibited the chloroquine-induced increase of autophagosomes. These findings indicate that mTOR inhibition or oxidant stress in cardiac myocytes upregulates PI3K-dependent autophagosome formation, while chloroquine results in autophagosome accumulation by blocking their degradation. To confirm that chloroquine causes autophagosome accumulation by lysosomal blockade, we evaluated the effects of chloroquine on HL-1 cardiac myocytes with Bafilomycin A1. Bafilomycin A1 is a macrolide antibiotic that was characterized initially for its selective inhibition of vacuolar-type proton ATPase.47-49 At nanomolar concentrations, it disrupts the vesicular proton gradient and ultimately increases the pH of acidic vesicles. This disruption prevents the fusion of autophagosomes with lysosomes, resulting in the accumulation of autophagosomes. In HL-1 cardiac myocytes, 50 nM Bafilomycin A1 increased the accumulation of autophagosomes (Fig. 4). Concurrent treatment with chloroquine and Bafilomycin A1 did not have an additive effect, indicating that they both target the late phase of autophagy. Given that chloroquine is relatively nontoxic, we reasoned that it could be used to measure autophagic flux in cardiac myocytes in vivo.
Figure 4.
Comparison of chloroquine and Bafilomycin A1 effects on autophagosome accumulation in cardiac myocytes. GFP-LC3-expressing HL-1 cardiac myocytes were incubated in the indicated conditions in serum free media. Represented is the percentage of cells with numerous punctate GFP-LC3 labeled vacuoles per total cells scored. Results are mean ± SE of three or more independent experiments. Saline (control), rapamycin (Rp, 5 μM), H2O2 (H2O2, 10−6 M), Bafilomycin A1(Baf, 50 nM), chloroquine (Cq, 3 μM), 3-MA (10 mM). *p < 0.05 . **p < 0.01. #p < 0.1. p = N.S Baf vs.Baf+Cq.
Chloroquine enhanced cardiac autophagic flux in vivo
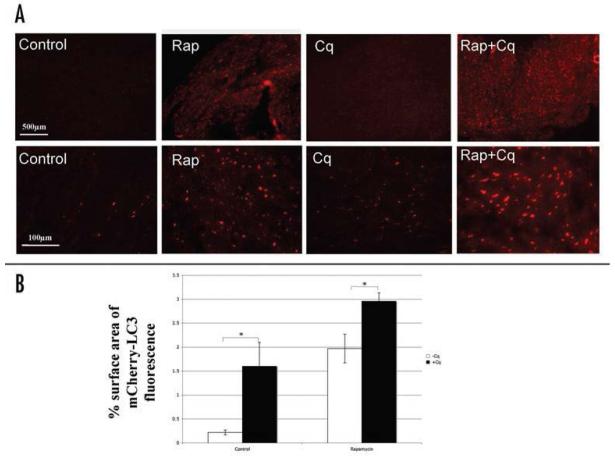
To assess whether chloroquine can be used for measuring autophagic flux in heart in vivo, we generated a line of transgenic mice expressing mCherry-tagged LC3 (mCherry-LC3) under the control of the cardiomyocyte-specific alpha-myosin heavy chain promoter. As noted earlier, prior studies have demonstrated that LC3 overexpression, both in vivo and in vitro,35,36 does not alter basal levels of autophagy. Transgenic mice expressing mCherry-LC3 were randomly assigned to receive rapamycin (2 mg/kg) or no treatment, with concurrent administration of chloroquine (10mg/kg) or saline via i.p. injection. Administration of rapamycin for 4 hr resulted in an increase in the abundance of mCherry-LC3 labeled puncta, while relatively little red fluorescence was observed in controls (Fig. 5). Nontransgenic mice showed almost no autofluorescence in the red channel (data not shown). The addition of chloroquine induced a modest increase in the abundance of mCherry-LC3 labeled puncta throughout the myocardium in controls, and a dramatic increase in hearts treated with rapamycin, indicating that the increase in autophagosomes after rapamycin administration reflects increased autophagic flux rather than impaired fusion with lysosomes.
Figure 5.
