Abstract
Recent controversies associated with the structure of the M2 protein from influenza A virus and the binding site of drug molecules amantadine and rimantadine motivated the comparison here of the drug binding to three viral porins including the M2 proteins from influenza A and B as well as the viral protein ‘u’ from HIV-1. While the M2 protein from influenza B does not normally bind amantadine, chimeras with the M2 protein from influenza A show blockage by amantadine. Similarly, Vpu does not normally bind rimantadine, but the single site mutation A18H converts a non-specific channel to a selective proton channel that is sensitive to rimantadine. The comparison of structures and amino acid sequences shows that the membrane protein sample environment can have a significant influence on the structural result. While a bilayer surface bound amphipathic helix has been characterized for AM2, such a helix may be possible for BM2 although it has evaded structural characterization in detergent micelles. A similar amphipathic helix seems less likely for Vpu. Even though the A18H Vpu mutant forms rimantadine sensitive proton channels, the binding of drug and its influence on the protein structure appears to be very different from that for the M2 proteins. Indeed, drug binding and drug resistance in these viral porins appears to result from a complex set of factors.
Keywords: M2 protein, Influenza A virus, Influenza B virus, Viral protein u, HIV, PISEMA, Solid-state NMR, Membrane protein
1. Introduction
Recently, there has been interest in the A18H mutant of Vpu, viral protein ‘u’ of HIV-1, that induces highly selective H+ conductance not present in the wild type and that is blocked by rimantadine [1,2]. This mutation generates the proton-conductance signature sequence HxxxW found in two M2 proteins from influenza virions [3]. While the influenza AM2 conductance is amantadine and rimantadine sensitive, influenza BM2 conductance is not [4]. These three proteins have essentially no sequence homology except for the HxxxW motif. Here, we compare the sequences, and what is known about the structures of these three proteins for the domain responsible for the H+ selective conductance. We will also discuss mechanisms for drug binding and drug resistance.
Of these three proteins, AM2 has been the subject of numerous conductance, structural, and computational studies. As with Vpu and BM2, there are no full-length structures, although for BM2 two overlapping constructs have been used to develop the first full-length structural model [5]. A variety of membrane mimetic environments have been used for the structural and functional studies. Most of the structural efforts have focused on the transmembrane (TM) domain, a single helix that as a tetramer for AM2 and BM2 conducts protons across the viral membranes, an essential function for the life cycle of the virus[3,6-9]. Based on detailed conductance studies by Pinto and coworkers [10], the minimal sequence that accounts for the conductance properties of AM2 near neutral pH includes not only the TM helix, but an additional sequence following the TM helix. This latter sequence is known to form a bilayer-bound amphipathic helix [11,12]. The drugs, amantadine and rimantadine, were effective against influenza A until recently when the influenza A strains became dominated by an S31N mutation displaying drug resistance. A variety of other naturally occurring mutants also display drug resistance. While there is no direct proton-conductance function associated with Vpu and HIV-1, the TM domain has been associated with non-specific ion conductance and the ability to enhance virion release from infected cells [13-15], such that a chimera of Vpu including the TM domain of AM2 appears to function normally in HIV-1 and displays proton selective currents [2,15].
The results discussed here reflect data obtained from a variety of technologies utilizing different membrane mimetic environments for membrane proteins. These environments have structural, dynamical, and functional implications that cannot be ignored if the goal is to describe the native state. Unfortunately, we are not yet able to characterize membrane proteins in their native membrane environment, but it is possible to characterize proteins in lipid bilayers, even liquid crystalline environments. Such environments are similar to native membranes in many respects, including a well-defined hydrophobic thickness, a complex interfacial region, and extreme dielectric and water concentration gradients. Yet it is important to acknowledge that synthetic bilayers and other model membrane environments may fall far short of complex native membranes.
2. Materials and methods
This paper is primarily a review paper; however, there are a few previously unpublished results that are included and hence this Materials and methods section.
2.1. Sample preparation
15N-amantadine•HCl was synthesized according to the literature procedure [16]. 15N-labeled acetonitrile (Isotec, Miamisburg, Ohio) was used to provide the 15N source. The final product was verified by Mass and 1H solution NMR spectroscopy. The M2 TM domain peptide, NH2-Ser22-Ser-Asp-Pro-Leu-Val-Val-Ala-Ala30-Ser-Ile-Ile-Gly-Ile-Leu-His37-Leu-Ile-Leu40-Trp-Ile-Leu-Asp-Arg-Leu46-COOH, was chemically synthesized by solid-phase synthesis on an Applied Biosystems 430A Synthesizer using 15N-labeled FMOC amino acids obtained from Isotec and Cambridge Isotope Labs (Cambridge, Mass). The peptide was purified and characterized as described previously [17].
Oriented samples of the 15N-labeled peptide in hydrated DMPC bilayers were prepared by first co-dissolving M2 TM domain (10 mg) and DMPC (100 mg) in 5 ml TFE (trifluoroethanol). TFE was removed by rotary evaporation and dried under high vacuum. Fifteen milliliters of 20 mM CBP (citrate-borate-phosphate) buffer (~37 °C, pH 8.0) with 1 mM EDTA (ethylenediamine tetra-acetic acid) was added to the dried mixture and shaken at 37 °C. This lipid suspension was bath sonicated for 10 min intermittently. The sonicated suspension was loaded into a 3-kDa MW cutoff dialysis bag. The dialysis bag was placed in 1 L of 20 mM CBP buffer overnight to adjust the pH of the M2 TM domain/DMPC liposomes. For an M2 TM domain sample with 10 mM amantadine, 46.9 mg (250 μmol) amantadine hydrochloride (Fisher Scientific, GA) in 5 ml CBP buffer was added to an M2 TM domain loaded vesicle suspension (20 ml). The suspension was incubated at room temperature overnight and pelleted in 2.5 h by ultracentrifugation at 196,000×g. The pH value of the pellet was inferred from a measurement of the supernatant. The pellet was agitated at 37 °C for 1 h until fluid. For M2 TM domain studies with amantadine, proteoliposomes were prepared first without amantadine and then amantadine was added to a 1-ml suspension of liposomes at the desired protein:drug ratio and incubated overnight before sample preparations. This thick fluid was spread onto 50 glass slides (5.7×12.0 mm) (Marienfeld Glassware, Lauda-Königshofen, Germany) and dried in a humidity (70–75% relative humidity) chamber using an N2 atmosphere. The partially dehydrated slides were stacked together, inserted into a glass tube, and rehydrated in a 96% relative humidity (saturated K2SO4) chamber at 40 °C for one week. Finally the glass tube was sealed with wax.