The effect of chloroquine on autophagic flux in vivo. Transgenic mice expressing mCherry-LC3 mice were randomly assigned to be exposed to chloroquine (Cq, 10 mg/kg), or rapamycin (2 mg/kg) or rapamycin plus chloroquine by i.p. injection. (A) Representative photographs of myocardium in mCherry-LC3 mice 4 hr after the indicated treatments. (B) Represented is the percentage surface area of mCherry-LC3 fluorescence per microscopic field. Results are mean ± SE of three independent experiments. *p < 0.001.
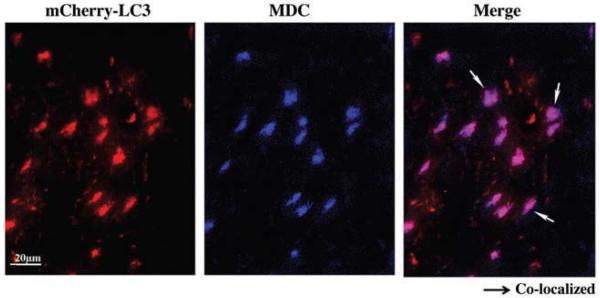
The foregoing results indicated that mCherry-LC3 reflects autophagy in the myocardium, and that chloroquine administration can provide additional information about autophagic flux. However, transgenic animals are not universally available, and in this instance, mCherry-LC3 expression is cardiac-restricted. It would be desirable to develop a means to assess autophagy in vivo that would be generally applicable. To this end, we evaluated monodansylcadaverine (MDC), which is known to label acidic endosomes, lysosomes, and autophagosomes. 50,51 We reasoned that under conditions that upregulate autophagy, MDC-labeled structures would similarly be upregulated. Using the mice described above, we injected MDC (1.5 mg/kg i.p.) 1 hr before sacrifice. As shown in Figure 6, MDC labels a subset of mCherry-LC3-positive structures, presumably autophagolysosomes. We did not detect any instances of MDC labeling of structures that were not also labeled with mCherry, suggesting that this reagent is specific and suitable for in vivo assessment of autophagy. The administration of chloroquine did not block MDC labeling, suggesting that MDC accumulation is independent of lysosomal pH, since (as shown in Fig. 3) chloroquine attenuated LysoTracker Red uptake almost as efficiently as Bafilomycin A1.
Figure 6.
Detection of autophagosomes by MDC labeling. Transgenic mice expressing mCherry-LC3 were treated with rapamycin and chloroquine as described in Figure 5. One hr before sacrifice, monodansylcadaverine (MDC, 1.5 mg/kg i.p.) was injected. After sacrifice, the heart was removed and prepared for imaging. Shown is a high magnification image (representative of three hearts), demonstrating colocalization of MDC with mCherry-LC3 (arrows). We did not observe MDC staining that did not overlap with mCherry-LC3, although we did observe some mCherry-LC3 puncta that did not colocalize with MDC.
Discussion
The need for research in cardiac autophagy has increased in recent years as growing evidence has indicated the involvement of this highly conserved mechanism in cardiac pathogenesis.13-20 Traditionally, autophagy has been evaluated by electron microscopy with recognition of characteristic structures. However, this method is difficult to quantify and provides no information about flux, or transit of material through the autophagic pathway. In this study, we monitored autophagy by the incorporation of fluorescent LC3 into autophagosomal membranes. In addition, we used here the maximal projection of Z-stacks to measure autophagy, which provided a more complete assessment of number of LC3 labeled structures than is possible with 2D imaging or electron microscopy.
High autophagosome content could reflect increased formation or decreased clearance of autophagosomes. For that reason, static measurements which score the number of autophagosomes present at a single time point may be misleading in certain circumstances, such as ischemia/reperfusion, where lysosomal clearance is impaired.17 Therefore, determination of autophagic flux is a more accurate representation of autophagy.12,52,53 We recently reported a method to measure flux in cell culture,12 but that method was not applied in vivo. Comparison of autophagosome number in the presence and absence of lysosomal inhibitors enables one to distinguish between increased autophagosome formation and decreased clearance, and thus can provide a quantitative index of flux.
In this study, we established a useful method with chloroquine for measuring autophagic flux-autophagosome generation and degeneration-in heart both in vitro and in vivo. Administration of chloroquine to HL-1 cells enhanced the accumulation of AVs induced by mTOR inhibition or oxidant stress, which was inhibited by 3-methyladenine, which blocks the initiation of autophagy. Chloroquine did not have an additive increase in AVs induced by Bafilomycin A1, indicating that both agents block autophagosome-lysosome fusion.24,25. Consistent with this, chloroquine attenuated LysoTracker Red uptake into lysosomes, although less potently than Bafilomycin A1.
Chloroquine was selected for evaluation in vivo because it has already been shown to be suitable for use in vivo and because it is quite inexpensive compared to Bafilomycin A1. We observed autophagosome accumulation in cell culture with as little as 3 μM chloroquine, which was below the 10 μM concentration reported to suppress cytokine release.42,43 Under these conditions we observed no cytotoxicity with chloroquine, suggesting that it can be used without concern for confounding cellular effects.
Due to the limited availability of the GFP-LC3 transgenic mice, we generated our own line, using mCherry-LC3 driven by the cardiac-restricted alpha myosin heavy chain promoter. For studies of cardiac autophagy, this eliminates the potentially confusing contribution of endothelial cells and fibroblasts present in numbers equal to cardiomyocytes. Of several founder lines, we chose the lowest-expressing line, as the high-expressing lines occasionally developed large fluorescent aggregates that were morphologically inconsistent with autophagosomes. The use of mCherry-LC3 offers two advantages over GFP-LC3: it retains fluorescence even in the acidic environment of the lysosome, and there is very little red autofluorescence.
In transgenic mice expressing mCherry-LC3, rapamycin administration caused an increase in the abundance of mCherry-LC3-labeled AVs in the myocardium. This was further enhanced by the coadministration of chloroquine, indicating that the increase in AVs after rapamycin administration was due to increased flux, not diminished clearance. These findings clearly indicate that chloroquine is useful in evaluation of autophagic flux in vivo. Moreover, these studies show that chloroquine can be used to suppress the late phase of autophagy, which may be of therapeutic value in certain conditions.
MDC has been criticized for its lack of specificity, yet we found that it colocalized perfectly with mCherry-LC3, and that MDC labeling increased in parallel with mCherry-LC3 puncta. While GFP-LC3 fluorescence is lost in the acidic environment of the lysosome, mCherry fluorescence is stable even at acid pH, thus allowing detection of both early and late autophagosomes.54 Thus it is conceivable that the colocalization of MDC with many (but not all) LC3-mCherry puncta is an indication of the number of autophagolysosomes. If true, MDC would represent an accurate indicator of flux, since it would only label autophagosomes that have fused with lysosomes. However, MDC has also been suggested to incorporate into membranes based on lipid characteristics independent of pH.55 If the incorporation is related to the double-membrane structure of the autophagosome, then MDC incorporation would be expected even when chloroquine is used to block autophagosome-lysosome fusion, as is the observation in our studies. Taken together, this study describes a method to measure autophagic flux in vivo that can be used in nontransgenic animals. It is hoped that use of this methodology will expand our understanding of autophagy in physiologic and pathologic settings.
The role of autophagy in cardiac pathology is contradictory, in part because of the difficulty of interpreting increased numbers of autophagosomes which could be due to increased formation or decreased clearance. The use of chloroquine in these studies may offer a better understanding of autophagy and its role-protective or maladaptive-in cardiac pathologic processes such as ischemia/reperfusion injury or heart failure. This enhanced understanding of autophagy may lead to the development of novel strategies to prevent cardiac diseases.
Material and Methods
Reagents
Chloroquine, rapamycin, Bafilomycin A1, and 3-Methyladenine (3-MA) were purchased from EMD Biosciences. Monodansylcadaverine (MDC) was from Sigma. pEGFP-C1 was obtained from BD Biosciences.