2.2. PISEMA spectroscopy
All PISEMA spectra were acquired at 600 MHz utilizing low-E probes at the NHMFL except for the full-length M2 PISEMA spectrum that was obtained at 400 MHz as described previously [11]. The PISEMA spectra of the M2 TM domain were obtained with the following parameters: 800 μs cross-polarization contact time, 4 ms acquisition time, 6 s recycle delay, and 1H decoupling with the SPINAL scheme [18]. Spectra were typically acquired with 32 t1 increments resulting in total acquisition times ranging from 6 h to 3 days. All experiments were conducted at either 30 or 40 °C, well above the gel to liquid crystalline phase transition for the bilayer systems employed. Spectra were processed using in-house scripts written for NMRPipe [19]. Processing scripts included the following: zero filling of t2 to 1024 points, linear prediction of t1 to 128 points; exponential multiplication window function for t1 and t2 domains with 164 Hz Lorentzian line broadening to enhance signal to noise; shifted sine bell curve for both t1 and t2 domains; Lorentz-to-Gauss window function to reduce linewidths; Fourier transformation of both t1 and t2 time domains; correction of the 15N–1H-dipolar coupling dimension by a scaling factor of 0.816 (sin54.7°). 15N-chemical shifts were referenced to liquid ammonia at 0 ppm via a saturated solution of 15NH4NO3 at 26 ppm.
3. Results and discussion
3.1. Sequence analysis of viral ion channels
All three proteins are single pass membrane proteins with a TM domain flanked by hydrophilic domains (Fig. 1). The focus here will be on the TM helix and the subsequent amphipathic sequence. With the exception of a few key functional residues lining the channel pore, amino acid composition of the TM helices is largely hydrophobic. For such a composition, the possibilities for hydrogen bonds and electrostatic contacts at the helix–helix interface are rare. TM helical association is therefore largely determined by weak van der Waals interactions. In fact, stronger inter-helical interactions might considerably hinder the dynamics important for channel function. The amphipathic sequence has a net positive charge and its amphipathic composition suggests a helical structure.
Fig. 1.
N-terminal sequences of AM2 (Udorn/72), BM2 (Lee/40), and Vpu (HIV-1 isolate HTLVIIIB) extended membrane domains. The italicized residues in BM2 and Vpu are from AM2 and the histidine and tryptophan residues in the HxxxW are highlighted in red.
The TM domains of AM2 and BM2 feature the proton channel signature sequence, HxxxW [3]. Otherwise, the TM sequences of AM2 and BM2 have only occasional sequence identity and yet both of these TM domains function to transport protons. Sequence determinants for oligomeric assemblies of membrane proteins have been recognized as having helical repeat motifs, such as AxxxG, GxxxG, SxxxS, and GxxxGxxG, composed of small residues at every fourth or seventh position, which allow for tight packing at helical interfaces [20]. Such packing generates the potential of CαH hydrogen bonding and significant inter-helical electrostatic interactions to complement the non-specific van der Waals interactions [20]. Both AM2 and BM2 have sequences characteristic of oligomeric proteins and yet alanine and glycine residues in the AxxxG sequence of AM2 are not at the helix–helix interface. Similarly, the SxxSxxxS sequence of BM2 places three serine residues in a narrow 60° arc of a helical wheel and are not positioned at the helix–helix interface, but rather they line the aqueous pore [5,21]. Consequently, the lipid facing and helix interfacial residues are typically bulky hydrophobic residues such as valine, leucine, and isoleucine, implying relatively poor stability for the tetrameric TM domain. Indeed, the plasticity of the AM2 TM domain has been extensively discussed in the literature [22] and in a detergent environment this tetrameric structure is not stable enough for a solution NMR structural characterization. [23]
For Vpu, an AxxAxxxAxxxA sequence places four alanine residues in an 80° arc of a helical wheel, and if we assume that the Trp22 Cα faces the pore (as the corresponding tryptophan residues do in AM2 and BM2), then the Ala residues are not facing into the pore but are largely at the helix–helix interface. This may explain why the Vpu TM domain oligomeric structure is more stable even in a detergent environment than the AM2 TM domain [24]. The stability of the Vpu TM domain is also seen in bicelle spectra where the NMR linewidths are much narrower than the line widths observed for the AM2 TM domain in bilayers [25]. The pore of Vpu is very hydrophobic with only 4 hydrophilic residues (i.e. 4 Trp22 residues, assuming that it is tetrameric and that tryptophan is considered hydrophilic), while AM2 has 12 hydrophilic residues and BM2 has 20 such residues per tetramer. BM2 and Vpu are not sensitive to the channel blocking drugs, amantadine and rimantadine, while the AM2 conductance is effectively blocked by amantadine, which binds with high affinity and induces a significant structural change to the protein [10,26,27]. Interestingly, the A18H mutation converts Vpu to a highly selective proton channel that has amantadine sensitivity and also doubles the number of hydrophilic residues facing the pore [25]. BM2 can also be made amantadine sensitive by forming a chimera in which the N-terminal half of the TM helix is replaced with the corresponding sequence from AM2 (Fig. 1) [28]. This chimera decreases the number of hydrophilic residues in the BM2 TM domain from 20 to 12. Consequently, it appears that amantadine binding requires a balance between hydrophilic and hydrophobic residues in the pore (Fig. 2). Amantadine and rimantadine are largely hydrocarbon structures (Fig. 2) with a primary amine and consequently it is not surprising that increased hydrophilicity in BM2 could inhibit drug binding.