Cell culture and transfection
Cells of the murine atrial-derived cardiac cell line HL-130-32 (kind gift of Dr. W. Claycomb) were plated onto tissue culture chamber slides (Nalge Nunc International) or 35 mm glass-bottomed culture dishes (MatTek Co.) and cultured in Claycomb medium (JRH Biosciences) supplemented with 10% fetal bovine serum, 0.1 mM norepinephrine, 2 mM L-glutamine, 100 U mL-1penicillin, 100 U mL-1streptomycin, and 0.25 μg mL-1amphotericin B. Cells were transfected with the indicated vectors using the transfection reagent Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, achieving at least 40–60% transfection efficiency.17,33
Activation of autophagy in vivo
Cardiac-specific expressing mCherry-LC3 transgenic mice were created in the FVB/N strain by pronuclear injection of murine alpha myosin heavy chain promoter driven mCherry-LC3 transgene in front of the human growth hormone poly adenylation signal. The mice were created at the Scripps Research Institute mouse genetics core. The administration of rapamycin, chloroquine, MDC or saline was done via i.p. injections at the indicated dose. Four hours after injection, animals were sacrificed and cardiac tissue was harvested immediately. The tissues were fixed in 10% formalin for preparation of paraffin-embedded sections and examined for cardiac autophagy under widefield fluorescence microscopy. Mice were treated in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All animal protocols were approved by the Animal Care Committee of the San Diego State University in San Diego, CA.
Widefield fluorescence microscopy
Cells and tissues were observed through a Nikon TE300 fluorescence microscope (Nikon) equipped with a 4× lens and a 20× lens (0.3 NA, Nikon), a 60× Plan Apo objective (1.3 NA oil immersion lens; Nikon), a Z-motor (ProScanII, Prior Scientific), a cooled CCD camera (Orca-ER, Hamamatsu) and automated excitation and emission filter wheels controlled by a LAMBDA 10-2 (Sutter Instrument) operated by MetaMorph 6.2r4 (Molecular Devices Co.). Fluorescence was excited through an excitation filter for FITC (HQ480/×40), Texas Red (D560/x40). Fluorescent light was collected via a polychroic beamsplitter (61002 bs) and an emission filter for fluorescein isothiocyanate (HQ535/50m), and Texas Red (D630/60m). All filters were from Chroma. Acquired wide field Z-stacks were routinely deconvolved using 10 iterations of a 3D blind deconvolution by MetaMorph 6.2r4. Unless stated otherwise, representative images shown are maximum projections of Z-stacks taken with 0.20 mm increments capturing total cellular volume.
Quantification of autophagic flux
Microtubule-associated protein light chain 3 (LC3), an 18-kDa mammalian homolog of autophagy-related protein 8 (Atg8) in yeast, is processed and conjugated to the nascent autophagosome membrane at the initiation of autophagy.34 Therefore, autophagosomes were quantified via fluorescence imaging of GFP-LC3 in cells. HL-1 cells were transfected with GFP-LC3, and 48 hr after transfection, cells were subjected to the indicated experimental conditions. Cells were fixed with 4% formaldehyde in phosphate-buffered saline, pH 7.4, for 15 min. To quantify the number of GFP-LC3 puncta in a single cell, Z-stack images of single GFP-LC3-expressing cells were captured using 60× oil objective and then background fluorescence was removed by applying a mean filter (4-pixel) to each stack image.12 Maximal projection images were produced, and the number of GFP-LC3 puncta per cell was determined using IMAGEJ software (NIH, Bethesda, MD). For population analysis, cells were inspected at 60× magnification and classified as either having predominantly diffuse GFP-LC3 fluorescence (<0–30 GFP-LC3 dots/cells) or having numerous punctate GFP-LC3 structures (>30 GFP-LC3 dots/cells), representing autophagosomes (AVs). Previous studies in this cell line established a clear bimodal distribution, allowing us to use the threshold of 30 dots as a cutoff.12 At least 200 cells were scored in each of three or more independent experiments. To quantify the autophagic flux in vivo, percentage of surface area covered by mCherry-LC3 fluorescence was measured using IMAGEJ software (http://rsb.info.nih.gov/ij).