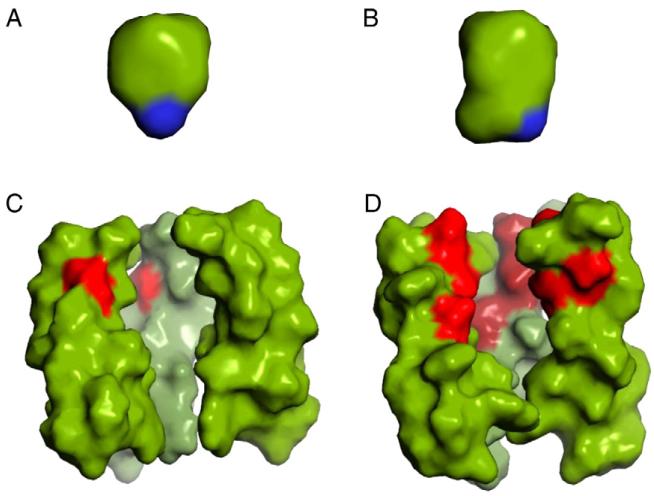
Fig. 2.
Comparison of the amphipathic drugs, amantadine (A) and rimantadine (B), with the pores from AM2 (C, PDB 2H95) and BM2 (D, PDB 2KIX) on the same scale. The AM2 and BM2 structures are represented by only 3 of the 4 helices so that a view into the pore can be achieved. Furthermore, only residues Leu26-Ile42 (AM2 numbering) are displayed. The amino group of the drugs is colored blue, the AM2 drug binding site is primarily hydrophobic indicated by green (left) while BM2 (right) channel pore is lined with polar residues in red.
For AM2, the conductance properties of the full-length protein are nearly identical (at least at neutral pH) to the TM domain if the additional residues of the amphipathic sequence are included [10]. Consequently, we refer to constructs for AM2 that include residues 22–62 or 18–60 as the ‘conductance domain’. The sequence following the TM helix in each of these proteins displays a preponderance of positively charges residues (Fig. 1). Within the 19 residues following the key tryptophan residue (Trp41 in AM2, Trp23 in BM2 and Trp22 in Vpu), each of these proteins has 6 positive charges (assuming histidine is counted as a positive charge). The 19-residue sequences have a net positive charge of 4 for AM2 and Vpu and a net positive charge of 6 for BM2. This is consistent with the positive inside rule and presumably reflecting interactions between the protein and the negatively charged lipids on the viral interior.
3.2. Structural perspective
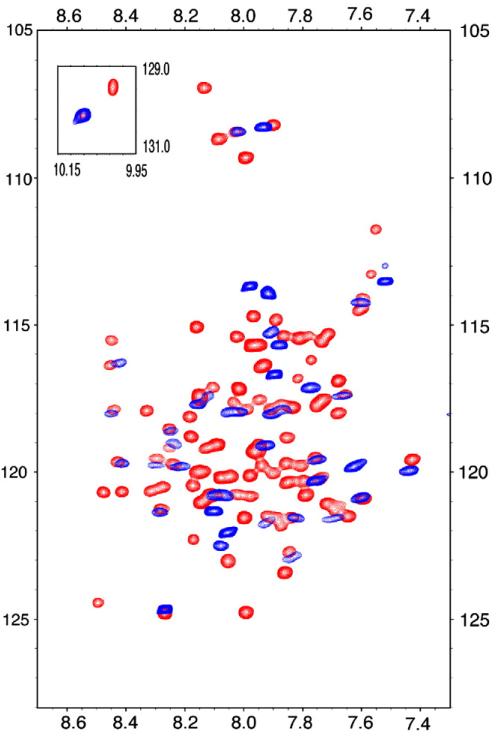
These viral porins provide some of the best examples of the ‘divide-and-conquer’ strategy [29,30] for the structural characterization of membrane proteins, in which individual functional domains are separately studied. The usefulness of this approach for these proteins is partly due to the fact that these proteins have identifiable functional domains. Substantiated with functional assays, this strategy can help overcome challenges associated with spectroscopy and crystallography studies of full-length membrane proteins. The structures of isolated domains of BM2, Vpu, and AM2 have provided significant insights into the structures and functions of the full-length proteins [31-35]. As an example, solution NMR HSQC spectra of the full-length and conductance domains of AM2 are compared in Fig. 3. The near perfect overlap of these two spectra and the appropriate dispersion of resonances for a largely helical protein are yet another justification that this divide-and-conquer approach is an effective and useful strategy for the structural characterization of such proteins with definable functional domains.
Fig. 3.
Overlay of solution NMR HSQC spectra of the AM2 conductance domain (residues 22-62) (blue) and the full-length protein (red) in LPPG micelles at pH 4. The conductance domain of AM2 folds independently of other domains, thus providing the basis for the divide-and-conquer approach for characterizing viral ion channels structures.