Determining lysosomal activity
Acidic organelles were labeled with the acidotropic dye LysoTracker Red (50 nM, Molecular Probes) diluted in culture medium for 5 min according to the manufacturer’s instructions. Labeled cells were imaged by fluorescence microscopy at 20× magnification using an excitation filter for Texas Red (D560/×40).
Statistics
The probability of statistically significant differences between two experimental groups was determined by Student’s t-test. Values are expressed as mean ± SEM of at least three independent experiments unless stated otherwise.
Acknowledgements
This work was supported by NIH grant P01-HL85577 (Dr. Gottlieb), and by a grant from the Ministry of Education, Science, and Culture of Japan (Dr. Iwai-Kanai).
References
- 1.Cuervo AM. Autophagy: Many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 3.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: Cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–48. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 4.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue Z, Jin S, Yang C, Levine AJ, Heinz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–84. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 8.Hara T, Nakamura K, Matui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–89. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez M, Master S, Singh S, Taylor G, Colombo M, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Myobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Rothermel BA, Hill JA. Myocyte autophagy in heart disease: Friend or foe? Autophagy. 2007;3:632–4. doi: 10.4161/auto.4913. [DOI] [PubMed] [Google Scholar]
- 11.Rabkin SW. Nitric oxide-induced cell death in the heart: The role of autophagy. Autophagy. 2007;3-4:347–49. doi: 10.4161/auto.4054. [DOI] [PubMed] [Google Scholar]
- 12.Brady NR, Hamacher-Brady A, Yuan H, Gottlieb RA. The autophagic response to nutrient deprivation in the HL-1 cardiac myocytes is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J. 2007;274:3184–97. doi: 10.1111/j.1742-4658.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto S, Sawada K, Shimomura H, Kawamura K, James TN. On the nature of cell death during remodeling of hypertrophied human myocardium. J Mol Cell Cardiol. 2000;32:161–75. doi: 10.1006/jmcc.1999.1064. [DOI] [PubMed] [Google Scholar]
- 14.Knaapen MW, Davis MJ, De Bie M, Haven AJ, Martiner W, Kockx MM. Apoptotic versus autophagic cell death in heart failure. Cardio Res. 2001;51:304–12. doi: 10.1016/s0008-6363(01)00290-5. [DOI] [PubMed] [Google Scholar]
- 15.Kostin S, Pool L, Elsasser A, Hein S, Drexler HC, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn WP, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–24. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 16.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Misote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 17.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 18.Akazawa H, Komazaki S, Shimomura H, Terasaki F, Zou Y, Takano H, Nagai T, Komuro I. Diphtheria toxin-induced autophagic cardiomyocyte death plays a pathogenic role in mouse model of heart failure. J Biol Chem. 2004;279:41095–103. doi: 10.1074/jbc.M313084200. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–93. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentim I, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits Beclin 1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–52. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 21.Rosamond W, Flegal K, Friday G, Furie K, Go A, Greenlund K, Haase N, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O’Donnell CJ, Roger V, Rumsfeld J, Sorlie P, Steinberger J, Thom T, Wasserthiel-Smoller S, Hong Y. American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2007 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2007;115:e69–171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 22.O’Neill PM, Bray PG, Hawley SR, Ward SA, Park BK. 4-Aminoquinolines-past, present, and future: A chemical perspective. Pharmacol Ther. 1998;77:29–58. doi: 10.1016/s0163-7258(97)00084-3. [DOI] [PubMed] [Google Scholar]
- 23.Kremer JM. Rational use of new and existing disease-modifying agents in rheumatoid arthritis. Ann Intern Med. 2001;134:695–706. doi: 10.7326/0003-4819-134-8-200104170-00013. [DOI] [PubMed] [Google Scholar]