3.2.1. The transmembrane domain
Two drug binding sites have been proposed for influenza A M2, one distal to the HxxxW motif in the N-terminal half of the pore based on TM domain studies,[26,36-39] and the other proximal to this motif on the lipidic face of the TM helices based on solution NMR studies of the longer conductance domain (Fig. 4) [23]. In addition, these two binding sites differ in their time-averaged orientation for the amantadine C-N bond: being approximately parallel to the bilayer normal for the pore binding site and approximately perpendicular to the bilayer normal for the lipid/M2 interfacial site. Amantadine and rimantadine belong to a class of amphiphilic drugs with high affinity for the membrane interfacial region, with partition coefficients as high as ~37 in POPC lipid bilayers measured by solution NMR diffusion experiments [40]. Solid-state NMR, neutron diffraction, and molecular dynamics simulations have reported similar results [41].
Fig. 4.
Comparison of the two drug binding sites described for M2 proteins. (A) Rimantadine binding site in the conductance domain of AM2 based on the solution NMR structure in DHPC micelles. The drug is near the bilayer interfacial region interacting with the lipid and a hydrophobic pocket on the external structure of the protein (PDB 2RLF) [23]. (B) Amantadine modeled into the pore of the TM domain of AM2 (based on data used for PDB 2H95) [26,37,38].
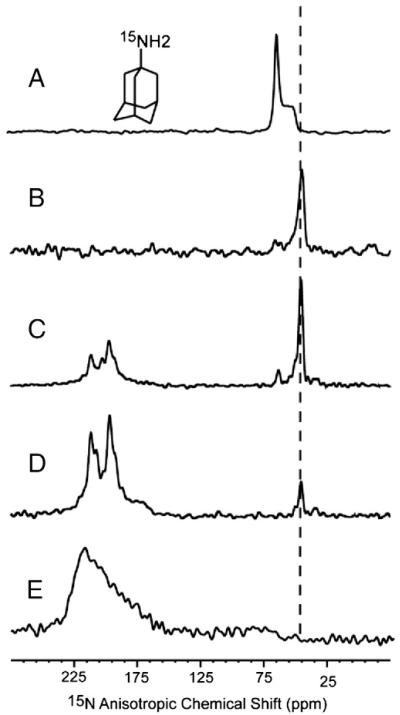
The debate over which drug binding site is biologically relevant appears to have been resolved with the recent measurement of distances between amantadine and the TM domain by Cady and coworkers using solid-state NMR where they identified the high affinity site as the pore binding site and the lipid interface site as a low affinity site [37]. In addition, functional studies of mutants have provided convincing evidence [42] and we have also obtained data on amantadine binding to the AM2 TM domain by isotopically labeling the amino group of amantadine with 15N and by using 15N-Leu-labeled AM2 TM domain through a series of 15N cross-polarization experiments with different peptide to amantadine ratios in DMPC liquid crystalline lipid bilayers. Fig. 5 shows the spectra of 15N-labeled amantadine with and without the 15N-labeled AM2 TM domain. All of the spectra were obtained from samples at pH 8.0 and 30 °C. The spectrum of amantadine in (unoriented) liposomes (Fig. 5A) shows a typical powder pattern for an 15NH2 group. As we have shown previously the hydrocarbon portion of this drug is buried in the hydrophobic core of the bilayer, while the amino group interacts with the interfacial region of the lipid bilayer, with the C-N vector pointing away from the middle of the bilayer [40,41]. In Fig. 5B the spectrum of the same sample uniformly aligned with the bilayer normal parallel to the magnetic field is shown. Here, in the lipid environment the amantadine resonance aligns with the δ∥ edge of the powder pattern indicating that the time-averaged orientation of the C-N bond in amantadine is parallel to the bilayer normal, in agreement with molecular dynamics simulations [41]. Upon the addition of the AM2 TM domain in a ratio of 8 drug molecules per tetramer the time-averaged alignment for amantadine is still parallel with the bilayer normal (Fig. 5C) as expected for the pore binding site, but not the lipid/M2 interfacial site. In the former case ~12.5% of the drug molecules are bound to the pore, which would have the C-N bond approximately parallel to the bilayer normal and in the latter case ~50% of the drug molecules would be bound to the protein and have their C-N bond perpendicular to the bilayer normal, resulting in 50% of the amantadine 15N signal near δ⊥. In this sample, most of the amantadine signal is from lipid bilayer bond amantadine as in Fig. 5B. The narrow AM2 TM domain signals clearly show that the protein is bound with amantadine, as the resonances are between 192 and 214 ppm. The unbound protein has very broad resonances between 165 and 225 ppm (Fig. 5E). A weak amantadine signal is observed near the δ⊥ edge of the powder pattern, which could either indicate a small amount of amantadine bound to the alternative AM2 binding site or a small amount of unoriented material in the sample (a situation that is common). When the molar ratio is reduced to 1:1 for the tetramer (Fig. 5D) the only amantadine signal is at δ∥ once again indicating that amantadine is in the pore binding site. At this stoichiometry if binding were occurring at the lipid/protein interface not only would we observe a different amantadine 15N frequency (close to δ∞), but we should see a break in the four fold symmetry of the TM domain resonances (since on average there would be 1 amantadine-bound M2 monomer to 3 unbound monomers); neither of which is observed. Indeed, almost all of the AM2 TM domain signal is in the form of the bound state, an indication that the stoichiometry for amantadine binding is 1:1 and clearly far removed from the 1:4 stoichiometry that would be required for the alternative binding site.
Fig. 5.
15N static spectra by cross-polarization of (A) randomly oriented bilayer sample with amantadine; (B) uniformly aligned sample of (A); (C) uniformly aligned bilayer sample with 5-site 15N leucine-labeled AM2-TM domain, at a 1:8 molar ratio of M2 TM domain to amantadine; (D) a sample similar to (C) but with a 1:1 molar ratio; (E) a sample similar to (C) but without amantadine. All the samples were prepared at pH 8.0 in DMPC bilayers; the oriented sample was uniformly aligned with the bilayer normal parallel to the static magnetic field.