- 24.Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981;90:665–69. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy. 2007;3:154–7. doi: 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- 26.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA. 1978;75:3327–31. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sewell RB, Barham SS, LaRusso NF. Effect of chloroquine on the form and function of hepatocyte lysosomes: Morphologic modifications and physiologic alternations related to the biliary excretion of lipids and proteins. Gastroenterology. 1983;85:1146. [PubMed] [Google Scholar]
- 28.Stein M, Bell MJ, Ang LC. Hydroxychloroquine neuromyotoxicity. J Rheumatol. 2000;27:2927–31. [PubMed] [Google Scholar]
- 29.Murakami N, Oyama F, Gu Y, McLennan IS, Nonaka I, Ihara Y. Accumulation of tau in autophagic vacuoles in chloroquine myopathy. J Neuropathol Exp Neurol. 1998;57:664–73. doi: 10.1097/00005072-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–84. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: Use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–9. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 32.George CH, Rogers SA, Bertrand BM, Tunwell RE, Thomas NL, Steele DS, Cox EV, Pepper C, Hazeel CJ, Claycomb WC, Lai FA. Alternative splicing of ryanodine receptors modulates cardiomyocyte Ca2+ signaling and susceptibility to apoptosis. Circ Res. 2007;100:874–83. doi: 10.1161/01.RES.0000260804.77807.cf. [DOI] [PubMed] [Google Scholar]
- 33.Iwai-Kanai E, Hasegawa K, Sawamura T, Fujita M, Yanazume T, Toyokuni S, Adachi S, Kihara Y, Sasayama S. Activation of lectin-like oxidized low-density lipoprotein receptor-1 induces apoptosis in cultured neonatal rat cardiac myocytes. Circulation. 2001;104:2948–54. doi: 10.1161/hc4901.100381. [DOI] [PubMed] [Google Scholar]
- 34.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized on autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–62. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 38.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansion in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 39.Chang CL, Liao JJ, Huang WP, Lee H. Lysophosphatidic acid inhibits serum deprivation-induced autophagy in human prostate cancer PC-3 cells. Autophagy. 2007;3:268–70. doi: 10.4161/auto.3909. [DOI] [PubMed] [Google Scholar]
- 40.Araki T, Hayashi M, Saruta T. Anion-exchange blocker enhances cytoplasmic vacuole formation and cell death in serum-deprived mouse kidney epithelial cells in mice. Cell Biol Int. 2006;30:93–100. doi: 10.1016/j.cellbi.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Mitchener JS, Shelburne JD, Bradford WD, Hawkins HK. Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am J Pathol. 1976;83:485–91. [PMC free article] [PubMed] [Google Scholar]
- 42.Hong Z, Jiang Z, Liangxi W, Guofu D, Ping L, Yongling L, Wendong P, Minghai W. Chloroquine protects mice from challenge with CpG ODN and LPS by decreasing proinflammatory cytokine release. Int Immunopharmacol. 2004;4:223–34. doi: 10.1016/j.intimp.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Weber SM, Levitz SM. Chloroquine interferes with lipopolysaccharide-induced TNF-α gene expression by a nonlysosomotropic mechanism. J Immunol. 2000;165:1534–40. doi: 10.4049/jimmunol.165.3.1534. [DOI] [PubMed] [Google Scholar]
- 44.Petiot A, Ogier-Denis E, Blommaart EFC, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 45.Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–92. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–6. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 47.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–6. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dröse S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993;32:3902–6. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 49.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–40. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munafo DB, Colombo MI. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001;114:3619–29. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 51.Yan CH, Yang YP, Qin ZH, Gu ZL, Reid P, Liang ZQ. Autophagy is involved in cytotoxic effects of crotoxin in human breast cancer cell line MCF-7 cells. Acta Pharmacol Sin. 2007;28:540–8. doi: 10.1111/j.1745-7254.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- 52.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, Molgo J, Diaz J, Lavandero S, Harper F, Pierron G, di Stefano D, Rizzuto R, Szabadkai G, Kroemer G. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029–39. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 54.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 55.Niemann A, Takatsuki A, Elsasser HP. The lysosomotropic agent monodansylcadaverine also acts as a solvent polarity probe. J Histochem Cytochem. 2000;48:251–8. doi: 10.1177/002215540004800210. [DOI] [PubMed] [Google Scholar]