A final note on these spectra is that the amantadine in the pore binding site is undergoing considerable dynamics although the time-averaged C-N bond is parallel to the bilayer normal. This is consistent with NMR observations that the symmetry of the tetramer is not broken by amantadine binding [26] as would be expected if the amino group were bound to a single helix, instead such interactions appear to be extensively averaged over all four helices. This result is also consistent with the molecular dynamics simulations that show rapid dynamics for bound amantadine and its alternating interactions with the four helices [43]. Therefore, we conclude that the stoichiometry of amantadine binding is one drug molecule per tetramer. Furthermore, the orientation of the drug in the bound state is such that the time-averaged orientation of the C-N bond is approximately parallel to the bilayer normal. These results confirm those of Cady et al. [37] in characterizing the pore binding site as the high affinity site, while not eliminating the possibility of a weak binding site at the lipidic interface. Interestingly, molecular dynamics simulations indicate that the C-N vector points toward the bulk water while bound in a lipid bilayer [41] but toward the middle of the bilayer while bound to the pore of the AM2 TM domain [38].
There is little doubt that Schnell and Chou [23] observed a binding site for rimantadine on the exterior of the tetramer just as there is little doubt that rimantadine was not binding in the pore. This solution NMR structure was obtained in a DHPC detergent micelle environment, which does not have a well-defined hydrophobic thickness, unlike a lipid bilayer. Consequently, it is possible to have helices packed in a tight bundle with only a small helix tilt angle with respect to the bundle axis in micelles while in bilayers the lengthy hydrophobic helix is forced to tilt with respect to the bilayer normal [12,44]. As a result there is no cavity in the pore of adequate size to accommodate the drug in DHPC micelles. The other structures of the AM2 TM domain all have considerably larger helical tilt angles and all display cavities appropriate for amantadine binding in the pore. In the recent structure of the BM2 TM domain [5] where the hydrophobic sequence is even longer than that in AM2, the solution NMR structure suggests an alternative mechanism by which the pore can be closed and amantadine access prevented; the formation of a coiled-coil structure resulting in tight packing of the helices [5]. The overall conclusion is that in addition to having an appropriate hydrophilic/hydrophobic environment in the pore for amantadine binding it is also necessary to have a substantial tilt of the helices to open a cavity large enough for the drug. Some of the naturally occurring amantadine-resistant mutants may function to impede drug binding by reducing the size of the cavity, such as S31N and A30T [38].
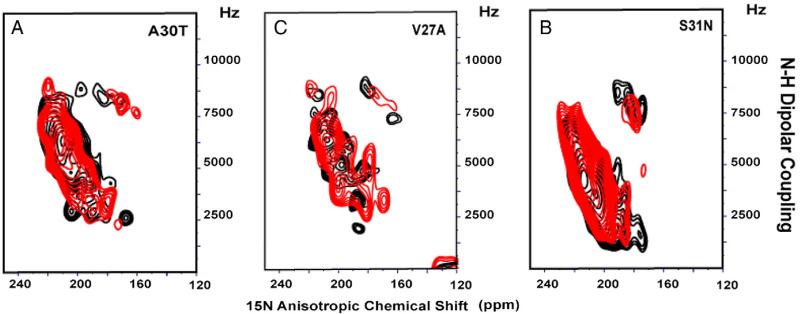
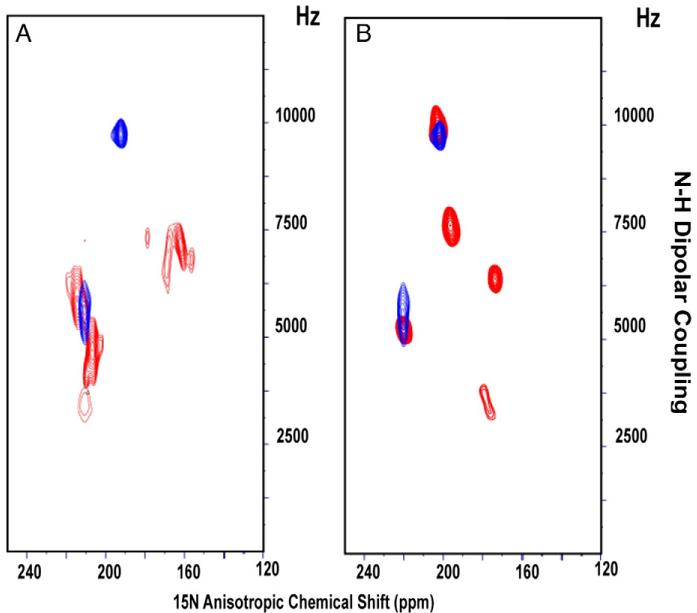
In order to further investigate the binding site of amantadine in M2 channels, we prepared mutants of the AM2 TM domain corresponding to the naturally occurring drug-resistant mutations, V27S, V27A, A30T, and S31N. As we noted above, spectra of aligned AM2 samples are very sensitive to amantadine binding; as a result, a direct inference about drug binding can be made by comparing spectra in the absence and presence of amantadine. Fig. 6 shows that the PISEMA spectra of 5 15N-labeled leucine residues in the M2 TM domain do not change significantly in the presence (red) or absence (black) of amantadine for the S31N, V27A, and A30T mutants. These data suggest that the TM domain is functioning similarly to the full-length protein; i.e., the TM domain mutants confer drug resistance by eliminating amantadine binding. We have already suggested that this may be due to the increased hydrophilicity in these constructs. Note that in AM2, A30 and V27 correspond to sites in BM2 that have serine residues supporting the argument that increased hydrophilicity decreases amantadine binding. For V27S a different result is observed, in which amantadine clearly binds to the tetramer and furthermore the amide resonance frequencies for the bound state are very similar to those of the wild type (Fig. 7), suggesting that the backbone structural changes induced by the V27S mutations are minimal, both in the presence and absence of amantadine. Previously, Astrahan and coworkers [45] using surface plasmon resonance also observed that some amantadine-resistant M2 TM domain mutants bind amantadine. So far the V27S mutation has been assayed for drug sensitive proton conductance using the full-length protein [46] while drug binding (shown here) has been assessed using the TM domain, so there is the possibility that the full-length protein does not bind amantadine and therefore, the possibility exists that for this mutant the TM domain maybe a poor model for the full-length protein with regard to drug binding. We have suggested above that amantadine resistance may be caused by increased hydrophilicity in the pore or a decrease in the size of the binding cavity. Now we see that V27S binds amantadine while V27A does not. In addition, the BM2 chimera that binds amantadine includes S9V (residue 27 in AM2) and WT BM2 with Ser9 does not bind amantadine. Hence, the mechanisms governing amantadine binding and resistance are complex, apparently with multiple mechanisms at play for achieving amantadine resistance and the prevention of amantadine binding.
Fig. 6.
PISEMA spectra of 5-site 15N Leu-labeled amantadine-resistant mutants of AM2 TM domain with (red) and without amantadine (black). (A) A30T; (B) S31N; and (C) V27A.
Fig. 7.
PISEMA spectra of the V27S mutant. (A) Comparison of V27S with and without amantadine; 2-site 15N Ile (33, 42 )-labeled V27S mutant of AM2 TM domain with (blue) and without (red) amantadine. (B) Comparison of V27S and WT AM2 in the presence of amantadine; 2-site 15N Ile V27S AM2 TM domain with amantadine (blue—as in panel A) superimposed with 5 site 15N Ile (32, 33, 35, 39, 42) WT AM2 TM domain with amantadine (red).
The A18H mutation converts Vpu from a non-selective channel to a highly selective H+ channel that is sensitive to rimantadine. In both micelle and bicelle samples, the structurally characterized TM helix length is increased by this mutation and consequently the helical tilt increases from 30° to 41° [25]. While it is tempting to assume that Vpu functions like the AM2 tetramer, the binding of rimantadine in the pore of the Vpu TM domain induces only modest changes in the anisotropic chemical shifts and 15N-1H dipolar couplings (less than 1 ppm and less than 0.1 kHz, respectively, except for His18, the counterpart to His37 in AM2). These minimal changes in frequency mean that there is no change in helix tilt and essentially no change in backbone structure upon rimantadine binding. In contrast, AM2 TM domain displays significant changes in the C-terminal half of the TM helix, including changes in the anisotropic chemical shifts and dipolar interactions of 30 ppm and 3 kHz, respectively, resulting in a substantial change in helix tilt from 31° to 20° [26]. As an aside, Vpu TM domain data were obtained from bicelles and therefore for a comparison to the aligned bilayers the range of spin interaction changes should be doubled. Clearly, the drug binding mechanisms for AM2 and Vpu are very different. Potentially, the increased stability of the Vpu TM domain tetramer is due to the numerous small residues being at the helix–helix interface resulting in a structure that is not susceptible to the influence of drug binding. In addition, the lack of a glycine residue (Gly34 in AM2, which is Ile15 in Vpu) would also hinder the formation of a kink in the TM helix. The greatest influence by rimantadine on the Vpu TM domain resonances is at His18, suggesting that the drug may be interacting directly with this residue, contrary to the situation in AM2, where the amantadine binding cavity is well removed from His37.
3.2.2. The positively charged sequence
Apart from TM domains, amphipathic helices are common motifs encountered in membrane proteins even though there are exceptionally few amphipathic helical structures interacting with lipid bilayers deposited into the PDB. The clustering of polar and hydrophobic residues on opposite faces of helices provides domains that can simultaneously interact with polar head groups as well as the hydrocarbon core of the lipid bilayer. Due to this bimodal interaction with the membrane interior and aqueous environment, amphipathic helices can have a major role in membrane protein stability and function. They have also been implicated in membrane localization, curvature sensing, and interaction with soluble proteins. Despite the significance of the amphipathic helix motif, the scarcity of these structures may be due to the sensitivity of these helices to surface charge, surface curvature, and numerous other details of the membrane interfacial region, all of which combine to make amphipathic helices one of the most challenging aspects of membrane protein structure biology.
Spectra of uniformly 15N-labeled and 15N Phe (Fig. 8)-labeled full-length AM2 protein reconstituted in DMPC:DMPG bilayers provides convincing evidence for a bilayer surface bound helix between residues 47 and 55 [11,47]. This sequence includes four of the five phenylalanine residues in the full-length protein. The PISEMA spectrum [11] shows that all but one of the resonances is in the vicinity of a helical wheel characterized by a 100° tilt with respect to the bilayer normal. The importance of 100° versus 80° tilt is that the C-terminus (with carbonyl oxygens not involved in helical hydrogen bonds) is more exposed to the bilayer surface at 100° vs. 80°. Furthermore, the resonances of the hydrophobic portion of this wheel showed resistance to deuterium amide exchange after prolonged exposure indicating that these residues are deeply buried in the hydrocarbon core of the bilayer [11]. However, in the recent solution NMR structure of the AM2 conductance domain in DHPC micelles, these helices formed a water soluble tetrameric bundle that showed no hindrance to H/D exchange [23]. We suspect that this is due to the inability of the detergent micelle environment to provide a suitable model of the amphipathic bilayer surface for these helices or that monomeric detergents have stabilized a water soluble tetramer bundle of this amphipathic helix.
Fig. 8.
PISEMA spectra of 15N Phe-labeled full-length M2 protein in DMPC:DMPG (4:1) bilayers. A simulated PISA wheel for a helix tilted at 100° with respect to the bilayer normal is superimposed.
The DHPC micelle solution structure of the BM2 TM domain (residues 1–33) features a TM helix through residue Gln30 [5]. The solution NMR structure of BM2 residues 26–109 anchored to LMPG micelles provides a helical structure for residues 44–103 and consequently most of the post-TM sequence that has a net positive charge (residues 26–41) is not structurally defined in these samples [5]. While a hypothetical helical wheel for BM2 residues 28–41 suggests an amphipathic helix, the fraction of the wheel that is hydrophobic is less than that for AM2 with two lysines, K32 and K38, penetrating into the hydrophobic surface and thereby requiring these residues to snorkel to the hydrophilic region of the bilayer interface[48,49]. A cluster of hydrophobic residues, Leu28, Ile31, Val35, and Ile39, make an arc of 80° on the helical wheel (with Lys 32 and Lys 38 the arc would extend to 120°). In addition, this hydrophobic surface appears to be a continuation of the lipid facing hydrophobic surface of the TM helix, and therefore, we would anticipate that this region forms a very similar structure to that of AM2 [11] with the exception that the amphipathic helix may not be so buried in the hydrophobic region of the bilayer. While this region was not structured in the solution NMR structure, detergent sample preparation was required to solubilize this domain in the 26–109 construct, it is possible that in bilayer preparations this region could be helical and associated with the bilayer interface.
As with AM2 and BM2, Vpu appears to form a weak amphipathic helix between residues Ile26 and Gln35 where Ile25 or Ile26 appear to be the end of the TM helix [25]. The amphipathic sequence is less than three turns in length and has a net charge of only +2. Consequently, the argument for a post-TM amphipathic helix is weakest for Vpu, consistent with the overall structural and functional features of this channel suggesting that it is very different from AM2 and BM2.
4. Conclusions
These three viral porins have relatively short sequences and perform multiple viral functions. While AM2 and BM2 form proton channels, wild-type Vpu has no such specificity, but with a single site mutation, A18H, it forms proton specific channels that are sensitive to rimantadine. Consequently, the focus of this paper was on the structural comparison of these three viral porins. The ‘divide-and- conquer’ approach for the viral porins with multiple functional domains has been very successful for studies of the transmembrane and conductance domains of M2 and Vpu. However, through these studies a sensitivity to the protein’s environment has been demonstrated, especially for M2. The helical interface for M2 is composed of large hydrophobic side chains with few opportunities for specific interactions to stabilize the helical bundle, while a portion of the Vpu helical interface is composed of alanine residues leading to a more stable helical complex. Consequently, the structural dependence on the protein environment has been more significant for M2 than for Vpu. Importantly these comparisons once again, emphasize that membrane protein structures are determined not just by their amino acid sequences and their intra-protein interactions, but also by their environments and the protein’s interactions with their environments, as recognized by Christian Anfinsen [50].
In the A18H mutant of Vpu the TM helix tilt appears to be greater than for AM2 and, while rimantadine binds, the mechanism by which it binds appears to be very different from that for AM2, where drug binding induces a helix kink and much more significant shifts in the PISEMA resonances. The recent debate over the biologically relevant drug binding site in AM2 appears to have been resolved by the measurement of distances to the drug [37] but also by our characterization of the drug:tetramer stoichiometry of 1:1 and drug orientation in the binding site. What is apparently far more complex is the mechanism by which mutations induce drug resistance; indeed, in M2 there may be a mix of steric, chemical, hydrophobic, and dynamic influences that result in such resistance.
Acknowledgments
The authors are grateful to the staff of the National High Magnetic Field Laboratory NMR facility (A. Blue) and the staff of Bioanalytical Synthesis and Services Laboratory (H. Hendricks and U. Goli) for their expertise, assistance, and maintenance of instruments essential for this work. This work was supported by the National Institutes of Health, AI 023007. The experiments were largely performed at the National High Magnetic Field Laboratory, supported by Cooperative Agreement (DMR-0084173) between the National Science Foundation and the State of Florida.
References
- [1].Hout DR, Gomez LM, Pacyniak E, Miller JM, Hill MS, Stephens EB. A single amino acid substitution within the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein renders simian-human immunodeficiency virus (SHIV(KU-1bMC33)) susceptible to rimantadine. Virology. 2006;348:449–461. doi: 10.1016/j.virol.2005.12.025. [DOI] [PubMed] [Google Scholar]
- [2].Hout DR, Gomez ML, Pacyniak E, Gomez LM, Fegley B, Mulcahy ER, Hill MS, Culley N, Pinson DM, Nothnick W, Powers MF, Wong SW, Stephens EB. Substitution of the transmembrane domain of Vpu in simian–human immunodeficiency virus (SHIVKU1bMC33) with that of M2 of influenza A results in a virus that is sensitive to inhibitors of the M2 ion channel and is pathogenic for pig-tailed macaques. Virology. 2006;344:541–559. doi: 10.1016/j.virol.2005.08.022. [DOI] [PubMed] [Google Scholar]
- [3].Pinto LH, Lamb RA. Influenza virus proton channels. Photochem. Photobiol. Sci. 2006;5:629–632. doi: 10.1039/b517734k. [DOI] [PubMed] [Google Scholar]
- [4].Mould JA, Paterson RG, Takeda M, Ohigashi Y, Venkataraman P, Lamb RA, Pinto LH. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- [5].Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat. Struct. Mol. Biol. 2009 doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- [7].Sakaguchi T, Tu Q, Pinto LH, Lamb RA. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl Acad. Sci. USA. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Balannik V, Lamb RA, Pinto LH. The oligomeric state of the active BM2 ion channel protein of influenza B virus. J. Biol. Chem. 2008;283:4895–4904. doi: 10.1074/jbc.M709433200. [DOI] [PubMed] [Google Scholar]
- [9].Mould JA, Paterson RG, Takeda M, Ohigashi Y, Venkataraman P, Lamb RA, Pinto LH. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell. 2003;5:175–184. doi: 10.1016/s1534-5807(03)00190-4. [DOI] [PubMed] [Google Scholar]
- [10].Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl Acad. Sci. USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Duong-Ly KC, Nanda V, DeGrado WF, Howard KP. The conformation of the pore region of the M2 prooton channel depends on lipid bilayer environment. Protein Sci. 2005;14:856–861. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- [14].Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Subczynski WK, Wojas KJ, Pezeshk V, Pezeshk A. Partitioning and localization of spin labeled amantadine in lipid bilayers: an EPR study. J. Pharm. Sci. 1998;87:1249–1254. doi: 10.1021/js970381n. [DOI] [PubMed] [Google Scholar]
- [17].Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: an intrinsic property. J. Mol. Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- [18].Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- [19].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [20].MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- [21].Ma C, Soto CS, Ohigashi Y, Taylor A, Bournas V, Glawe B, Udo MK, Degrado WF, Lamb RA, Pinto LH. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J. Biol. Chem. 2008;283:15921–15931. doi: 10.1074/jbc.M710302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim. Biophys. Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- [25].Park SH, Opella SJ. Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs. Protein Sci. 2007;16:2205–2215. doi: 10.1110/ps.073041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys. J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moffat JC, Vijayvergiya V, Gao PF, Cross TA, Woodbury DJ, Busath DD. Proton transport through influenza A virus M2 protein reconstituted in vesicles. Biophys. J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ohigashi Y, Ma C, Jing X, Balannick V, Pinto LH, Lamb RA. An amantadine-sensitive chimeric BM2 ion channel of influenza B virus has implications for the mechanism of drug inhibition. Proc. Natl Acad. Sci. USA. 2009;106:18775–18779. doi: 10.1073/pnas.0910584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bordag N, Keller S. a-Helical transmembrane peptides: a “divide and conquer” approach to membrane proteins. Chem. Phys. Lipids. 2010;163:1–26. doi: 10.1016/j.chemphyslip.2009.07.009. [DOI] [PubMed] [Google Scholar]
- [30].Hu J, Sharma M, Qin H, Gao FP, Cross TA. Ligand binding in the conserved interhelical loop of CorA, a magnesium transporter from mycobacterium tuberculosis. J. Biol. Chem. 2009;284:15619–15628. doi: 10.1074/jbc.M901581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat. Struct. Mol. Biol. 2009;16:1267–1271. doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marassi FM, Ma C, Gratkowski H, Straus SK, Strebel K, Oblatt-Montal M, Montal M, Opella SJ. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc. Natl Acad. Sci. USA. 1999;96:14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002;11:546–557. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc. Natl Acad. Sci. USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- [36].Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yi M, Cross TA, Zhou HX. A secondary gate as a mechanism for inhibition of the M2 proton channel by amantadine. J. Phys. Chem. B. 2008;112:7977–7979. doi: 10.1021/jp800171m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the m2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J, Schnell JR, Chou JJ. Amantadine partition and localization in phospholipid membrane: a solution NMR study. Biochem. Biophys. Res. Commun. 2004;324:212–217. doi: 10.1016/j.bbrc.2004.09.039. [DOI] [PubMed] [Google Scholar]
- [41].Li C, Yi M, Hu J, Zhou HX, Cross TA. Solid-state NMR and MD simulations of the antiviral drug amantadine solubilized in DMPC bilayers. Biophys. J. 2008;94:1295–1302. doi: 10.1529/biophysj.107.112482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc. Natl Acad. Sci. USA. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yi M, Cross TA, Zhou HX. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc. Natl Acad. Sci. USA. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].de Planque MR, Greathouse DV, Koeppe RE, II, Schafer H, Marsh D, Killian JA. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A 2H NMR and ESR study using designed transmembrane alpha-helical peptides and gramicidin A. Biochemistry. 1998;37:9333–9345. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- [45].Astrahan P, Kass I, Cooper MA, Arkin IT. A novel method of resistance for influenza against a channel-blocking antiviral drug. Proteins. 2004;55:251–257. doi: 10.1002/prot.20018. [DOI] [PubMed] [Google Scholar]
- [46].Balannik V, Carnevale V, Fiorin G, Levine BG, Lamb RA, Klein ML, Degrado WF, Pinto LH. Functional studies and modeling of pore-lining residue mutants of the influenza a virus M2 ion channel. Biochemistry. 2010;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Expression and initial structural insights from solid-state NMR of the M2 proton channel from influenza A virus. Biochemistry. 2002;41:11294–11300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- [48].Stranberg E, Morein S, Rijkers DTS, Liskamp RMJ, van der Wel PCA, Killian JA. Lipid dependence of membrane anchoring properties and snorkeling ehavior of aromatic and charged residues in transmembrane peptides. Biochemistry. 2002;41:7190–7198. doi: 10.1021/bi012047i. [DOI] [PubMed] [Google Scholar]
- [49].de Planque MR, Kruijtzer JA, Liskamp RM, Marsh D, Greathouse DV, Koeppe RE, II, de Kruijff B, Killian JA. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- [50].Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]